Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma
Abstract
1. Introduction
2. Results
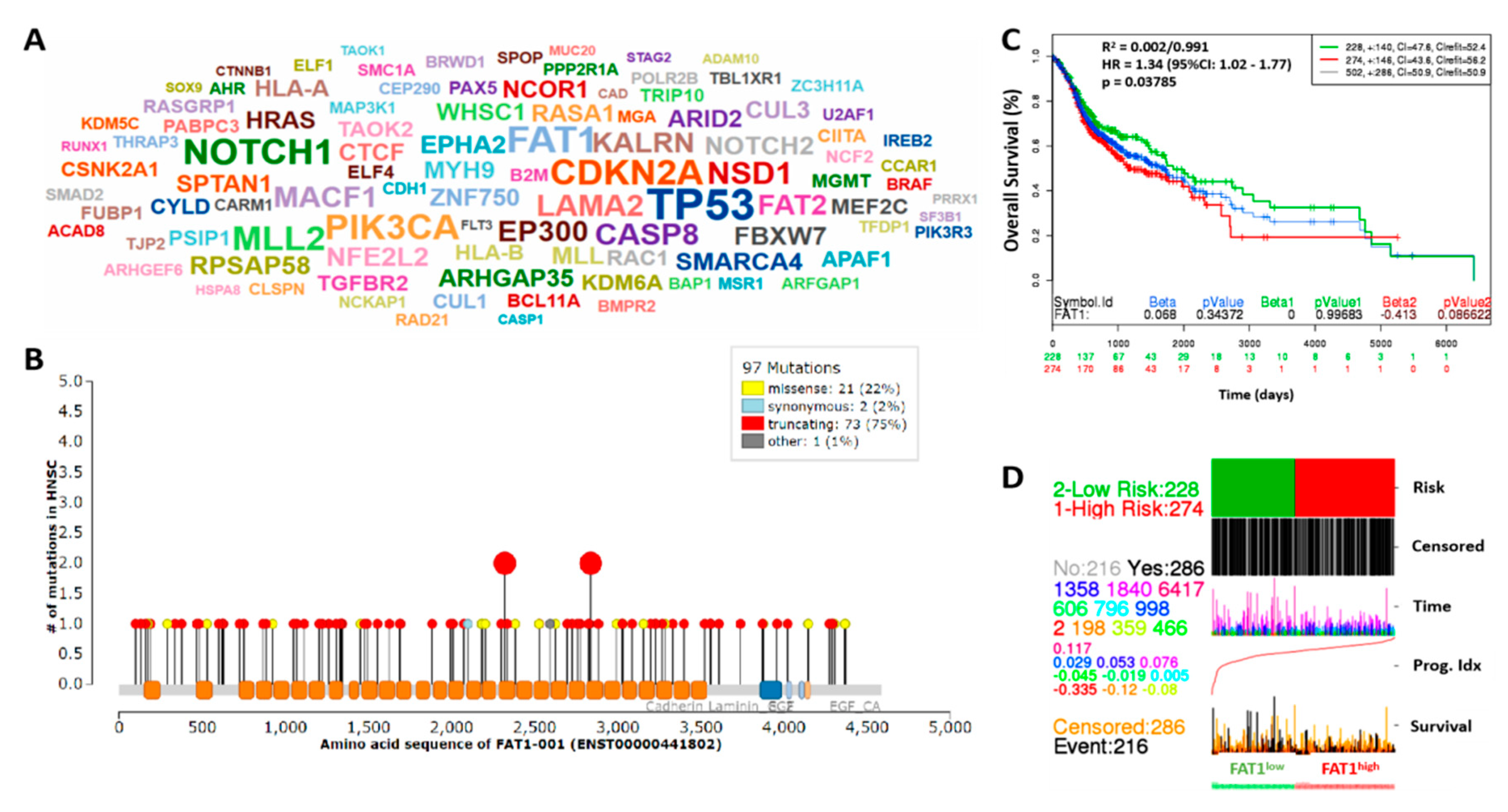
2.1. High FAT1 Expression Drives HNSC, Defines High Risk Population and is Associated with Poor Prognosis
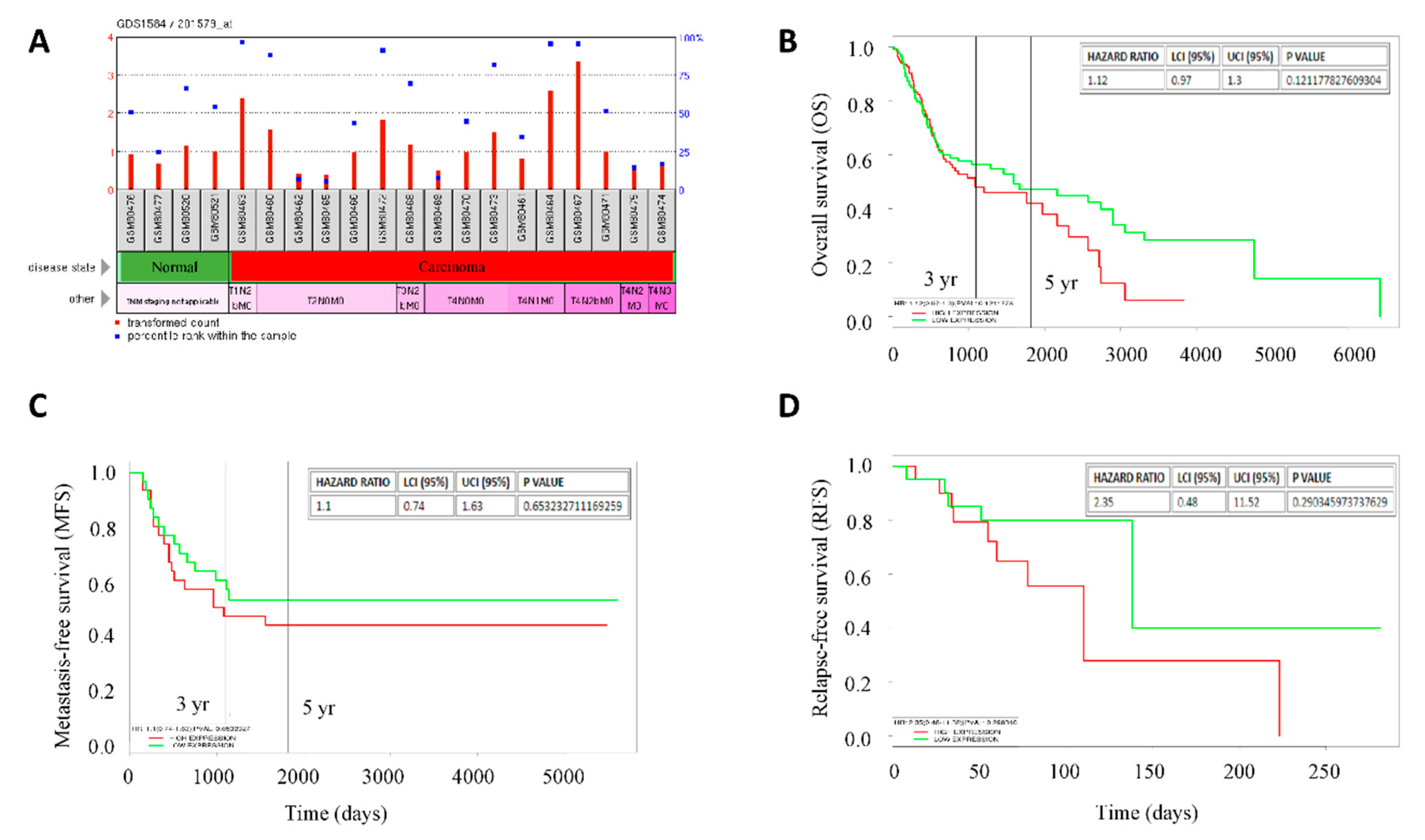
2.2. High FAT1 Expression Is Positively Correlated with Disease Progression and Poor Clinical Outcome in Patients with HNSC
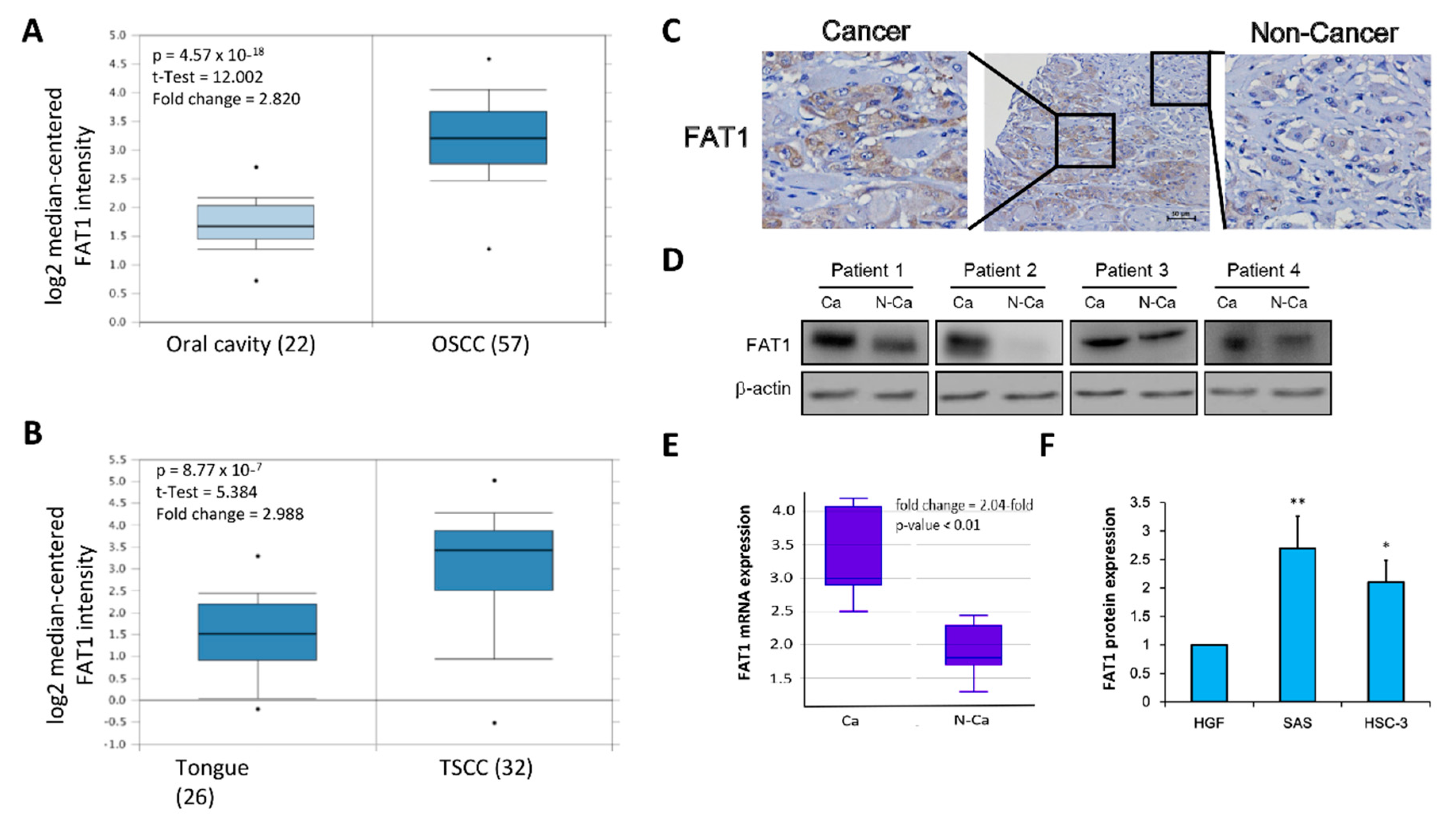
2.3. FAT1 Is Aberrantly-Expressed in OSCC Clinical Samples and Cell Lines
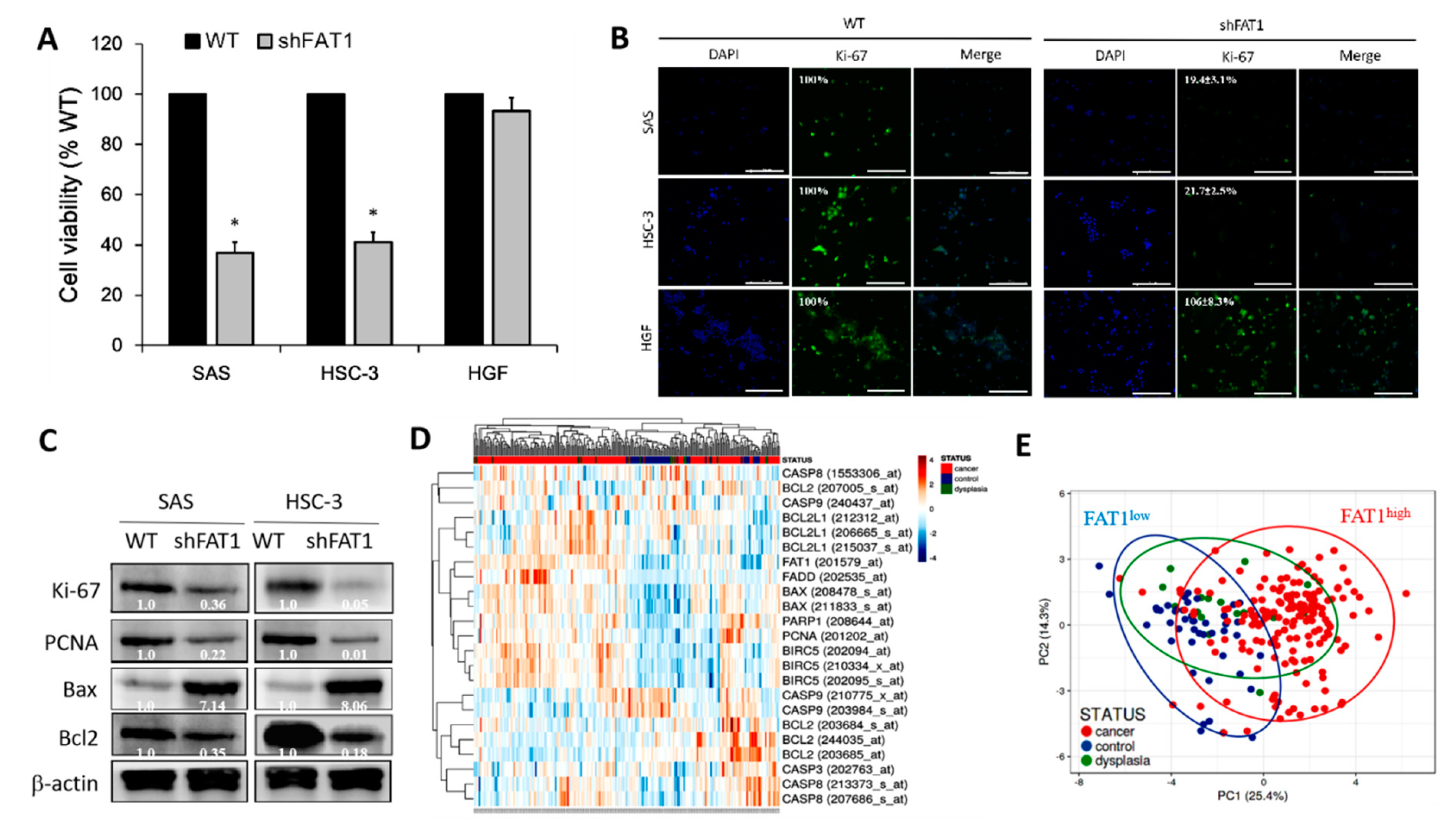
2.4. Loss-of-FAT1 Function Impairs OSCC Cell Proliferation and Enhances Cell Death
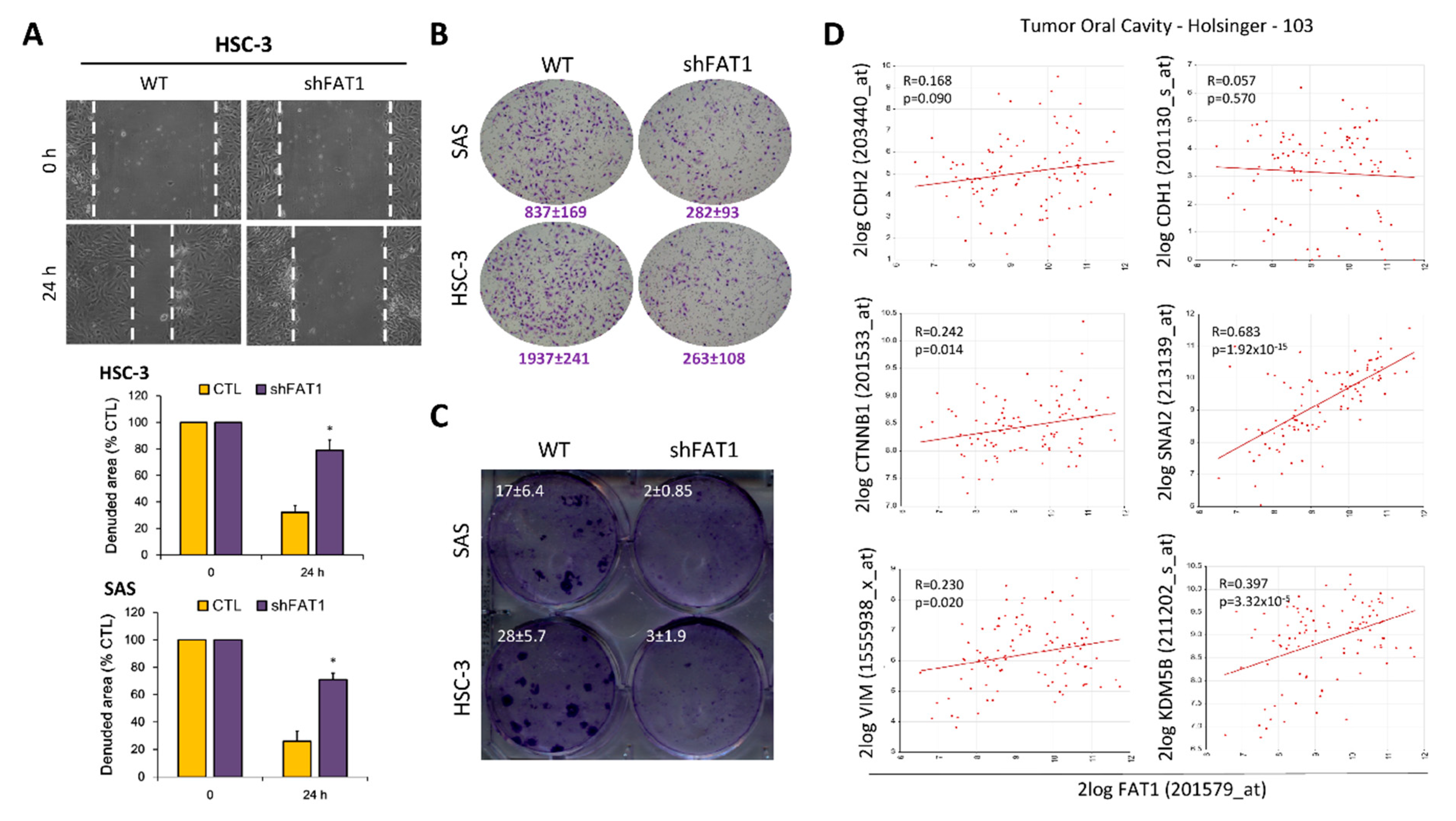
2.5. Loss-of-FAT1 Function Attenuates the Oncogenicity and Metastatic Phenotype of OSCC Cells
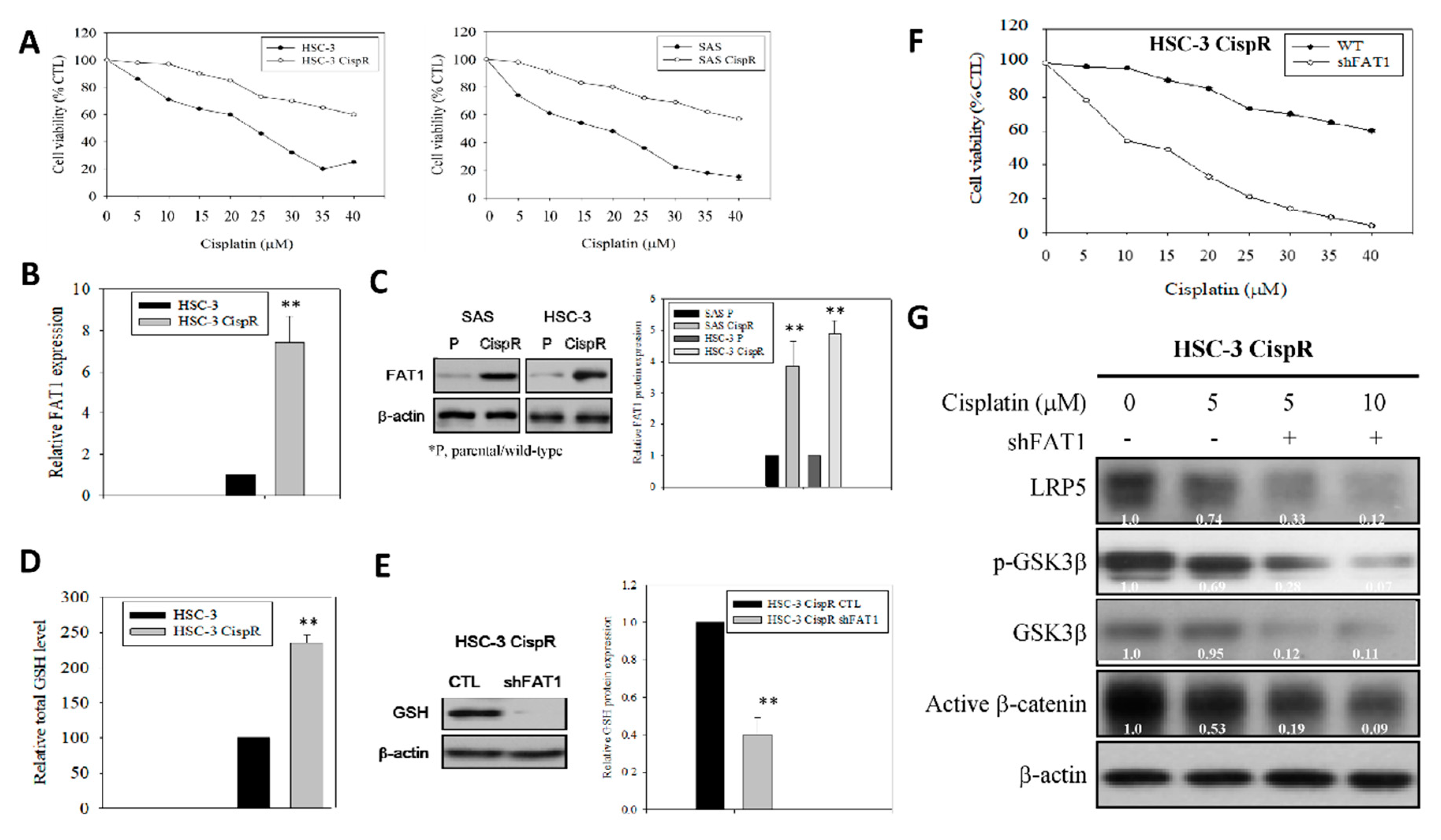
2.6. Molecular Targeting of FAT1 Re-Sensitizes Cisplatin-Resistant OSCC Cells to Cisplatin through Enhanced Oxidative Stress and Deregulation of LRP/Wnt Signaling
3. Discussion
4. Materials and Methods
4.1. Reagents and Drugs
4.2. OSCC Tissue Specimens
4.3. Cell Lines and Cell Culture
4.4. Mutation Status Analyses
4.5. Immunohistochemistry
4.6. shFAT1 Transfection and Establishment of Stable Knockdown Cell Lines
4.7. Cell Migration and Invasion Assays
4.8. Western Blot Analysis
4.9. Cell Viability and Proliferation Assay
4.10. Establishment of Cisplatin-Resistant (CispR) OSCC Cells
4.11. Measurement of Intracellular Glutathione (GSH) Levels
4.12. Real-time Polymerase Chain Reaction (qRT-PCR)
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France; Available online: https://gco.iarc.fr/today (accessed on 8 November 2019).
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality world-wide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- LeHew, C.W.; Weatherspoon, D.J.; Peterson, C.E.; Goben, A.; Reitmajer, K.; Sroussi, H.; Kaste, L.M. The Health System and Policy Implications of Changing Epidemiology for Oral Cavity and Oropharyngeal Cancers in the United States from 1995 to 2016. Epidemiol. Rev. 2017, 39, 132–147. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Shang, Q.C.; Hu, Z.H.; Liu, Y.; Li, B.; Wang, B.; Wang, A.H. Biomarkers in molecular epidemiology study of oral squamous cell carcinoma in the era of precision medicine. Cancer Transl. Med. 2017, 3, 214–218. [Google Scholar]
- National Comprehensive Cancer Network. NCCN Practice Guidelines in Oncology: Head and Neck Cancers. 2013. Available online: http://www.nccn.org/professionals/physician_gls/f_guidelines.asp (accessed on 13 September 2019).
- Chinn, S.B.; Myers, J.N. Oral Cavity Carcinoma: Current Management, Controversies, and Future Directions. J. Clin. Oncol. 2015, 33, 3269–3276. [Google Scholar] [CrossRef] [PubMed]
- Tangthongkum, M.; Kirtsreesakul, V.; Supanimitjaroenporn, P.; Leelasawatsuk, P. Treatment outcome of advance staged oral cavity cancer: Concurrent chemoradiotherapy compared with primary surgery. Eur. Arch. Otorhinolaryngol. 2017, 274, 2567–2572. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.W.; Pouliot, L.M.; Hall, M.D.; Gottesman, M.M. Cisplatin resistance: A cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012, 64, 706–721. [Google Scholar] [CrossRef] [PubMed]
- Mascitti, M.; Rubini, C.; De Michele, F.; Balercia, P.; Girotto, R.; Troiano, G.; Muzio, L.L.; Santarelli, A. American Joint Committee on Cancer staging system 7th edition versus 8th edition: Any improvement for patients with squamous cell carcinoma of the tongue? Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2018, 126, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef]
- Belli, C.; Trapani, D.; Viale, G.; D’Amico, P.; Duso, B.A.; Della Vigna, P.; Orsi, F.; Curigliano, G. Targeting the microenvironment in solid tumors. Cancer Treat. Rev. 2018, 65, 22–32. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; De Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Harjunpää, H.; Asens, M.L.; Guenther, C.; Fagerholm, S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Katoh, M. Function and cancer genomics of FAT family genes (Review). Int. J. Oncol. 2012, 41, 1913–1918. [Google Scholar] [CrossRef] [PubMed]
- Skouloudaki, K.; Puetz, M.; Simons, M.; Courbard, J.R.; Boehlke, C.; Hartleben, B.; Engel, C.; Moeller, M.J.; Englert, C.; Bollig, F.; et al. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc. Natl. Acad. Sci. USA 2009, 106, 8579–8584. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhai, Y.; Kong, P.; Cui, H.; Yan, T.; Yang, J.; Qian, Y.; Ma, Y.; Wang, F.; Li, H.; et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017, 397, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Lin, L.H.; Yu, S.Y.; Kao, S.Y.; Chang, K.W.; Cheng, H.W.; Liu, C.J. FAT1 somatic mutations in head and neck carcinoma are associated with tumor progression and survival. Carcinogenesis 2018, 39, 1320–1330. [Google Scholar] [CrossRef]
- Li, Z.; Razavi, P.; Li, Q.; Toy, W.; Liu, B.; Ping, C.; Hsieh, W.; Sanchez-Vega, F.; Brown, D.N.; Da Cruz Paula, A.F.; et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 2018, 34, 893–905.e8. [Google Scholar] [CrossRef] [PubMed]
- Madan, E.; Dikshit, B.; Gowda, S.H.; Srivastava, C.; Sarkar, C.; Chattopadhyay, P.; Sinha, S.; Chosdol, K. FAT1 is a novel upstream regulator of HIF1α and invasion of high grade glioma. Int. J. Cancer 2016, 139, 2570–2582. [Google Scholar] [CrossRef]
- Valletta, D.; Czech, B.; Spruss, T.; Ikenberg, K.; Wild, P.; Hartmann, A.; Weiss, T.S.; Oefner, P.J.; Müller, M.; Bosserhoff, A.K.; et al. Regulation and function of the atypical cadherin FAT1 in hepatocellular carcinoma. Carcinogenesis 2014, 35, 1407–1415. [Google Scholar] [CrossRef]
- de Bock, C.E.; Ardjmand, A.; Molloy, T.J.; Bone, S.M.; Johnstone, D.; Campbell, D.M.; Shipman, K.L.; Yeadon, T.M.; Holst, J.; Spanevello, M.D.; et al. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia 2012, 26, 918–926. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.E.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018, 132, 587–597. [Google Scholar] [CrossRef]
- Alenzi, F.Q. Links between apoptosis, proliferation and the cell cycle. Br. J. Biomed. Sci. 2004, 61, 99–102. [Google Scholar] [CrossRef]
- Barbieri, B.D.; Marçola, M.; Rocha, C.; Okamoto, O.K. Glutathione depletion overcomes chemotherapy resistance in aggressive medulloblastoma stem-like cells [abstract]. In Proceedings of the AACR International Conference held in cooperation with the Latin American Cooperative Oncology Group (LACOG) on Translational Cancer Medicine, São Paulo, Brazil, 4–6 May 2017. [Google Scholar]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxid. Med. Cell Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Celeste Simon, M. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Sadeqzadeh, E.; de Bock, C.E.; Thorne, R.F. Sleeping giants: Emerging roles for the fat cadherins in health and disease. Med. Res. Rev. 2014, 34, 190–221. [Google Scholar] [CrossRef]
- Saburi, S.; Hester, I.; Goodrich, L.; McNeill, H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development 2012, 139, 1806–1820. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, J.; Liang, X.; Chen, J.; Hong, J.; Li, L.; He, Q.; Cai, X. History and progression of Fat cadherins in health and disease. Oncol. Targets Ther. 2016, 9, 7337–7343. [Google Scholar] [CrossRef]
- Dikshit, B.; Irshad, K.; Madan, E.; Aggarwal, N.; Sarkar, C.; Chandra, P.S.; Gupta, D.K.; Chattopadhyay, P.; Sinha, S.; Chosdol, K. FAT1 acts as an upstream regulator of oncogenic and inflammatory pathways, via PDCD4, in glioma cells. Oncogene 2013, 32, 3798–3808. [Google Scholar] [CrossRef]
- Pileri, P.; Campagnoli, S.; Grandi, A.; Parri, M.; De Camilli, E.; Song, C.; Ganfini, L.; Lacombe, A.; Naldi, I.; Sarmientos, P.; et al. FAT1: A potential target for monoclonal antibody therapy in colon cancer. Br. J. Cancer 2016, 115, 40–51. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Cao, G.Y.; Wang, S.P.; Chen, Y.; Liu, G.D.; Gao, Y.J.; Hu, J.P. POU2F1 promotes growth and metastasis of hepatocellular carcinoma through the FAT1 signaling pathway. Am. J. Cancer Res. 2017, 7, 1665–1679. [Google Scholar]
- Kang, M.H.; Jeong, G.S.; Smoot, D.T.; Ashktorab, H.; Hwang, C.M.; Kim, B.S.; Kim, H.S.; Park, Y.Y. Verteporfin inhibits gastric cancer cell growth by suppressing adhesion molecule FAT1. Oncotarget 2017, 8, 98887–98897. [Google Scholar] [CrossRef]
- Matsui, T.; Ota, T.; Ueda, Y.; Tanino, M.; Odashima, S. Isolation of a highly metastatic cell line to lymph node in human oral squamous cell carcinoma by orthotopic implantation in nude mice. Oral Oncol. 1998, 34, 253–256. [Google Scholar] [CrossRef]
- Takahashi, K.; Kanazawa, H.; Akiyama, Y.; Tazaki, S.; Takahara, M.; Muto, T.; Tanzawa, H.; Sato, K.I. Establishment and characterization of a cell line (SAS) from poorly differentiated human squamous cell carcinoma of the tongue. J. Jpn. Stomatol. Soc. 1989, 38, 20–28. [Google Scholar] [CrossRef]
- Tadateru, A.; Tarou, I.; Tachikawa, T. Establishment and charactalization of human oral squamous cell carcinoma cell line with highly lymph node metastatic potential. J. Jpn. Soc. Oral. Oncol. 2001, 13, 301–306. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. TP53: An oncogene in disguise. Cell Death Differ. 2015, 22, 1239–1249. [Google Scholar] [CrossRef]
- Shen, L.; Shi, Q.; Wang, W. Double agents: Genes with both oncogenic and tumor-suppressor functions. Oncogenesis 2018, 7, 25. [Google Scholar] [CrossRef]
- Muller, H.J. Further studies on the nature and causes of gene mutations. Int. Congr. Genet. 1932, 6, 231–255. [Google Scholar]
- Yang, X.; Chen, M.W.; Terry, S.; Vacherot, F.; Chopin, D.K.; Bemis, D.L.; Kitajewski, J.; Benson, M.C.; Guo, Y.; Buttyan, R. A human- and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res. 2005, 65, 5263–5271. [Google Scholar] [CrossRef]
- Tahir, S.A.; Yang, G.; Goltsov, A.; Song, K.D.; Ren, C.; Wang, J.; Chang, W.; Thompson, T.C. Caveolin-1-LRP6 signaling module stimulates aerobic glycolysis in prostate cancer. Cancer Res. 2013, 73, 1900–1911. [Google Scholar] [CrossRef]
- Li, S.; Li, C.; Jin, S.; Liu, J.; Xue, X.; Eltahan, A.S.; Sun, J.; Tan, J.; Dong, J.; Liang, X.J. Overcoming resistance to cisplatin by inhibition of glutathione S-transferases (GSTs) with ethacraplatin micelles in vitro and in vivo. Biomaterials 2017, 144, 119–129. [Google Scholar] [CrossRef]
- Dong, S.C.; Sha, H.H.; Xu, X.Y.; Hu, T.M.; Lou, R.; Li, H.; Wu, J.Z.; Dan, C.; Feng, J. Glutathione S-transferase π: A potential role in antitumor therapy. Drug Des. Devel. Ther. 2018, 12, 3535–3547. [Google Scholar] [CrossRef]
- Matés, J.M.; Segura, J.A.; Alonso, F.J.; Márquez, J. Intracellular redox status and oxidative stress: Implication for cell proliferation, apoptosis, and carcinogenesis. Arch. Toxicol. 2008, 82, 273–299. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, T.-N.; Huang, C.-M.; Huang, C.-S.; Huang, M.-S.; Yeh, C.-T.; Chao, T.-Y.; Bamodu, O.A. Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma. Cancers 2019, 11, 1883. https://doi.org/10.3390/cancers11121883
Hsu T-N, Huang C-M, Huang C-S, Huang M-S, Yeh C-T, Chao T-Y, Bamodu OA. Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma. Cancers. 2019; 11(12):1883. https://doi.org/10.3390/cancers11121883
Chicago/Turabian StyleHsu, Tung-Nien, Chih-Ming Huang, Chin-Sheng Huang, Mao-Suan Huang, Chi-Tai Yeh, Tsu-Yi Chao, and Oluwaseun Adebayo Bamodu. 2019. "Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma" Cancers 11, no. 12: 1883. https://doi.org/10.3390/cancers11121883
APA StyleHsu, T.-N., Huang, C.-M., Huang, C.-S., Huang, M.-S., Yeh, C.-T., Chao, T.-Y., & Bamodu, O. A. (2019). Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma. Cancers, 11(12), 1883. https://doi.org/10.3390/cancers11121883






