Gathering Novel Circulating Exosomal microRNA in Osteosarcoma Cell Lines and Possible Implications for the Disease
Abstract
:1. Introduction
2. Results
2.1. RNA Profiles of Osteosarcoma Cell Lines and Osteosarcoma Cell-Derived Exosomes
2.2. Validation of the Candidates’ Novel microRNAs
2.3. Candidate Novel microRNA Expression Circulating in Osteosarcoma Samples
3. Discussion
4. Materials and Methods
4.1. Ethics Approval
4.2. Cell Lines and Reagents
4.3. Exosome Purification
4.4. Small RNA Library Construction and Sequencing
4.5. Small RNA-Seq in Silico Analysis
4.6. Real-time PCR Validation and Digital PCR of Novel microRNAs
4.7. miRNA Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating Micrornas as Stable Blood-Based Markers for Cancer Detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of Micrornas in Serum: A Novel Class of Biomarkers for Diagnosis of Cancer and Other Diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating Micrornas in Breast Cancer: Novel Diagnostic and Prognostic Biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sholl, L.M. Next-Generation Sequencing from Liquid Biopsies in Lung Cancer Patients: Advances in Comprehensive Biomarker Testing. J. Thorac. Oncol. 2017, 12, 1464–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarver, A.L.; Phalak, R.; Thayanithy, V.; Subramanian, S. S-Med: Sarcoma Microrna Expression Database. Lab. Investig. 2010, 90, 753–761. [Google Scholar] [CrossRef] [Green Version]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sultmann, H. Serum Micrornas as Non-Invasive Biomarkers for Cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef] [Green Version]
- Ottaviani, G.; Jaffe, N. The Etiology of Osteosarcoma. Cancer Treat. Res. 2009, 152, 15–32. [Google Scholar]
- Marina, N.; Gebhardt, M.; Teot, L.; Gorlick, R. Biology and Therapeutic Advances for Pediatric Osteosarcoma. Oncologist 2004, 9, 422–441. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pelle, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid Biopsy of Cancer: A Multimodal Diagnostic Tool in Clinical Oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Raimondi, L.; de Luca, A.; Gallo, A.; Costa, V.; Russelli, G.; Cuscino, N.; Manno, M.; Raccosta, S.; Carina, V.; Bellavia, D.; et al. Osteosarcoma Cell-Derived Exosomes Affect Tumor Microenvironment by Specific Packaging of Micrornas. Carcinogenesis 2019. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabieres, C. Real-Time Liquid Biopsy in Cancer Patients: Fact or Fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondi, L.; de Luca, A.; Costa, V.; Amodio, N.; Carina, V.; Bellavia, D.; Tassone, P.; Pagani, S.; Fini, M.; Alessandro, R.; et al. Circulating Biomarkers in Osteosarcoma: New Translational Tools for Diagnosis and Treatment. Oncotarget 2017, 8, 100831–100851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Seebacher, N.A.; Hornicek, F.J.; Xiao, T.; Duan, Z. Application of Liquid Biopsy in Bone and Soft Tissue Sarcomas: Present and Future. Cancer Lett. 2018, 439, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Liu, Q.; Tian, J.; He, H.B.; Luo, W. Angiogenesis Process in Osteosarcoma: An Updated Perspective of Pathophysiology and Therapeutics. Am. J. Med. Sci. 2019, 357, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Mohseny, A.B.; Cai, Y.; Kuijjer, M.; Xiao, W.; van den Akker, B.; de Andrea, C.E.; Jacobs, R.; Dijke, P.T.; Hogendoorn, P.C.; Cleton-Jansen, A.M. The Activities of Smad and Gli Mediated Signalling Pathways in High-Grade Conventional Osteosarcoma. Eur. J. Cancer 2012, 48, 3429–3438. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B. Aligning Short Sequencing Reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11–17. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. Mirdeep2 Accurately Identifies Known and Hundreds of Novel Microrna Genes in Seven Animal Clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Coulter, S.J. Mitigation of the Effect of Variability in Digital Pcr Assays through Use of Duplexed Reference Assays for Normalization. Biotechniques 2018, 65, 86–91. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting Effective Microrna Target Sites in Mammalian Mrnas. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative Html5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
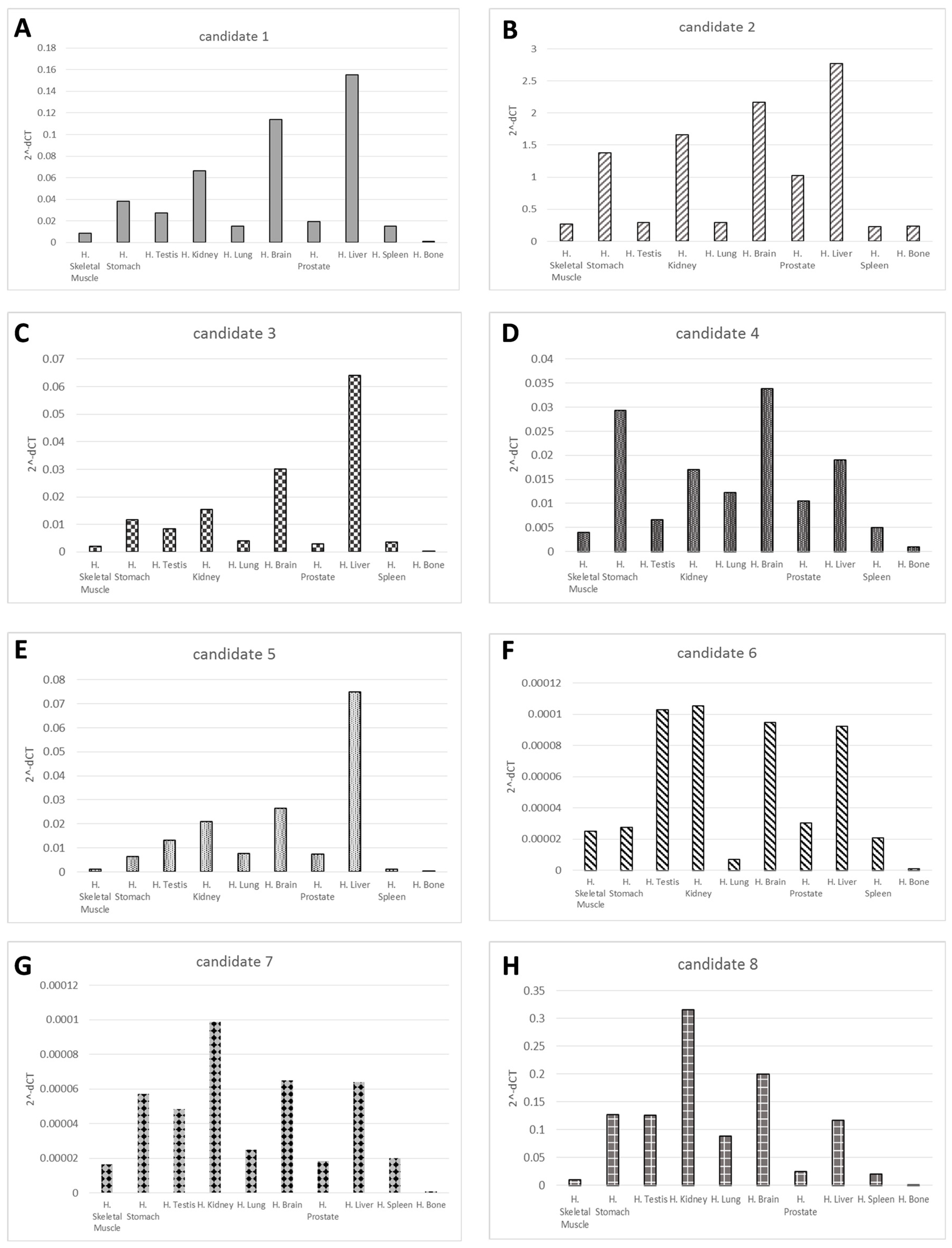
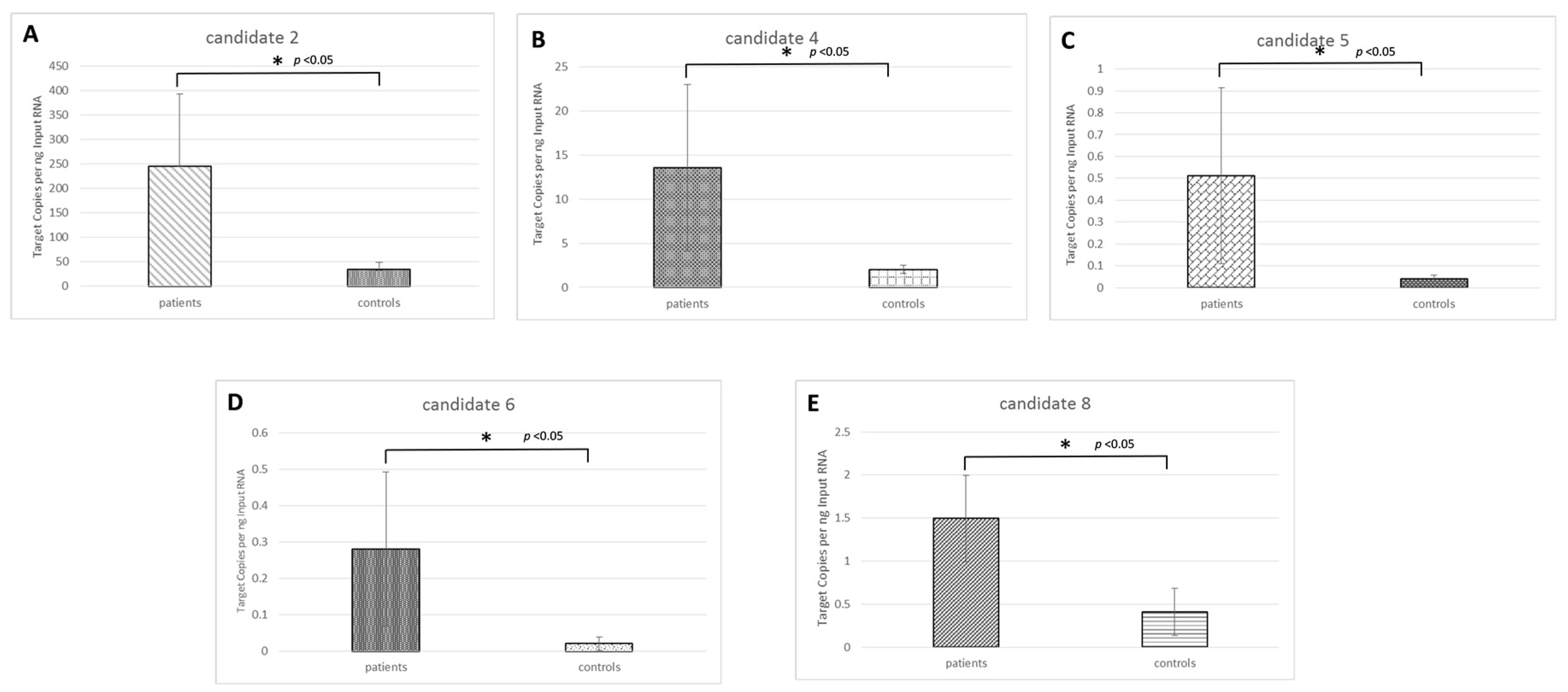
| Novel ID | Location (hg19) | Strand | Mature Sequence |
|---|---|---|---|
| Candidate 1 | chr9:136204572..136204654 | − | CCCCACACUGCUAAAUUUGAC |
| Candidate 2 | chr3:14436198..14436257 | + | GGAAUAACGGGUGCUGUAGGCU |
| Candidate 3 | chr9:89037929..89037970 | − | CCCCUCACUGCUAAAUUUGAC |
| Candidate 4 | chrX:102411053..102411132 | − | CCAUCUGUGGGAUUAUGACUGA |
| Candidate 5 | chr16:33962833..33962878 | + | UGCGCAGUGGCAGUAUCGUAGCC |
| Candidate 6 | chr8:56821958..56822010 | − | UAUGUGCCUAGUGGCUGCUGUCU |
| Candidate 7 | chr13:27259470..27259536 | + | UCUGGGCAACAAGGUGAGACC |
| Candidate 8 | chr9:89037927..89037972 | − | AUGGAUUUUUGGAAAUAGGAGA |
| Patient ID | Gender | Age | Site of Origin | Histologic Subtype | metastasis | Treatments |
|---|---|---|---|---|---|---|
| Patient 1 | M | 14 | sx prox Tibia | High Grade Surface OS | No | Chir |
| Patient 2 | M | 17 | dx distal Femur | OS G3 | No | CHT (CDDP/ADM MTX HD × 2) |
| Patient 3 | M | 18 | dx prox Tibia | OS condrobastic G3 | No | CHT (CDDP/ADM MTX HD × 2) |
| Patient 4 | M | 19 | sx Tibia | OS fibroblastic | Lung | CHT (CDDP/ADM MTX HD × 2) |
| Patient 5 | M | 16 | dx Pelvis | OS G3 | Lung | CHT (CDDP/ADM MTX HD × 2) |
| Candidate 2 | KEGG pathway | p-value | Genes |
| FoxO signaling pathway | 6.37 × 10−5 | SMAD2;SMAD4;CCND2;MAPK1;PIK3R1;IGF1 | |
| TGF-beta signaling pathway | 1.208 × 10−3 | SMAD2;SMAD4;MAPK1;ACVR1B;BMPR1B;SKP1 | |
| Hippo signaling pathway | 1.29 × 10−3 | FZD1;SMAD2;PRKCI;SMAD4;CCND2;NF2 | |
| Proteoglycans in cancer | 1.41 × 10−3 | FZD1;SMAD2;PDCD4;MAPK1;PIK3R1;IGF1 | |
| Hepatocellular carcinoma | 1.75 × 10−3 | FZD1;SMAD2;SMAD4;CDK6;MAPK1;PIK3R1 | |
| Glioma | 3.09 × 10−3 | CDK6;MAPK1;PIK3R1;IGF1;CALM1 | |
| Pancreatic cancer | 3.09 × 10−3 | SMAD2;SMAD4;CDK6;MAPK1;PIK3R1 | |
| Breast cancer | 3.35 × 10−3 | FZD1;JAG2;CDK6;MAPK1;PIK3R1;IGF1;ESR1 | |
| Candidate 4 | KEGG pathway | p-value | Genes |
| Cell adhesion molecules | 3.61 × 10−4 | NLGN3;CNTNAP1;CD4;NLGN2;PECAM1;NRXN3 | |
| Focal adhesion | 6.73 × 10−4 | PPP1CB;PPP1CC;RAP1A;COL4A4;ITGB8;MAPK1;CRK | |
| TGF-beta signaling pathway | 1.08 × 10−3 | SRC;MAPK1;CRK;CD44;PFN2 | |
| Renal cell carcinoma | 1.12 × 10−3 | PPP1CB;PPP1CC;FGF7;ITGB8;MAPK1;PIP4K2C;CRK | |
| Rap1 signaling pathway | 1.32 x 10−3 | PPP1CB;PPP1CC;PRKAB2;RAPGEF1;MAPK1 | |
| ErbB signaling pathway | 4.50 × 10−3 | ZFYVE16;SP1;MAPK1;INHBA;RGMB | |
| VEGF signaling pathway | 8.46 × 10−3 | PPP1CB;PPP1CC;RAP1A;MAPK1 | |
| Proteoglycans in cancer | 9.37 × 10−3 | RAP1A;RAPGEF1;MAPK1;CRK | |
| Candidate 5 | KEGG pathway | p-value | Genes |
| Bladder cancer | 1.22 × 10−2 | VEGFA | |
| VEGF signaling pathway | 1.75 × 10−2 | VEGFA | |
| Renal cell carcinoma | 2.05 × 10−2 | VEGFA | |
| Pancreatic cancer | 2.22 × 10−2 | VEGFA | |
| HIF-1 signaling pathway | 2.69 × 10−2 | VEGFA | |
| Focal adhesion | 2.69 × 10−2 | VEGFA | |
| Proteoglycans in cancer | 2.96 × 10−2 | VEGFA | |
| Candidate 6 | KEGG pathway | p-value | Genes |
| Proteoglycans in cancer | 2.45 × 10−3 | PPP1R12A;ARHGEF12;FZD5;ERBB4;KDR;IGF1R | |
| MAPK signaling pathway | 3.93 × 10−3 | GABRB2;CACNA1C;GABARAP;TRAK2 | |
| Focal adhesion | 1.13 × 10−2 | PDGFRA;CACNB4;ERBB4;KDR;CACNA1C;IGF1R | |
| Rap1 signaling pathway | 5.11 × 10−2 | PDGFRA;PPP1R12A;KDR;PARVA;IGF1R | |
| Candidate 8 | KEGG pathway | p-value | Genes |
| Hippo signaling pathway | 2,00 × 10−4 | PPP1CB;LATS2;DLG3;FZD4;TCF7;YWHAZ;TGFBR1; | |
| TGF-beta signaling pathway | 2.14 × 10−3 | CREBBP;TCF7;BAIAP2;TGFBR1;WASF3 | |
| Breast cancer | 5.62 × 10−3 | CREBBP;RPS6KB1;NEO1;ACVR2B;TGFBR1 | |
| Colorectal cancer | 7.87 × 10−3 | HDAC4;BECN1;RPS6KB1;ADCY2;CALM2;TGFBR1 | |
| Prostate cancer | 9.07 × 10−3 | CREBBP;FZD4;TCF7;ADCY2;CALM2 | |
| Gastric cancer | 1.09 × 10−2 | RPS6KB1;FZD4;NCOA3;TCF7;PGR;LRP6 | |
| Wnt signaling pathway | 2.19 × 10−2 | NRP1;CREBBP;CDC27;ADCY2;TGFBR1;CREB5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuscino, N.; Raimondi, L.; De Luca, A.; Carcione, C.; Russelli, G.; Conti, L.; Baldi, J.; Conaldi, P.G.; Giavaresi, G.; Gallo, A. Gathering Novel Circulating Exosomal microRNA in Osteosarcoma Cell Lines and Possible Implications for the Disease. Cancers 2019, 11, 1924. https://doi.org/10.3390/cancers11121924
Cuscino N, Raimondi L, De Luca A, Carcione C, Russelli G, Conti L, Baldi J, Conaldi PG, Giavaresi G, Gallo A. Gathering Novel Circulating Exosomal microRNA in Osteosarcoma Cell Lines and Possible Implications for the Disease. Cancers. 2019; 11(12):1924. https://doi.org/10.3390/cancers11121924
Chicago/Turabian StyleCuscino, Nicola, Lavinia Raimondi, Angela De Luca, Claudia Carcione, Giovanna Russelli, Laura Conti, Jacopo Baldi, Pier Giulio Conaldi, Gianluca Giavaresi, and Alessia Gallo. 2019. "Gathering Novel Circulating Exosomal microRNA in Osteosarcoma Cell Lines and Possible Implications for the Disease" Cancers 11, no. 12: 1924. https://doi.org/10.3390/cancers11121924
APA StyleCuscino, N., Raimondi, L., De Luca, A., Carcione, C., Russelli, G., Conti, L., Baldi, J., Conaldi, P. G., Giavaresi, G., & Gallo, A. (2019). Gathering Novel Circulating Exosomal microRNA in Osteosarcoma Cell Lines and Possible Implications for the Disease. Cancers, 11(12), 1924. https://doi.org/10.3390/cancers11121924






