Changes of O6-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse—A Meta-Analysis Type Literature Review
Abstract
1. Introduction
1.1. Epigenetic Changes
1.2. Glioblastoma Multiforme and Gene Methylation
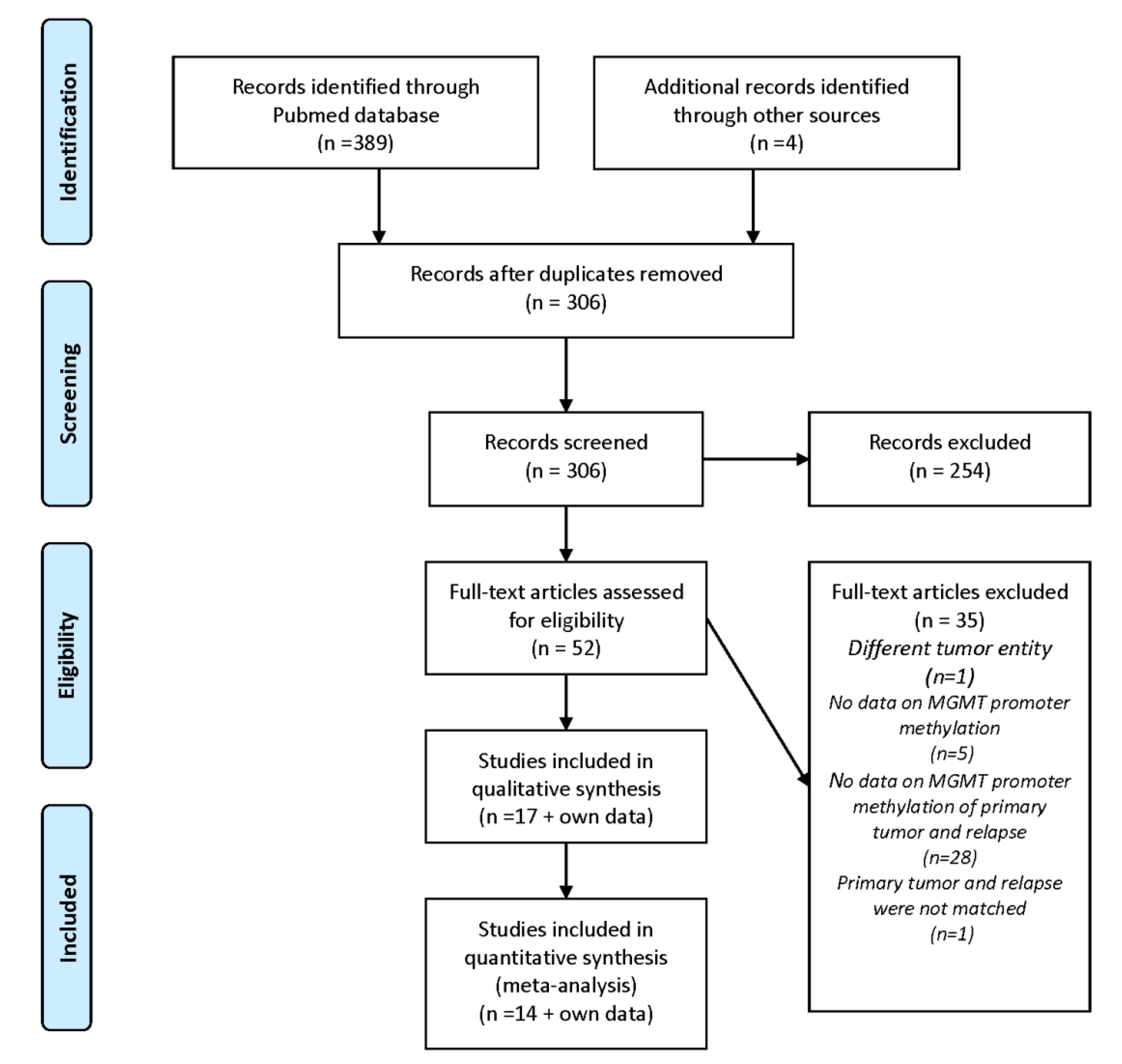
1.3. Rationale and Methodology
2. Initial and Incidental Observations of MGMT Promoter Methylation Changes
3. Systematic Clinical Studies on MGMT Promoter Methylation Changes
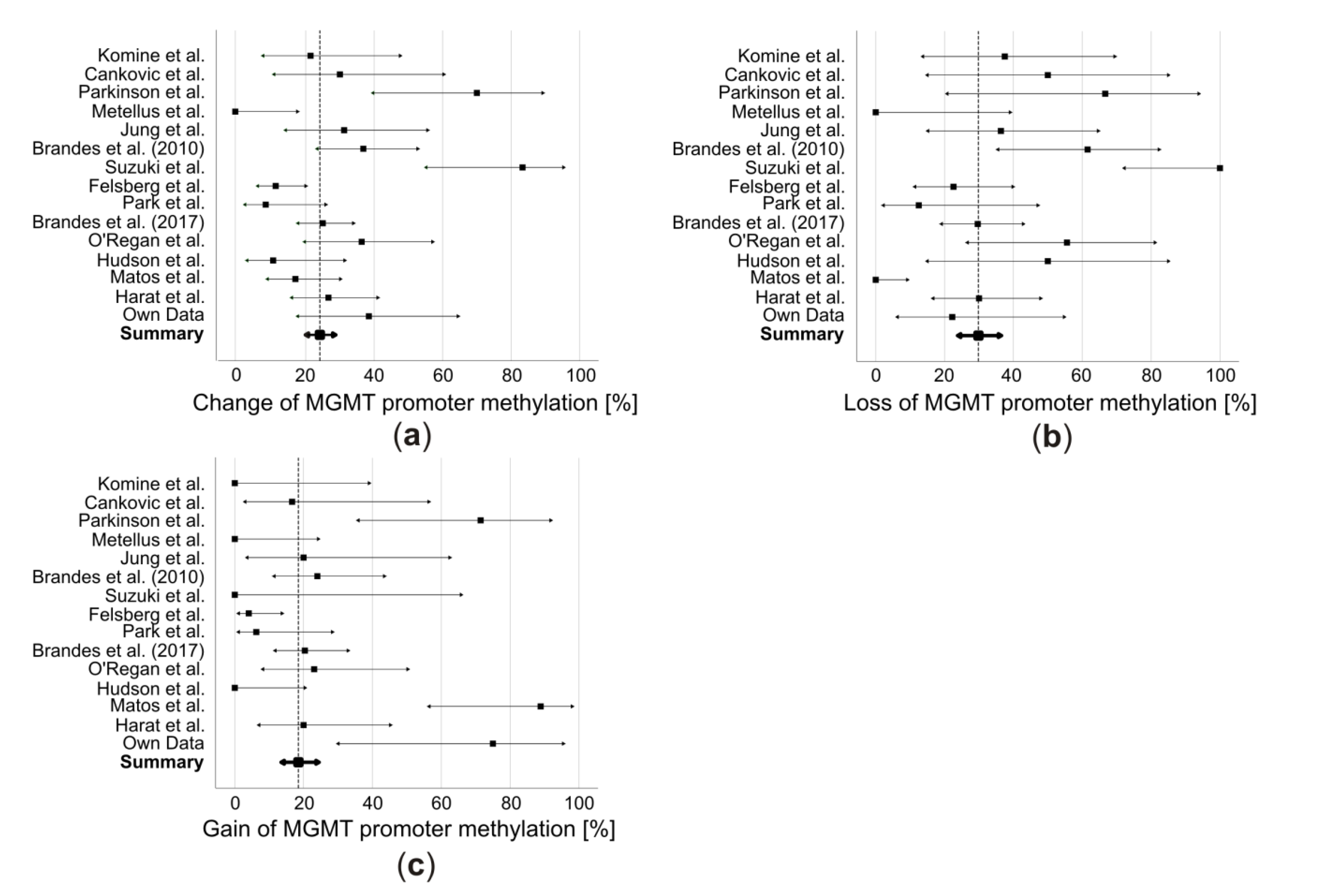
4. Combined-Analysis
5. Potential Reasons for MGMT Promoter Methylation Changes and Their Clinical Implications
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Waddington, C.H. The epigenotype. Endeavour 1942, 1, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Nicoglou, A.; Merlin, F. Epigenetics: A way to bridge the gap between biological fields. Stud. Hist. Philos. Biol. Biomed. Sci. 2017, 66, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G.; Baylin, S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.P. Use of restriction enzymes to study eukaryotic dna methylation II. symmetry of methylated sites supports semi-conservative copying of methylation pattern. J. Mol. Biol. 1978, 118, 49–60. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nature reviews. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [PubMed]
- Dabrowski, M.J.; Wojtas, B. Global DNA Methylation Patterns in Human Gliomas and Their Interplay with Other Epigenetic Modifications. Int. J. Mol. Sci. 2019, 20, 3478. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Natsume, A. Overview of DNA methylation in adult diffuse gliomas. Brain Tumor Pathol. 2019, 36, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Gusyatiner, O.; Hegi, M.E. Glioma epigenetics: From subclassification to novel treatment options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Hottinger, A.F.; Pacheco, P.; Stupp, R. Tumor treating fields: A novel treatment modality and its use in brain tumors. Neuro-Oncology 2016, 18, 1338–1349. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Kesari, S.; Steinberg, D.M.; Toms, S.A.; Taylor, L.P.; Lieberman, F.; Silvani, A.; Fink, K.L.; et al. Maintenance Therapy with Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA 2015, 314, 2535–2543. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, S.; Song, C.; Zha, Y.; Li, L. The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients’ survival: A meta-analysis. World J. Surg. Oncol. 2016, 14, 261. [Google Scholar] [CrossRef]
- Weller, M.; Stupp, R.; Reifenberger, G.; Brandes, A.A.; Van Den Bent, M.J.; Wick, W.; Hegi, M.E. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat. Rev. Neurol. 2010, 6, 39–51. [Google Scholar] [CrossRef]
- Dunn, J.; Baborie, A.; Alam, F.; Joyce, K.; Moxham, M.; Sibson, R.; Crooks, D.; Husband, D.; Shenoy, A.; Brodbelt, A.; et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br. J. Cancer 2009, 101, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Kaina, B.; Christmann, M.; Naumann, S.; Roos, W.P. MGMT: Key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair 2007, 6, 1079–1099. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Tabatabai, G.; Kastner, B.; Felsberg, J.; Steinbach, J.P.; Wick, A.; Schnell, O.; Hau, P.; Herrlinger, U.; Sabel, M.C.; et al. MGMT Promoter Methylation Is a Strong Prognostic Biomarker for Benefit from Dose-Intensified Temozolomide Rechallenge in Progressive Glioblastoma: The DIRECTOR Trial. Clin. Cancer Res. 2015, 21, 2057–2064. [Google Scholar] [CrossRef]
- Malmström, A.; Grønberg, B.H.; Marosi, C.; Stupp, R.; Frappaz, D.; Schultz, H.; Abacioglu, U.; Tavelin, B.; Lhermitte, B.; Hegi, M.E.; et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol. 2012, 13, 916–926. [Google Scholar] [CrossRef]
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012, 13, 707–715. [Google Scholar] [CrossRef]
- Wick, W.; Osswald, M.; Wick, A.; Winkler, F. Treatment of glioblastoma in adults. Ther. Adv. Neurol. Disord. 2018, 11, 1756286418790452. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Herrlinger, U.; Rieger, J.; Koch, D.; Loeser, S.; Blaschke, B.; Kortmann, R.D.; Steinbach, J.P.; Hundsberger, T.; Wick, W.; Meyermann, R.; et al. Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT-03. J. Clin. Oncol. 2006, 24, 4412–4417. [Google Scholar] [CrossRef]
- Glas, M.; Happold, C.; Rieger, J.; Wiewrodt, D.; Bähr, O.; Steinbach, J.P.; Wick, W.; Kortmann, R.; Reifenberger, G.; Weller, M.; et al. Long-term survival of patients with glioblastoma treated with radiotherapy and lomustine plus temozolomide. J. Clin. Oncol. 2009, 27, 1257–1261. [Google Scholar] [CrossRef]
- Herrlinger, U.; Tzaridis, T.; Mack, F.; Steinbach, J.P.; Schlegel, U.; Sabel, M.; Hau, P.; Kortmann, R.D.; Krex, D.; Grauer, O.; et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): A randomised, open-label, phase 3 trial. Lancet 2019, 393, 678–688. [Google Scholar] [CrossRef]
- Vlassenbroeck, I.; Califice, S.; Diserens, A.C.; Migliavacca, E.; Straub, J.; Di Stefano, I.; Moreau, F.; Hamou, M.F.; Renard, I.; Delorenzi, M.; et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J. Mol. Diagn. 2008, 10, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Felsberg, J.; Rapp, M.; Loeser, S.; Fimmers, R.; Stummer, W.; Goeppert, M.; Steiger, H.J.; Friedensdorf, B.; Reifenberger, G.; Sabel, M.C. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin. Cancer Res. 2009, 15, 6683–6693. [Google Scholar] [CrossRef] [PubMed]
- Osuka, S.; Van Meir, E.G. Overcoming therapeutic resistance in glioblastoma: The way forward. J. Clin. Investig. 2017, 127, 415–426. [Google Scholar] [CrossRef]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef]
- Brandes, A.A.; Franceschi, E.; Paccapelo, A.; Tallini, G.; De Biase, D.; Ghimenton, C.; Danieli, D.; Zunarelli, E.; Lanza, G.; Silini, E.M.; et al. Role of MGMT Methylation Status at Time of Diagnosis and Recurrence for Patients with Glioblastoma: Clinical Implications. Oncologist 2017, 22, 432–437. [Google Scholar] [CrossRef]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.I.; Hagemann, C.; Löhr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. OncoTargets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef]
- Agarwal, S.; Suri, V.; Sharma, M.C.; Sarkar, C. Therapy and progression--induced O6-methylguanine-DNA methyltransferase and mismatch repair alterations in recurrent glioblastoma multiforme. Indian J. Cancer 2015, 52, 568–573. [Google Scholar]
- Komine, C.; Watanabe, T.; Katayama, Y.; Yoshino, A.; Yokoyama, T.; Fukushima, T. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003, 13, 176–184. [Google Scholar] [CrossRef]
- Cankovic, M.; Mikkelsen, T.; Rosenblum, M.L.; Zarbo, R.J. A simplified laboratory validated assay for MGMT promoter hypermethylation analysis of glioma specimens from formalin-fixed paraffin-embedded tissue. Lab. Investig. 2007, 87, 392–397. [Google Scholar] [CrossRef]
- Metellus, P.; Coulibaly, B.; Nanni, I.; Fina, F.; Eudes, N.; Giorgi, R.; Barrie, M.; Chinot, O.; Fuentes, S.; Dufour, H.; et al. Prognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: A prospective patient cohort. Cancer 2009, 115, 4783–4794. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Nakada, M.; Hayashi, Y.; Nakada, S.; Sawada-Kitamura, S.; Furuyama, N.; Suzuki, T.; Kamide, T.; Hayashi, Y.; Yano, S.; et al. Epithelioid glioblastoma changed to typical glioblastoma: The methylation status of MGMT promoter and 5-ALA fluorescence. Brain Tumor Pathol. 2011, 28, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Hudson, A.L.; Parker, N.R.; Khong, P.; Parkinson, J.F.; Dwight, T.; Ikin, R.J.; Zhu, Y.; Chen, J.; Wheeler, H.R.; Howell, V.M. Glioblastoma Recurrence Correlates With Increased APE1 and Polarization Toward an Immuno-Suppressive Microenvironment. Front. Oncol. 2018, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Matos, B.; Bostjancic, E.; Matjasic, A.; Popovic, M.; Glavac, D. Dynamic expression of 11 miRNAs in 83 consecutive primary and corresponding recurrent glioblastoma: Correlation to treatment, time to recurrence, overall survival and MGMT methylation status. Radiol. Oncol. 2018, 52, 422–432. [Google Scholar] [CrossRef]
- Parkinson, J.F.; Wheeler, H.R.; Clarkson, A.; McKenzie, C.A.; Biggs, M.T.; Little, N.S.; Cook, R.J.; Messina, M.; Robinson, B.G.; McDonald, K.L. Variation of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J. Neuro-Oncol. 2008, 87, 71–78. [Google Scholar] [CrossRef]
- Christmann, M.; Nagel, G.; Horn, S.; Krahn, U.; Wiewrodt, D.; Sommer, C.; Kaina, B. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: A comparative study on astrocytoma and glioblastoma. Int. J. Cancer 2010, 127, 2106–2118. [Google Scholar] [CrossRef]
- Jung, T.Y.; Jung, S.; Moon, K.S.; Kim, I.Y.; Kang, S.S.; Kim, Y.H.; Park, C.S.; Lee, K.H. Changes of the O6-methylguanine-DNA methyltransferase promoter methylation and MGMT protein expression after adjuvant treatment in glioblastoma. Oncol. Rep. 2010, 23, 1269–1276. [Google Scholar] [CrossRef]
- Brandes, A.A.; Franceschi, E.; Tosoni, A.; Bartolini, S.; Bacci, A.; Agati, R.; Ghimenton, C.; Turazzi, S.; Talacchi, A.; Skrap, M.; et al. O6-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: Clinical implications. Neuro-Oncology 2010, 12, 283–288. [Google Scholar] [CrossRef]
- Felsberg, J.; Thon, N.; Eigenbrod, S.; Hentschel, B.; Sabel, M.C.; Westphal, M.; Schackert, G.; Kreth, F.W.; Pietsch, T.; Löffler, M.; et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int. J. Cancer 2011, 129, 659–670. [Google Scholar] [CrossRef]
- Park, C.K.; Kim, J.E.; Kim, J.Y.; Song, S.W.; Kim, J.W.; Choi, S.H.; Kim, T.M.; Lee, S.H.; Kim, I.H.; Park, S.H. The Changes in MGMT Promoter Methylation Status in Initial and Recurrent Glioblastomas. Transl. Oncol. 2012, 5, 393–397. [Google Scholar] [CrossRef]
- O’Regan, C.J.; Kearney, H.; Beausang, A.; Farrell, M.A.; Brett, F.M.; Cryan, J.B.; Loftus, T.E.; Buckley, P.G. Temporal stability of MGMT promoter methylation in glioblastoma patients undergoing STUPP protocol. J. Neurooncol. 2018, 137, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Barresi, V.; Caffo, M.; De Luca, G.; Giuffr, G. O-6-methylguanine-DNA methyltransferase promoter methylation can change in glioblastoma recurrence due to intratumor heterogeneity. Glioma 2018, 1, 208–213. [Google Scholar] [CrossRef]
- Harat, M.; Blok, M.; Harat, A.; Soszynska, K. The impact of adjuvant radiotherapy on molecular prognostic markers in gliomas. OncoTargets Ther. 2019, 12, 2215–2224. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nakada, M.; Yoshida, Y.; Nambu, E.; Furuyama, N.; Kita, D.; Hayashi, Y.; Hayashi, Y.; Hamada, J.I. The correlation between promoter methylation status and the expression level of O6-methylguanine-DNA methyltransferase in recurrent glioma. Jpn. J. Clin. Oncol. 2011, 41, 190–196. [Google Scholar] [CrossRef][Green Version]
- Switzeny, O.J.; Christmann, M.; Renovanz, M.; Giese, A.; Sommer, C.; Kaina, B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin. Epigenet. 2016, 8, 49. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Wilson, E.B. Probable Inference, the Law of Succession, and Statistical Inference. J. Am. Stat. Assoc. 1927, 22, 209–212. [Google Scholar] [CrossRef]
- Grasbon-Frodl, E.M.; Kreth, F.W.; Ruiter, M.; Schnell, O.; Bise, K.; Felsberg, J.; Reifenberger, G.; Tonn, J.C.; Kretzschmar, H.A. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int. J. Cancer 2007, 121, 2458–2464. [Google Scholar] [CrossRef]
- Parker, N.R.; Hudson, A.L.; Khong, P.; Parkinson, J.F.; Dwight, T.; Ikin, R.J.; Zhu, Y.; Cheng, Z.J.; Vafaee, F.; Chen, J.; et al. Intratumoral heterogeneity identified at the epigenetic, genetic and transcriptional level in glioblastoma. Sci. Rep. 2016, 6, 22477. [Google Scholar] [CrossRef]
- Hamilton, M.G.; Roldan, G.; Magliocco, A.; McIntyre, J.B.; Parney, I.; Easaw, J.C. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J. Neurooncol. 2011, 102, 255–260. [Google Scholar] [CrossRef]
- Della Puppa, A.; Persano, L.; Masi, G.; Rampazzo, E.; Sinigaglia, A.; Pistollato, F.; Denaro, L.; Barzon, L.; Palù, G.; Basso, G.; et al. MGMT expression and promoter methylation status may depend on the site of surgical sample collection within glioblastoma: A possible pitfall in stratification of patients? J. Neurooncol. 2012, 106, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Barciszewska, A.-M.; Gurda, D.; Głodowicz, P.; Nowak, S.; Naskręt-Barciszewska, M.Z. A New Epigenetic Mechanism of Temozolomide Action in Glioma Cells. PLoS ONE 2015, 10, e0136669. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Curry, E.; Magnani, L.; Wilhelm-Benartzi, C.S.; Borley, J. Poised epigenetic states and acquired drug resistance in cancer. Nat. Rev. Cancer 2014, 14, 747. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.; Leder, K.; Hawkins-Daarud, A.; Swanson, K.; Ahmed, A.U.; Rockne, R.C.; Foo, J. Glioblastoma Recurrence and the Role of O6-Methylguanine-DNA Methyltransferase Promoter Methylation. JCO Clin. Cancer Inf. 2019, 3, 1–12. [Google Scholar] [CrossRef]
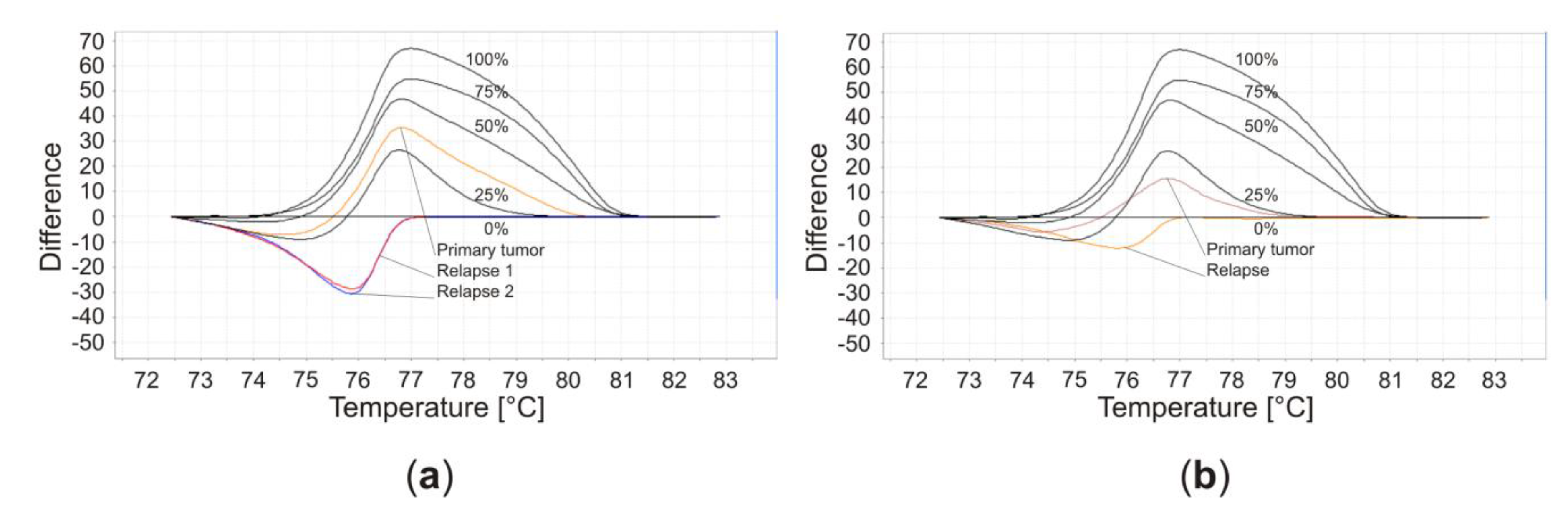
| Patient Characteristics | Tumor Characteristics | Therapy | Outcome | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Sex | Age [years] | ECOG | Tumor Volume [ccm] | Tumor Localization [hemisphere/lobe] | IDH Mutation | MGMT Promoter Methylation | Ki67 Staining [%] | Extent of Resection | RT | TMZ | Relapse | OS [months] | PFS [months] | |
| Primary Tumor | Relapse | ||||||||||||||
| 1 | Female | 69 | 1 | 33.6 | Right/occipital | No | Negative | Negative | 20 | Subtotal | Yes | Yes | Local | 18 | 3 |
| 2 | Female | 49 | 1 | 25.5 | Left/parietal | No | Positive | Positive | 35 | Subtotal | Yes | Yes | Local | 31 | 15 |
| 3 | Male | 42 | 0 | 54.1 | Left/parietal | No | Positive | Negative | 30 | Subtotal | Yes | Yes | Local | 45 | 12 |
| 4 | Female | 70 | 1 | 9.3 | Left/temporal | No | Positive | Positive | 10 | Subtotal | Yes | Yes | Local | 30 | 3 |
| 5 | Female | 59 | 0 | 8.2 | Left/temporal | No | Positive | Positive | 20 | Subtotal | Yes | Yes | Local | 14 | 4 |
| 6 | Male | 48 | 0 | 58.2 | Right/frontal | No | Positive | Positive | 30 | Total | No | Yes | Local | 34 | 22 |
| 7 | Male | 66 | 1 | 19.1 | Left/frontal | No | Negative | Positive | 20 | Subtotal | Yes | Yes | Local | 15 | 4 |
| 8 | Male | 58 | 3 | 89.9 | Right/occipital | No | Positive | Positive | 25 | Subtotal | Yes | Yes | Multifocal | 24 | 4 |
| 9 | Male | 49 | 0 | 54.8 | Left/frontal | Yes | Negative | Positive | 30 | Total | Yes | Yes | Local | 48 | 6 |
| 10 | Male | 60 | 1 | 79.2 | Left/temporal | Yes | Positive | Negative | 25 | Total | Yes | Yes | Local | 25 | 7 |
| 11 | Male | 22 | 3 | 2.2 | Left/frontal | Yes | Positive | Positive | 35 | Subtotal | Yes | Yes | Multifocal | 27 | 10 |
| 12 | Female | 47 | 1 | 2.3 | Left/frontal | No | Positive | Positive | 25 | Subtotal | Yes | Yes | Multifocal | 47 | 7 |
| 13 | Male | 74 | 0 | 60.7 | Left/temporal | No | Negative | Positive | 15 | Subtotal | Yes | No | Local | 12 | 7 |
| Total | Female: 5/38.5% Male: 8/61.5% | Median: 58 | Median: 1 | Median: 33.6 | Left: 10/76.9% Right: 3/23.1% Frontal: 5/38.5% Temporal: 4/30.8% Occipital: 2/15.4% Parietal: 2/15.4% | No: 10/76.9% Yes: 3/32.1% | Negative: 4/30.8% Positive: 9/69.2% | Negative: 3/23.1% Positive: 10/76.9% | Median: 25 | Total: 3/23.1% Subtotal: 10/76.9% | Yes: 12/92.3% No: 1/7.7% | Yes: 12/92.3% No: 1/7.7% | Local: 10/76.9% Multifocal: 3/23.1% | Median: 27 | Median: 7 |
| Patients’ Characteristics | MGMT-Status | Therapy | Outcome | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Authors | Tumorgrade | Total Patients | Male | Female | Median Age at Diagnosis [years] | Methylated Primary Tumor | Unmethylated Primary Tumor | Methylated Relapse | Unmethylated Relapse | MGMT Status in Relapse (Identical/Loss/Gain) | Extent of Tumor Resection (Total/Subtotal/Biopsy) | RT (RT Only/ Concomitant RT + TMZ) | Adjuvant TMZ Therapy | Median PFS [months] | Median OS [months] | Method and (Type of Tissue) |
| Komine et al. (2003) | LGG | 14 | 8 | 6 | 11 | 3 | 11/3/0 | MSP (PE) | ||||||||
| Cankovic et al. (2007) | LGG GBM | 10 | 4 | 6 | 3 | 7 | 7/2/1 | MSP (PE) | ||||||||
| Parkinson et al. (2007) | GBM | 10 | 8 | 2 | 55 | 3 | 7 | 6 | 4 | 3/2/5 | 8/0 | 9 | Promoter sequencing & MSP (FF) | |||
| Metellus et al. (2009) 1 | GBM | 18 | 6 | 12 | 6 | 12 | 18/0/0 | MSP & methyLight technique (FF) | ||||||||
| Christmann et al. (2010) 2,3 | LGG GBM | 9 | 2/0 | 7 | 1/1 | 8 | 7/2/0 | MSP (PE) | ||||||||
| Jung et al. (2010) | GBM | 16 | 7 | 9 | 53 | 11 | 5 | 9 | 7 | 11/4/1 | 15/1/0 | 14/2 | 16 | 19.5 | MSP (PE) | |
| Brandes et al. (2010) | GBM | 38 | 28 | 10 | 49 | 13 | 25 | 11 | 27 | 24/8/6 | 27/10/1 | 11/27 | 38 | 12.0 | 24.3 | MSP (PE) |
| Suzuki et al. (2010) | LGG GBM | 12 | 9 | 3 | 52 | 10 | 2 | 0 | 12 | 2/10/0 | 1/5 | 5 | 18.0 | MSP (PE & FF) | ||
| Tanaka et al. (2010) 2 | GBM | 1 | 1 | 0 | 55 | 1 | 0 | 0 | 1 | 0/1/0 | 1/0/0 | 1/0 | 0 | MSP | ||
| Felsberg et al. (2011) 3 | GBM | 80 | 58 | 22 | 57 | 31/3 | 49 | 26/3 | 54 | 71/7/2 | 40/33/6 | 16/64 | 56 | 9.1 | 18.3 | MSP & Pyro- sequencing (FF) |
| Park et al. (2012) | GBM | 24 | 15 | 9 | 60 | 8 | 16 | 8 | 16 | 22/1/1 | 9/14 | 24 | 8.0 | MSP & MS-MLPA (PE) | ||
| Brandes et al. (2017) | GBM | 108 | 69 | 39 | 51 | 54 | 54 | 49 | 59 | 81/16/11 | 50/42/16 | 0/108 | 24.4 | MSP & Pyro- sequencing (PE) | ||
| O’Regan et al. (2017) | GBM | 22 | 12 | 10 | 50 | 9 | 13 | 7 | 15 | 14/5/3 | 0/22 | 22 | Pyrosequencing (PE) | |||
| Hudson et al. (2018) | GBM | 19 | 12 | 7 | 60 | 4 | 15 | 2 | 17 | 17/2/0 | 0/19 | 19 | 7.0 | 15.0 | Pyrosequencing (FF) | |
| Matos et al. (2018) 4 | GBM | 47 | 38 | 9 | 46 | 1 | 39/0/8 | 47 | MSP (PE) | |||||||
| Barresi et al. (2018) 5 | GBM | 11 | 4 | 7 | 60 | 4 | 10 | 3 | 8 | 11/0/0 | 11/0/0 | 0/11 | 11 | 16.0 | 27.0 | MSP (PE) |
| Harat et al. (2019) 6 | LGG GBM | 45 | 45 | 30 | 15 | 27 | 18 | 33/9/3 | 14/20 | MS-MLPA (PE) | ||||||
| Own Data | GBM | 13 | 8 | 5 | 58 | 9 | 4 | 10 | 3 | 8/2/3 | 3/10/0 | 1/11 | 12 | 7.0 | 27.0 | HRM (FF) |
| Summary5 | 485 | 238 | 253 | 229 | 260 | 377/64/44 | ||||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feldheim, J.; Kessler, A.F.; Monoranu, C.M.; Ernestus, R.-I.; Löhr, M.; Hagemann, C. Changes of O6-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse—A Meta-Analysis Type Literature Review. Cancers 2019, 11, 1837. https://doi.org/10.3390/cancers11121837
Feldheim J, Kessler AF, Monoranu CM, Ernestus R-I, Löhr M, Hagemann C. Changes of O6-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse—A Meta-Analysis Type Literature Review. Cancers. 2019; 11(12):1837. https://doi.org/10.3390/cancers11121837
Chicago/Turabian StyleFeldheim, Jonas, Almuth F. Kessler, Camelia M. Monoranu, Ralf-Ingo Ernestus, Mario Löhr, and Carsten Hagemann. 2019. "Changes of O6-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse—A Meta-Analysis Type Literature Review" Cancers 11, no. 12: 1837. https://doi.org/10.3390/cancers11121837
APA StyleFeldheim, J., Kessler, A. F., Monoranu, C. M., Ernestus, R.-I., Löhr, M., & Hagemann, C. (2019). Changes of O6-Methylguanine DNA Methyltransferase (MGMT) Promoter Methylation in Glioblastoma Relapse—A Meta-Analysis Type Literature Review. Cancers, 11(12), 1837. https://doi.org/10.3390/cancers11121837





