Balancing STAT Activity as a Therapeutic Strategy
Abstract
:1. Introduction
2. Activating STAT3 and STAT5
3. IL-6 Family Cytokine Dysregulation in the TME
4. STAT3 Activation of a Mesenchymal/CSC Program in Cancer Cells
5. A STAT3-Generated Pro-Metastatic Immune Microenvironment
6. STAT5 in Cancer Cells
7. STAT5 in Treg-Associated Immunosuppression
8. Targeting STAT Activity
9. Balancing Opposing STAT-Activated Cytokine Signaling as a Therapeutic Strategy
10. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Lin, E.Y.; Nguyen, A.V.; Russell, R.G.; Pollard, J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001, 193, 727–740. [Google Scholar] [CrossRef]
- Vaupel, P. Tumor microenvironmental physiology and its implications for radiation oncology. Semin. Radiat. Oncol. 2004, 14, 198–206. [Google Scholar] [CrossRef]
- Jimenez-Sanchez, A.; Memon, D.; Pourpe, S.; Veeraraghavan, H.; Li, Y.; Vargas, H.A.; Gill, M.B.; Park, K.J.; Zivanovic, O.; Konner, J.; et al. Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell 2017, 170, 927–938.e920. [Google Scholar] [CrossRef]
- Johnson, B.E.; Mazor, T.; Hong, C.; Barnes, M.; Aihara, K.; McLean, C.Y.; Fouse, S.D.; Yamamoto, S.; Ueda, H.; Tatsuno, K.; et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014, 343, 189–193. [Google Scholar] [CrossRef]
- De Bruin, E.C.; McGranahan, N.; Mitter, R.; Salm, M.; Wedge, D.C.; Yates, L.; Jamal-Hanjani, M.; Shafi, S.; Murugaesu, N.; Rowan, A.J.; et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014, 346, 251–256. [Google Scholar] [CrossRef] [Green Version]
- McGranahan, N.; Favero, F.; de Bruin, E.C.; Birkbak, N.J.; Szallasi, Z.; Swanton, C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015, 7, 283ra254. [Google Scholar] [CrossRef]
- Natrajan, R.; Sailem, H.; Mardakheh, F.K.; Arias Garcia, M.; Tape, C.J.; Dowsett, M.; Bakal, C.; Yuan, Y. Microenvironmental Heterogeneity Parallels Breast Cancer Progression: A Histology-Genomic Integration Analysis. PLoS Med. 2016, 13, e1001961. [Google Scholar] [CrossRef]
- Azizi, E.; Carr, A.J.; Plitas, G.; Cornish, A.E.; Konopacki, C.; Prabhakaran, S.; Nainys, J.; Wu, K.; Kiseliovas, V.; Setty, M. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018, 174, 1293–1308.e1236. [Google Scholar] [CrossRef]
- Rybinski, B.; Yun, K. Addressing intra-tumoral heterogeneity and therapy resistance. Oncotarget 2016, 7, 72322–72342. [Google Scholar] [CrossRef] [Green Version]
- Mroz, E.A.; Tward, A.D.; Pickering, C.R.; Myers, J.N.; Ferris, R.L.; Rocco, J.W. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013, 119, 3034–3042. [Google Scholar] [CrossRef]
- Caswell, D.R.; Swanton, C. The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC Med. 2017, 15, 133. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Quezada, S.A.; Larkin, J.; Swanton, C. Translational implications of tumor heterogeneity. Clin. Cancer Res. 2015, 21, 1258–1266. [Google Scholar] [CrossRef]
- Plava, J.; Cihova, M.; Burikova, M.; Matuskova, M.; Kucerova, L.; Miklikova, S. Recent advances in understanding tumor stroma-mediated chemoresistance in breast cancer. Mol. Cancer 2019, 18, 67. [Google Scholar] [CrossRef]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323. [Google Scholar] [CrossRef]
- Wagner, K.U.; Rui, H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J. Mammary Gland. Biol. Neoplasia 2008, 13, 93–103. [Google Scholar] [CrossRef]
- Ahonen, T.J.; Xie, J.; LeBaron, M.J.; Zhu, J.; Nurmi, M.; Alanen, K.; Rui, H.; Nevalainen, M.T. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J. Biol. Chem. 2003, 278, 27287–27292. [Google Scholar] [CrossRef]
- Tan, S.H.; Dagvadorj, A.; Shen, F.; Gu, L.; Liao, Z.; Abdulghani, J.; Zhang, Y.; Gelmann, E.P.; Zellweger, T.; Culig, Z.; et al. Transcription factor Stat5 synergizes with androgen receptor in prostate cancer cells. Cancer Res. 2008, 68, 236–248. [Google Scholar] [CrossRef]
- Leong, P.L.; Xi, S.; Drenning, S.D.; Dyer, K.F.; Wentzel, A.L.; Lerner, E.C.; Smithgall, T.E.; Grandis, J.R. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene 2002, 21, 2846–2853. [Google Scholar] [CrossRef] [Green Version]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Hosui, A.; Kimura, A.; Yamaji, D.; Zhu, B.M.; Na, R.; Hennighausen, L. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-{beta} and STAT3 activation. J. Exp. Med. 2009, 206, 819–831. [Google Scholar] [CrossRef]
- Lesina, M.; Kurkowski, M.U.; Ludes, K.; Rose-John, S.; Treiber, M.; Klöppel, G.; Yoshimura, A.; Reindl, W.; Sipos, B.; Akira, S.; et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011, 19, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.M.; Jin, X.; Lin, H.J.; Huang, M.; Liu, R.; Reynolds, R.K.; Lin, J. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene 2001, 20, 7925–7934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.L.; Li, Z.W.; Lou, C.J.; Pang, D.; Zhang, Y.Q. Phospho-STAT5 expression is associated with poor prognosis of human colonic adenocarcinoma. Pathol. Oncol. Res. 2011, 17, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.H.; Qi, J.; Lin, S.Q.; Zhang, C.Y.; Liu, F.Y.; Xie, W.D.; Li, X. STAT3 targets ERR-alpha to promote epithelial-mesenchymal transition, migration and invasion in triple negative breast cancer cells. Mol. Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.X.; Huang, F.J.; Aldape, K.D.; Kang, S.H.; Liu, M.; Gershenwald, J.E.; Xie, K.; Sawaya, R.; Huang, S. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006, 66, 3188–3196. [Google Scholar] [CrossRef] [PubMed]
- Abdulghani, J.; Gu, L.; Dagvadorj, A.; Lutz, J.; Leiby, B.; Bonuccelli, G.; Lisanti, M.P.; Zellweger, T.; Alanen, K.; Mirtti, T.; et al. Stat3 promotes metastatic progression of prostate cancer. Am. J. Pathol. 2008, 172, 1717–1728. [Google Scholar] [CrossRef]
- Klupp, F.; Diers, J.; Kahlert, C.; Neumann, L.; Halama, N.; Franz, C.; Schmidt, T.; Lasitschka, F.; Warth, A.; Weitz, J.; et al. Expressional STAT3/STAT5 Ratio is an Independent Prognostic Marker in Colon Carcinoma. Ann. Surg. Oncol. 2015, 22 (Suppl. 3), S1548–S1555. [Google Scholar] [CrossRef]
- Real, P.J.; Sierra, A.; De Juan, A.; Segovia, J.C.; Lopez-Vega, J.M.; Fernandez-Luna, J.L. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene 2002, 21, 7611–7618. [Google Scholar] [CrossRef] [Green Version]
- Fan, Q.M.; Jing, Y.Y.; Yu, G.F.; Kou, X.R.; Ye, F.; Gao, L.; Li, R.; Zhao, Q.D.; Yang, Y.; Lu, Z.H.; et al. Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2014, 352, 160–168. [Google Scholar] [CrossRef]
- Baskiewicz-Masiuk, M.; Machalinski, B. The role of the STAT5 proteins in the proliferation and apoptosis of the CML and AML cells. Eur. J. Haematol. 2004, 72, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Morcinek, J.C.; Weisser, C.; Geissinger, E.; Schartl, M.; Wellbrock, C. Activation of STAT5 triggers proliferation and contributes to anti-apoptotic signalling mediated by the oncogenic Xmrk kinase. Oncogene 2002, 21, 1668–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmud, S.A.; Manlove, L.S.; Farrar, M.A. Interleukin-2 and STAT5 in regulatory T cell development and function. JAKSTAT 2013, 2, e23154. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Murphy, J.J. STAT5 in Cancer and Immunity. J. Interferon Cytokine Res. 2016, 36, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Toker, A.; Liu, Z.Q.; Ohashi, P.S. Turning the Tide Against Regulatory T Cells. Front. Oncol. 2019, 9, 279. [Google Scholar] [CrossRef]
- Shou, J.; Zhang, Z.; Lai, Y.; Chen, Z.; Huang, J. Worse outcome in breast cancer with higher tumor-infiltrating FOXP3+ Tregs: A systematic review and meta-analysis. BMC Cancer 2016, 16, 687. [Google Scholar] [CrossRef]
- Lim, C.P.; Cao, X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2006, 2, 536–550. [Google Scholar] [CrossRef]
- Bromberg, J. Stat proteins and oncogenesis. J. Clin. Investig. 2002, 109, 1139–1142. [Google Scholar] [CrossRef]
- Clevenger, C.V. Roles and Regulation of Stat Family Transcription Factors in Human Breast Cancer. Am. J. Pathol. 2004, 165, 1449–1460. [Google Scholar] [CrossRef] [Green Version]
- Decker, T.; Kovarik, P.; Meinke, A. GAS elements: A few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 1997, 17, 121–134. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.; Kamimura, D.; Hirano, T. Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity 2019, 50, 812–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinrich, P.C.; Behrmann, I.; Haan, S.; Hermanns, H.M.; Muller-Newen, G.; Schaper, F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003, 374, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raz, R.; Lee, C.K.; Cannizzaro, L.A.; d’Eustachio, P.; Levy, D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 1999, 96, 2846–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirito, K.; Osawa, M.; Morita, H.; Shimizu, R.; Yamamoto, M.; Oda, A.; Fujita, H.; Tanaka, M.; Nakajima, K.; Miura, Y.; et al. A functional role of Stat3 in in vivo megakaryopoiesis. Blood 2002, 99, 3220–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akira, S. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 2000, 19, 2607–2611. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Cui, Y.; Watford, W.T.; Bream, J.H.; Yamaoka, K.; Hissong, B.D.; Li, D.; Durum, S.K.; Jiang, Q.; Bhandoola, A.; et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 1000–1005. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Song, D.; Li, H.; Yang, Y.; Ma, X.; Deng, S.; Ren, C.; Shu, X. Negative regulators of STAT3 signaling pathway in cancers. Cancer Manag. Res. 2019, 11, 4957–4969. [Google Scholar] [CrossRef]
- Kershaw, N.J.; Murphy, J.M.; Lucet, I.S.; Nicola, N.A.; Babon, J.J. Regulation of Janus kinases by SOCS proteins. Biochem. Soc. Trans. 2013, 41, 1042–1047. [Google Scholar] [CrossRef]
- Wingelhofer, B.; Neubauer, H.A.; Valent, P.; Han, X.; Constantinescu, S.N.; Gunning, P.T.; Muller, M.; Moriggl, R. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia 2018, 32, 1713–1726. [Google Scholar] [CrossRef] [Green Version]
- Orlova, A.; Wingelhofer, B.; Neubauer, H.A.; Maurer, B.; Berger-Becvar, A.; Keseru, G.M.; Gunning, P.T.; Valent, P.; Moriggl, R. Emerging therapeutic targets in myeloproliferative neoplasms and peripheral T-cell leukemia and lymphomas. Expert Opin. Targets 2018, 22, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef] [PubMed]
- Shahmarvand, N.; Nagy, A.; Shahryari, J.; Ohgami, R.S. Mutations in the signal transducer and activator of transcription family of genes in cancer. Cancer Sci. 2018, 109, 926–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Kee, W.H.; Seow, K.T.; Fung, W.; Cao, X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol. Cell Biol. 2000, 20, 7132–7139. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Witzig, T.E.; Gupta, M. A novel missense (M206K) STAT3 mutation in diffuse large B cell lymphoma deregulates STAT3 signaling. PLoS ONE 2013, 8, e67851. [Google Scholar] [CrossRef]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S.; et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef] [Green Version]
- James, C.; Ugo, V.; Le Couedic, J.P.; Staerk, J.; Delhommeau, F.; Lacout, C.; Garcon, L.; Raslova, H.; Berger, R.; Bennaceur-Griscelli, A.; et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005, 434, 1144–1148. [Google Scholar] [CrossRef]
- Panopoulos, A.D.; Zhang, L.; Snow, J.W.; Jones, D.M.; Smith, A.M.; El Kasmi, K.C.; Liu, F.; Goldsmith, M.A.; Link, D.C.; Murray, P.J.; et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood 2006, 108, 3682–3690. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Tanaka, T.; Narazaki, M.; Kishimoto, T. Targeting Interleukin-6 Signaling in Clinic. Immunity 2019, 50, 1007–1023. [Google Scholar] [CrossRef]
- Mutze, J.; Roth, J.; Gerstberger, R.; Hubschle, T. Nuclear translocation of the transcription factor STAT5 in the rat brain after systemic leptin administration. Neurosci. Lett. 2007, 417, 286–291. [Google Scholar] [CrossRef]
- Ribeiro, D.; Melao, A.; van Boxtel, R.; Santos, C.I.; Silva, A.; Silva, M.C.; Cardoso, B.A.; Coffer, P.J.; Barata, J.T. STAT5 is essential for IL-7-mediated viability, growth, and proliferation of T-cell acute lymphoblastic leukemia cells. Blood Adv. 2018, 2, 2199–2213. [Google Scholar] [CrossRef] [PubMed]
- Demoulin, J.B.; Uyttenhove, C.; Lejeune, D.; Mui, A.; Groner, B.; Renauld, J.C. STAT5 activation is required for interleukin-9-dependent growth and transformation of lymphoid cells. Cancer Res. 2000, 60, 3971–3977. [Google Scholar] [PubMed]
- Rochman, Y.; Kashyap, M.; Robinson, G.W.; Sakamoto, K.; Gomez-Rodriguez, J.; Wagner, K.U.; Leonard, W.J. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 19455–19460. [Google Scholar] [CrossRef] [PubMed]
- Okutani, Y.; Kitanaka, A.; Tanaka, T.; Kamano, H.; Ohnishi, H.; Kubota, Y.; Ishida, T.; Takahara, J. Src directly tyrosine-phosphorylates STAT5 on its activation site and is involved in erythropoietin-induced signaling pathway. Oncogene 2001, 20, 6643–6650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirito, K.; Watanabe, T.; Sawada, K.; Endo, H.; Ozawa, K.; Komatsu, N. Thrombopoietin regulates Bcl-xL gene expression through Stat5 and phosphatidylinositol 3-kinase activation pathways. J. Biol. Chem. 2002, 277, 8329–8337. [Google Scholar] [CrossRef]
- Scheeren, F.A.; Diehl, S.A.; Smit, L.A.; Beaumont, T.; Naspetti, M.; Bende, R.J.; Blom, B.; Karube, K.; Ohshima, K.; van Noesel, C.J.; et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5: Evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood 2008, 111, 4706–4715. [Google Scholar] [CrossRef]
- Maier, E.; Mittermeir, M.; Ess, S.; Neuper, T.; Schmiedlechner, A.; Duschl, A.; Horejs-Hoeck, J. Prerequisites for Functional Interleukin 31 Signaling and Its Feedback Regulation by Suppressor of Cytokine Signaling 3 (SOCS3). J. Biol. Chem. 2015, 290, 24747–24759. [Google Scholar] [CrossRef] [Green Version]
- Toniolo, P.A.; Liu, S.; Yeh, J.E.; Moraes-Vieira, P.M.; Walker, S.R.; Vafaizadeh, V.; Barbuto, J.A.; Frank, D.A. Inhibiting STAT5 by the BET bromodomain inhibitor JQ1 disrupts human dendritic cell maturation. J. Immunol. 2015, 194, 3180–3190. [Google Scholar] [CrossRef]
- Tripathi, P.; Kurtulus, S.; Wojciechowski, S.; Sholl, A.; Hoebe, K.; Morris, S.C.; Finkelman, F.D.; Grimes, H.L.; Hildeman, D.A. STAT5 is critical to maintain effector CD8+ T cell responses. J. Immunol. 2010, 185, 2116–2124. [Google Scholar] [CrossRef]
- Kieslinger, M.; Woldman, I.; Moriggl, R.; Hofmann, J.; Marine, J.C.; Ihle, J.N.; Beug, H.; Decker, T. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev. 2000, 14, 232–244. [Google Scholar]
- Kuroda, E.; Ho, V.; Ruschmann, J.; Antignano, F.; Hamilton, M.; Rauh, M.J.; Antov, A.; Flavell, R.A.; Sly, L.M.; Krystal, G. SHIP represses the generation of IL-3-induced M2 macrophages by inhibiting IL-4 production from basophils. J. Immunol. 2009, 183, 3652–3660. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Li, J.; Hannah, R.; Biddie, S.; Leal-Cervantes, A.I.; Kirschner, K.; Flores Santa Cruz, D.; Sexl, V.; Gottgens, B.; Green, A.R. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBO J. 2016, 35, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.R.; Ulirsch, J.C.; Wilmes, S.; Unal, E.; Moraga, I.; Karakukcu, M.; Yuan, D.; Kazerounian, S.; Abdulhay, N.J.; King, D.S.; et al. Functional Selectivity in Cytokine Signaling Revealed Through a Pathogenic EPO Mutation. Cell 2017, 168, 1053–1064 e1015. [Google Scholar] [CrossRef] [PubMed]
- Wingelhofer, B.; Maurer, B.; Heyes, E.C.; Cumaraswamy, A.A.; Berger-Becvar, A.; de Araujo, E.D.; Orlova, A.; Freund, P.; Ruge, F.; Park, J.; et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia 2018, 32, 1135–1146. [Google Scholar] [CrossRef] [Green Version]
- Page, B.D.; Khoury, H.; Laister, R.C.; Fletcher, S.; Vellozo, M.; Manzoli, A.; Yue, P.; Turkson, J.; Minden, M.D.; Gunning, P.T. Small molecule STAT5-SH2 domain inhibitors exhibit potent antileukemia activity. J. Med. Chem. 2012, 55, 1047–1055. [Google Scholar] [CrossRef]
- Elumalai, N.; Berg, A.; Rubner, S.; Blechschmidt, L.; Song, C.; Natarajan, K.; Matysik, J.; Berg, T. Rational development of Stafib-2: A selective, nanomolar inhibitor of the transcription factor STAT5b. Sci. Rep. 2017, 7, 819. [Google Scholar] [CrossRef]
- Unver, N.; McAllister, F. IL-6 family cytokines: Key inflammatory mediators as biomarkers and potential therapeutic targets. Cytokine Growth Factor Rev. 2018, 41, 10–17. [Google Scholar] [CrossRef]
- Sullivan, N.J.; Sasser, A.K.; Axel, A.E.; Vesuna, F.; Raman, V.; Ramirez, N.; Oberyszyn, T.M.; Hall, B.M. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 2009, 28, 2940–2947. [Google Scholar] [CrossRef]
- Putoczki, T.L.; Ernst, M. IL-11 signaling as a therapeutic target for cancer. Immunotherapy 2015, 7, 441–453. [Google Scholar] [CrossRef]
- Lu, J.; Ksendzovsky, A.; Yang, C.; Mehta, G.U.; Yong, R.L.; Weil, R.J.; Park, D.M.; Mushlin, H.M.; Fang, X.; Balgley, B.M.; et al. CNTF receptor subunit alpha as a marker for glioma tumor-initiating cells and tumor grade: Laboratory investigation. J. Neurosurg. 2012, 117, 1022–1031. [Google Scholar] [CrossRef]
- Li, X.; Yang, Q.; Yu, H.; Wu, L.; Zhao, Y.; Zhang, C.; Yue, X.; Liu, Z.; Wu, H.; Haffty, B.G.; et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014, 5, 788–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, W.C.; Lin, J.T.; Wu, C.Y.; Huang, S.P.; Lin, M.T.; Wu, A.S.; Huang, Y.J.; Wu, M.S. Serum interleukin-6 level but not genotype predicts survival after resection in stages II and III gastric carcinoma. Clin. Cancer Res. 2008, 14, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.P.; Yang, D.C.; Elliott, R.L.; Head, J.F. Reduction in serum IL-6 after vacination of breast cancer patients with tumour-associated antigens is related to estrogen receptor status. Cytokine 2000, 12, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.-C.; Chang, Y.-F. Serum interleukin-6 levels reflect the disease status of colorectal cancer. J. Surg. Oncol. 2003, 83, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Tawara, K.; Oxford, J.T.; Jorcyk, C.L. Clinical significance of interleukin (IL)-6 in cancer metastasis to bone: Potential of anti-IL-6 therapies. Cancer Manag. Res. 2011, 3, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Komoda, H.; Tanaka, Y.; Honda, M.; Matsuo, Y.; Hazama, K.; Takao, T. Interleukin-6 levels in colorectal cancer tissues. World J. Surg. 1998, 22, 895–898. [Google Scholar] [CrossRef]
- Ebbing, E.A.; van der Zalm, A.P.; Steins, A.; Creemers, A.; Hermsen, S.; Rentenaar, R.; Klein, M.; Waasdorp, C.; Hooijer, G.K.J.; Meijer, S.L.; et al. Stromal-derived interleukin 6 drives epithelial-to-mesenchymal transition and therapy resistance in esophageal adenocarcinoma. Proc. Natl. Acad. Sci. USA 2019, 116, 2237–2242. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.; Kumar, B.; Datta, J.; Teknos, T.N.; Kumar, P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol. Cancer Res. 2011, 9, 1658–1667. [Google Scholar] [CrossRef]
- Gyamfi, J.; Lee, Y.H.; Eom, M.; Choi, J. Interleukin-6/STAT3 signalling regulates adipocyte induced epithelial-mesenchymal transition in breast cancer cells. Sci. Rep. 2018, 8, 8859. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, J.; Zhang, W.; Zhang, R.; Ye, Y.; Liu, P.; Yu, W.; Wei, F.; Ren, X.; Yu, J. Interleukin-6 Trans-Signaling Pathway Promotes Immunosuppressive Myeloid-Derived Suppressor Cells via Suppression of Suppressor of Cytokine Signaling 3 in Breast Cancer. Front. Immunol. 2017, 8, 1840. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yu, S.; Li, A.; Xu, H.; Han, X.; Wu, K. Targeting interlukin-6 to relieve immunosuppression in tumor microenvironment. Tumour Biol. 2017, 39, 1010428317712445. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Noma, K.; Ohara, T.; Kashima, H.; Katsura, Y.; Sato, H.; Komoto, S.; Katsube, R.; Ninomiya, T.; Tazawa, H.; et al. Cancer-Associated Fibroblasts Affect Intratumoral CD8(+) and FoxP3(+) T Cells Via IL6 in the Tumor Microenvironment. Clin. Cancer Res. 2018, 24, 4820–4833. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Bournazou, E.; Sansone, P.; Berishaj, M.; Gao, S.P.; Daly, L.; Wels, J.; Theilen, T.; Granitto, S.; Zhang, X.; et al. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia 2013, 15, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tao, P.; Zhou, Q.; Li, J.; Yu, Z.; Wang, X.; Li, J.; Li, C.; Yan, M.; Zhu, Z.; et al. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget 2017, 8, 20741–20750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Q.; Daly, L.; Bromberg, J. The IL-6 feed-forward loop: A driver of tumorigenesis. Semin. Immunol. 2014, 26, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Smigiel, J.; Parvani, J.G.; Tamagno, I.; Polak, K.; Jackson, M.W. Breaking the oncostatin M feed-forward loop to suppress metastasis and therapy failure. J. Pathol. 2018, 245, 6–8. [Google Scholar] [CrossRef] [Green Version]
- Zarling, J.M.; Shoyab, M.; Marquardt, H.; Hanson, M.B.; Lioubin, M.N.; Todaro, G.J. Oncostatin M: A growth regulator produced by differentiated histiocytic lymphoma cells. Proc. Natl. Acad. Sci. USA 1986, 83, 9739–9743. [Google Scholar] [CrossRef]
- Auguste, P.; Guillet, C.; Fourcin, M.; Olivier, C.; Veziers, J.; Pouplard-Barthelaix, A.; Gascan, H. Signaling of type II oncostatin M receptor. J. Biol. Chem. 1997, 272, 15760–15764. [Google Scholar] [CrossRef]
- Mirmohammadsadegh, A.; Hassan, M.; Bardenheuer, W.; Marini, A.; Gustrau, A.; Nambiar, S.; Tannapfel, A.; Bojar, H.; Ruzicka, T.; Hengge, U.R. STAT5 phosphorylation in malignant melanoma is important for survival and is mediated through SRC and JAK1 kinases. J. Investig. Derm. 2006, 126, 2272–2280. [Google Scholar] [CrossRef]
- Lacreusette, A.; Nguyen, J.M.; Pandolfino, M.C.; Khammari, A.; Dreno, B.; Jacques, Y.; Godard, A.; Blanchard, F. Loss of oncostatin M receptor β in metastatic melanoma cells. Oncogene 2006, 26, 881–892. [Google Scholar] [CrossRef]
- Hassel, J.C.; Winnemoller, D.; Schartl, M.; Wellbrock, C. STAT5 contributes to antiapoptosis in melanoma. Melanoma Res. 2008, 18, 378–385. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Murray, J.I.; Watson, P.H. Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene 2014, 33, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Lapeire, L.; Hendrix, A.; Lambein, K.; Van Bockstal, M.; Braems, G.; Van Den Broecke, R.; Limame, R.; Mestdagh, P.; Vandesompele, J.; Vanhove, C.; et al. Cancer-associated adipose tissue promotes breast cancer progression by paracrine oncostatin M and Jak/STAT3 signaling. Cancer Res. 2014, 74, 6806–6819. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Louwagie, J.; Carvalho, B.; Terhaar Sive Droste, J.S.; Park, H.L.; Chae, Y.K.; Yamashita, K.; Liu, J.; Ostrow, K.L.; Ling, S.; et al. Promoter DNA methylation of oncostatin m receptor-beta as a novel diagnostic and therapeutic marker in colon cancer. PLoS ONE 2009, 4, e6555. [Google Scholar] [CrossRef] [PubMed]
- Queen, M.M.; Ryan, R.E.; Holzer, R.G.; Keller-Peck, C.R.; Jorcyk, C.L. Breast cancer cells stimulate neutrophils to produce oncostatin M: Potential implications for tumor progression. Cancer Res. 2005, 65, 8896–8904. [Google Scholar] [CrossRef]
- Junk, D.J.; Bryson, B.L.; Smigiel, J.M.; Parameswaran, N.; Bartel, C.A.; Jackson, M.W. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 2017, 36, 4001–4013. [Google Scholar] [CrossRef] [Green Version]
- Jahani-Asl, A.; Yin, H.; Soleimani, V.D.; Haque, T.; Luchman, H.A.; Chang, N.C.; Sincennes, M.-C.; Puram, S.V.; Scott, A.M.; Lorimer, I.A.J.; et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat. Neurosci. 2016, 19, 798–806. [Google Scholar] [CrossRef] [Green Version]
- Shien, K.; Papadimitrakopoulou, V.A.; Ruder, D.; Behrens, C.; Shen, L.; Kalhor, N.; Song, J.; Lee, J.J.; Wang, J.; Tang, X.; et al. JAK1/STAT3 Activation through a Proinflammatory Cytokine Pathway Leads to Resistance to Molecularly Targeted Therapy in Non-Small Cell Lung Cancer. Mol. Cancer 2017, 16, 2234–2245. [Google Scholar] [CrossRef]
- Smigiel, J.M.; Parameswaran, N.; Jackson, M.W. Potent EMT and CSC Phenotypes Are Induced By Oncostatin-M in Pancreatic Cancer. Mol. Cancer Res. 2017, 15, 478–488. [Google Scholar] [CrossRef] [Green Version]
- Tawara, K.; Bolin, C.; Koncinsky, J.; Kadaba, S.; Covert, H.; Sutherland, C.; Bond, L.; Kronz, J.; Garbow, J.R.; Jorcyk, C.L. OSM potentiates preintravasation events, increases CTC counts, and promotes breast cancer metastasis to the lung. Breast Cancer Res. 2018, 20, 53. [Google Scholar] [CrossRef]
- Doherty, M.R.; Cheon, H.; Junk, D.J.; Vinayak, S.; Varadan, V.; Telli, M.L.; Ford, J.M.; Stark, G.R.; Jackson, M.W. Interferon-beta represses cancer stem cell properties in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 13792–13797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, N.R.; Hegazy, A.N.; Owens, B.M.J.; Bullers, S.J.; Linggi, B.; Buonocore, S.; Coccia, M.; Gortz, D.; This, S.; Stockenhuber, K.; et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017, 23, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Diveu, C.; Venereau, E.; Froger, J.; Ravon, E.; Grimaud, L.; Rousseau, F.; Chevalier, S.; Gascan, H. Molecular and functional characterization of a soluble form of oncostatin M/interleukin-31 shared receptor. J. Biol. Chem. 2006, 281, 36673–36682. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Huang, G.; Wang, R.; Pan, Y.; He, Z.; Chu, X.; Song, H.; Chen, L. Cancer-associated fibroblasts treated with cisplatin facilitates chemoresistance of lung adenocarcinoma through IL-11/IL-11R/STAT3 signaling pathway. Sci. Rep. 2016, 6, 38408. [Google Scholar] [CrossRef]
- Putoczki, T.L.; Thiem, S.; Loving, A.; Busuttil, R.A.; Wilson, N.J.; Ziegler, P.K.; Nguyen, P.M.; Preaudet, A.; Farid, R.; Edwards, K.M.; et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 2013, 24, 257–271. [Google Scholar] [CrossRef]
- Yu, H.; Yue, X.; Zhao, Y.; Li, X.; Wu, L.; Zhang, C.; Liu, Z.; Lin, K.; Xu-Monette, Z.Y.; Young, K.H.; et al. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat. Commun. 2014, 5, 5218. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Yue, X.; Zhao, Y.; Zhang, C.; Li, J.; Liu, Z.; Liu, J.; Hu, W. Leukemia inhibitory factor promotes EMT through STAT3-dependent miR-21 induction. Oncotarget 2016, 7, 3777–3790. [Google Scholar] [CrossRef]
- Bohrer, L.R.; Chuntova, P.; Bade, L.K.; Beadnell, T.C.; Leon, R.P.; Brady, N.J.; Ryu, Y.; Goldberg, J.E.; Schmechel, S.C.; Koopmeiners, J.S.; et al. Activation of the FGFR-STAT3 pathway in breast cancer cells induces a hyaluronan-rich microenvironment that licenses tumor formation. Cancer Res. 2014, 74, 374–386. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, A.; Ojeda-Marquez, L.; Marrugal, A.; Yague, P.; Ponce-Aix, S.; Salinas, A.; Carnero, A.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. The FGFR4-388arg Variant Promotes Lung Cancer Progression by N-Cadherin Induction. Sci. Rep. 2018, 8, 2394. [Google Scholar] [CrossRef] [Green Version]
- Yao, C.; Su, L.; Shan, J.; Zhu, C.; Liu, L.; Liu, C.; Xu, Y.; Yang, Z.; Bian, X.; Shao, J.; et al. IGF/STAT3/NANOG/Slug Signaling Axis Simultaneously Controls Epithelial-Mesenchymal Transition and Stemness Maintenance in Colorectal Cancer. Stem Cells 2016, 34, 820–831. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Ji, J.; Jiang, J.; Cai, Q.; Wang, C.; Shi, M.; Yu, Y.; Zhu, Z.; Zhang, J. HGF-mediated crosstalk between cancer-associated fibroblasts and MET-unamplified gastric cancer cells activates coordinated tumorigenesis and metastasis. Cell Death Dis. 2018, 9, 867. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.C.; Liao, P.N.; Ho, A.S.; Lim, K.H.; Chang, J.; Su, Y.W.; Chen, C.G.; Chiang, Y.W.; Yang, B.L.; Lin, H.C.; et al. STAT3 exacerbates survival of cancer stem-like tumorspheres in EGFR-positive colorectal cancers: RNAseq analysis and therapeutic screening. J. Biomed. Sci. 2018, 25, 60. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Pan, C.; Sun, J.; Gilbert, C.; Drews-Elger, K.; Azzam, D.J.; Picon-Ruiz, M.; Kim, M.; Ullmer, W.; El-Ashry, D.; et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene 2015, 34, 3107–3119. [Google Scholar] [CrossRef]
- Ando, T.; Kudo, Y.; Iizuka, S.; Tsunematsu, T.; Umehara, H.; Shrestha, M.; Matsuo, T.; Kubo, T.; Shimose, S.; Arihiro, K.; et al. Ameloblastin induces tumor suppressive phenotype and enhances chemosensitivity to doxorubicin via Src-Stat3 inactivation in osteosarcoma. Sci. Rep. 2017, 7, 40187. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Xu, Z.; Wang, J.; Zhu, Z.; Lin, G.; Jiang, L.; Lu, X.; Zou, C. Matrine inhibits BCR/ABL mediated ERK/MAPK pathway in human leukemia cells. Oncotarget 2017, 8, 108880–108889. [Google Scholar] [CrossRef] [Green Version]
- Puram, S.V.; Yeung, C.M.; Jahani-Asl, A.; Lin, C.; de la Iglesia, N.; Konopka, G.; Jackson-Grusby, L.; Bonni, A. STAT3-iNOS Signaling Mediates EGFRvIII-Induced Glial Proliferation and Transformation. J. Neurosci. 2012, 32, 7806–7818. [Google Scholar] [CrossRef]
- Yuan, Z.L.; Guan, Y.J.; Chatterjee, D.; Chin, Y.E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 2005, 307, 269–273. [Google Scholar] [CrossRef]
- Limagne, E.; Thibaudin, M.; Euvrard, R.; Berger, H.; Chalons, P.; Vegan, F.; Humblin, E.; Boidot, R.; Rebe, C.; Derangere, V.; et al. Sirtuin-1 Activation Controls Tumor Growth by Impeding Th17 Differentiation via STAT3 Deacetylation. Cell Rep. 2017, 19, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Johnston, P.A.; Grandis, J.R. STAT3 signaling: Anticancer strategies and challenges. Mol. Interv. 2011, 11, 18–26. [Google Scholar] [CrossRef]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E. Stat3 as an Oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Avalle, L.; Pensa, S.; Regis, G.; Novelli, F.; Poli, V. STAT1 and STAT3 in tumorigenesis: A matter of balance. JAKSTAT 2012, 1, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Horvath, C.M.; Besser, D.; Lathem, W.W.; Darnell, J.E., Jr. Stat3 activation is required for cellular transformation by v-src. Mol. Cell Biol. 1998, 18, 2553–2558. [Google Scholar] [CrossRef] [PubMed]
- Dechow, T.N.; Pedranzini, L.; Leitch, A.; Leslie, K.; Gerald, W.L.; Linkov, I.; Bromberg, J.F. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc. Natl. Acad. Sci. USA 2004, 101, 10602–10607. [Google Scholar] [CrossRef] [Green Version]
- Gough, D.J.; Corlett, A.; Schlessinger, K.; Wegrzyn, J.; Larner, A.C.; Levy, D.E. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009, 324, 1713–1716. [Google Scholar] [CrossRef]
- Fukuda, A.; Wang, S.C.; Morris, J.P.t.; Folias, A.E.; Liou, A.; Kim, G.E.; Akira, S.; Boucher, K.M.; Firpo, M.A.; Mulvihill, S.J.; et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 2011, 19, 441–455. [Google Scholar] [CrossRef]
- Bryson, B.L.; Junk, D.J.; Cipriano, R.; Jackson, M.W. STAT3-mediated SMAD3 activation underlies Oncostatin M-induced Senescence. Cell Cycle 2017, 16, 319–334. [Google Scholar] [CrossRef] [Green Version]
- Rokavec, M.; Oner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.H.; Jeong, K.J.; Shin, S.C.; Kang, J.; Park, C.G.; Lee, H.Y. STAT3 mediates TGF-beta1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett. 2013, 336, 167–173. [Google Scholar] [CrossRef]
- Xiong, H.; Hong, J.; Du, W.; Lin, Y.-w.; Ren, L.-l.; Wang, Y.-c.; Su, W.-y.; Wang, J.-l.; Cui, Y.; Wang, Z.-h.; et al. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J. Biol. Chem. 2012, 287, 5819–5832. [Google Scholar] [CrossRef]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.X.; Wei, D.; Liu, M.; Gao, A.C.; Ali-Osman, F.; Sawaya, R.; Huang, S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene 2004, 23, 3550–3560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.H.; Jia, Y.; Guo, F.J.; Chen, J.; Zhang, X.W.; Cui, M.H. Phosphorylation of STAT3 at Tyr705 regulates MMP-9 production in epithelial ovarian cancer. PLoS ONE 2017, 12, e0183622. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Tsai, P.H.; Kandaswami, C.C.; Lee, P.P.; Huang, C.J.; Hwang, J.J.; Lee, M.T. Matrix metalloproteinase-9 cooperates with transcription factor Snail to induce epithelial-mesenchymal transition. Cancer Sci. 2011, 102, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.; Evans, A.J.; Rakha, E.A.; Green, A.R.; Ellis, I.O. Influence of E-cadherin expression on the mammographic appearance of invasive nonlobular breast carcinoma detected at screening. Radiology 2009, 253, 51–55. [Google Scholar] [CrossRef]
- Rakha, E.A.; Abd El Rehim, D.; Pinder, S.E.; Lewis, S.A.; Ellis, I.O. E-cadherin expression in invasive non-lobular carcinoma of the breast and its prognostic significance. Histopathology 2005, 46, 685–693. [Google Scholar] [CrossRef]
- Rakha, E.A.; Patel, A.; Powe, D.G.; Benhasouna, A.; Green, A.R.; Lambros, M.B.; Reis-Filho, J.S.; Ellis, I.O. Clinical and biological significance of E-cadherin protein expression in invasive lobular carcinoma of the breast. Am. J. Surg. Pathol. 2010, 34, 1472–1479. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, X.; Shang, M.; Zhang, Y.; Xia, B.; Niu, M.; Liu, Y.; Pang, D. Dysregulated expression of Slug, vimentin, and E-cadherin correlates with poor clinical outcome in patients with basal-like breast cancer. J. Surg. Oncol. 2013, 107, 188–194. [Google Scholar] [CrossRef]
- Markiewicz, A.; Welnicka-Jaskiewicz, M.; Seroczynska, B.; Skokowski, J.; Majewska, H.; Szade, J.; Zaczek, A.J. Epithelial-mesenchymal transition markers in lymph node metastases and primary breast tumors—Relation to dissemination and proliferation. Am. J. Transl. Res. 2014, 6, 793–808. [Google Scholar]
- Zhou, L.; Dicker, D.T.; Matthew, E.; El-Deiry, W.S.; Alpaugh, R.K. Circulating tumor cells: Silent predictors of metastasis. F1000Research 2017, 6. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, S.; Liu, Z.; Huang, J.; Pu, X.; Li, J.; Yang, D.; Deng, H.; Yang, N.; Xu, J. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS ONE 2015, 10, e0123976. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Mitra, A.; Brownlee, Z.; Xia, X.; Bellister, S.; Overman, M.J.; Kopetz, S.; Ellis, L.M.; Meng, Q.H.; Li, S. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res. 2015, 21, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Doherty, M.R.; Smigiel, J.M.; Junk, D.J.; Jackson, M.W. Cancer Stem Cell Plasticity Drives Therapeutic Resistance. Cancers 2016, 8, 8. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.F.; Jackson, E.L.; Woolfenden, A.E.; Lawrence, S.; Babar, I.; Vogel, S.; Crowley, D.; Bronson, R.T.; Jacks, T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005, 121, 823–835. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef]
- Hayashida, T.; Jinno, H.; Kitagawa, Y.; Kitajima, M. Cooperation of cancer stem cell properties and epithelial-mesenchymal transition in the establishment of breast cancer metastasis. J. Oncol. 2011, 2011, 591427. [Google Scholar] [CrossRef]
- Choi, S.; Yu, J.; Park, A.; Dubon, M.J.; Do, J.; Kim, Y.; Nam, D.; Noh, J.; Park, K.S. BMP-4 enhances epithelial mesenchymal transition and cancer stem cell properties of breast cancer cells via Notch signaling. Sci. Rep. 2019, 9, 11724. [Google Scholar] [CrossRef]
- Hung, J.J.; Kao, Y.S.; Huang, C.H.; Hsu, W.H. Overexpression of Aiolos promotes epithelial-mesenchymal transition and cancer stem cell-like properties in lung cancer cells. Sci. Rep. 2019, 9, 2991. [Google Scholar] [CrossRef] [PubMed]
- Reiman, J.M.; Knutson, K.L.; Radisky, D.C. Immune promotion of epithelial-mesenchymal transition and generation of breast cancer stem cells. Cancer Res. 2010, 70, 3005–3008. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, V.; Rezende, N.C.; Scotland, K.B.; Shaffer, S.M.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009, 18, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.H.; Wu, Q.; Chew, J.L.; Vega, V.B.; Zhang, W.; Chen, X.; Bourque, G.; George, J.; Leong, B.; Liu, J.; et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006, 38, 431–440. [Google Scholar] [CrossRef]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E., 3rd; Zhang, Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Kang, J.W.; Song, X.; Kim, B.K.; Yoo, Y.D.; Kwon, Y.T.; Lee, Y.J. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell Signal 2013, 25, 961–969. [Google Scholar] [CrossRef] [Green Version]
- Bareiss, P.M.; Paczulla, A.; Wang, H.; Schairer, R.; Wiehr, S.; Kohlhofer, U.; Rothfuss, O.C.; Fischer, A.; Perner, S.; Staebler, A.; et al. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013, 73, 5544–5555. [Google Scholar] [CrossRef]
- Jiang, J.; Li, Z.; Yu, C.; Chen, M.; Tian, S.; Sun, C. MiR-1181 inhibits stem cell-like phenotypes and suppresses SOX2 and STAT3 in human pancreatic cancer. Cancer Lett. 2015, 356, 962–970. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, B.H.; Zheng, S.S.; Gao, D.M.; Qiu, S.J.; Wu, W.Z.; Ren, Z.G. Coexpression of gene Oct4 and Nanog initiates stem cell characteristics in hepatocellular carcinoma and promotes epithelial-mesenchymal transition through activation of Stat3/Snail signaling. J. Hematol. Oncol. 2015, 8, 23. [Google Scholar] [CrossRef]
- Van den Hoogen, C.; van der Horst, G.; Cheung, H.; Buijs, J.T.; Lippitt, J.M.; Guzman-Ramirez, N.; Hamdy, F.C.; Eaton, C.L.; Thalmann, G.N.; Cecchini, M.G.; et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010, 70, 5163–5173. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martin, M.; Castellanos, A.; Attolini, C.S.; Berenguer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Mimeault, M.; Batra, S.K. Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J. Cell Mol. Med. 2013, 17, 30–54. [Google Scholar] [CrossRef]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Goldman, A.; Majumder, B.; Dhawan, A.; Ravi, S.; Goldman, D.; Kohandel, M.; Majumder, P.K.; Sengupta, S. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat. Commun. 2015, 6, 6139. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Gao, R.; Sei, E.; Brandt, R.; Hartman, J.; Hatschek, T.; Crosetto, N.; Foukakis, T.; Navin, N.E. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018, 173, 879–893 e813. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, H.; Yin, X.; Long, L.; Chen, X.; Feng, F.; Liu, Y.; Zhao, P.; Xu, Y.; Li, M.; et al. IL-6R/STAT3/miR-204 feedback loop contributes to cisplatin resistance of epithelial ovarian cancer cells. Oncotarget 2017, 8, 39154–39166. [Google Scholar] [CrossRef]
- Ganguly, D.; Fan, M.; Yang, C.H.; Zbytek, B.; Finkelstein, D.; Roussel, M.F.; Pfeffer, L.M. The critical role that STAT3 plays in glioma-initiating cells: STAT3 addiction in glioma. Oncotarget 2018, 9, 22095–22112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Cao, J.; Wu, J.; Sullivan, K.; Shen, J.; Ryu, B.; Xu, Z.; Wei, W.; Cui, R. Stat3-targeted therapies overcome the acquired resistance to vemurafenib in melanomas. J. Investig. Derm. 2013, 133, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Nagathihalli, N.S.; Castellanos, J.A.; Lamichhane, P.; Messaggio, F.; Shi, C.; Dai, X.; Rai, P.; Chen, X.; VanSaun, M.N.; Merchant, N.B. Inverse Correlation of STAT3 and MEK Signaling Mediates Resistance to RAS Pathway Inhibition in Pancreatic Cancer. Cancer Res. 2018, 78, 6235–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Ren, D.; Wu, X.; Lin, X.; Ye, L.; Lin, C.; Wu, S.; Zhu, J.; Peng, X.; Song, L. miR-1266 Contributes to Pancreatic Cancer Progression and Chemoresistance by the STAT3 and NF-kappaB Signaling Pathways. Mol. Nucleic Acids 2018, 11, 142–158. [Google Scholar] [CrossRef]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, F.; Cui, J.Y.; Chen, L.; Chen, Y.T.; Liu, B.W. CAFs enhance paclitaxel resistance by inducing EMT through the IL6/JAK2/STAT3 pathway. Oncol. Rep. 2018, 39, 2081–2090. [Google Scholar] [CrossRef]
- Gupta, P.B.; Fillmore, C.M.; Jiang, G.; Shapira, S.D.; Tao, K.; Kuperwasser, C.; Lander, E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011, 146, 633–644. [Google Scholar] [CrossRef]
- Korkaya, H.; Kim, G.I.; Davis, A.; Malik, F.; Henry, N.L.; Ithimakin, S.; Quraishi, A.A.; Tawakkol, N.; D’Angelo, R.; Paulson, A.K.; et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol. Cell 2012, 47, 570–584. [Google Scholar] [CrossRef]
- Lee, H.J.; Zhuang, G.; Cao, Y.; Du, P.; Kim, H.J.; Settleman, J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 2014, 26, 207–221. [Google Scholar] [CrossRef]
- Simon, G.R.; Somaiah, N. A tabulated summary of targeted and biologic therapies for non-small-cell lung cancer. Clin. Lung Cancer 2014, 15, 21–51. [Google Scholar] [CrossRef]
- Abell, K.; Watson, C.J. The Jak/Stat pathway: A novel way to regulate PI3K activity. Cell Cycle 2005, 4, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Avila, M.A.; Kagan, P.; Sultan, M.; Tachlytski, I.; Safran, M.; Ben-Ari, Z. Both MAPK and STAT3 signal transduction pathways are necessary for IL-6-dependent hepatic stellate cells activation. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-kappaB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, S.; Oishi, K.; Yoshimatsu, T.; Nakafuku, M.; Masuyama, N.; Gotoh, Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 2004, 6, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, M.A.; Patel, A.K.; Nakamura, R.E.; Yi, H.; Surapaneni, K.; Hackam, A.S. The Wnt/beta-catenin pathway cross-talks with STAT3 signaling to regulate survival of retinal pigment epithelium cells. PLoS ONE 2012, 7, e46892. [Google Scholar] [CrossRef]
- Tang, L.Y.; Heller, M.; Meng, Z.; Yu, L.R.; Tang, Y.; Zhou, M.; Zhang, Y.E. Transforming Growth Factor-beta (TGF-beta) Directly Activates the JAK1-STAT3 Axis to Induce Hepatic Fibrosis in Coordination with the SMAD Pathway. J. Biol. Chem. 2017, 292, 4302–4312. [Google Scholar] [CrossRef]
- Bui, Q.T.; Im, J.H.; Jeong, S.B.; Kim, Y.M.; Lim, S.C.; Kim, B.; Kang, K.W. Essential role of Notch4/STAT3 signaling in epithelial-mesenchymal transition of tamoxifen-resistant human breast cancer. Cancer Lett. 2017, 390, 115–125. [Google Scholar] [CrossRef]
- Canino, C.; Luo, Y.; Marcato, P.; Blandino, G.; Pass, H.I.; Cioce, M. A STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget 2015, 6, 12637–12653. [Google Scholar] [CrossRef]
- Alt-Holland, A.; Sowalsky, A.G.; Szwec-Levin, Y.; Shamis, Y.; Hatch, H.; Feig, L.A.; Garlick, J.A. Suppression of E-cadherin function drives the early stages of Ras-induced squamous cell carcinoma through upregulation of FAK and Src. J. Investig. Derm. 2011, 131, 2306–2315. [Google Scholar] [CrossRef]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef]
- D’Amico, S.; Shi, J.; Martin, B.L.; Crawford, H.C.; Petrenko, O.; Reich, N.C. STAT3 is a master regulator of epithelial identity and KRAS-driven tumorigenesis. Genes Dev. 2018, 32, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Saxena, S.; Awaji, M.; Singh, R.K. Tumor-Associated Neutrophils in Cancer: Going Pro. Cancers 2019, 11, 564. [Google Scholar] [CrossRef]
- Yang, C.; He, L.; He, P.; Liu, Y.; Wang, W.; He, Y.; Du, Y.; Gao, F. Increased drug resistance in breast cancer by tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling pathway. Med. Oncol. 2015, 32, 352. [Google Scholar] [CrossRef]
- Webb, N.J.; Myers, C.R.; Watson, C.J.; Bottomley, M.J.; Brenchley, P.E. Activated human neutrophils express vascular endothelial growth factor (VEGF). Cytokine 1998, 10, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cong, X.; Gao, H.; Lan, X.; Li, Z.; Wang, W.; Song, S.; Wang, Y.; Li, C.; Zhang, H.; et al. Tumor-associated neutrophils induce EMT by IL-17a to promote migration and invasion in gastric cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 6. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Xu, J.Y.; Shi, X.Y.; Huang, W.; Ruan, T.Y.; Xie, P.; Ding, J.L. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. 2013, 93, 844–854. [Google Scholar] [CrossRef]
- Pignatelli, J.; Bravo-Cordero, J.J.; Roh-Johnson, M.; Gandhi, S.J.; Wang, Y.; Chen, X.; Eddy, R.J.; Xue, A.; Singer, R.H.; Hodgson, L.; et al. Macrophage-dependent tumor cell transendothelial migration is mediated by Notch1/MenaINV-initiated invadopodium formation. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Pignatelli, J.; Goswami, S.; Jones, J.G.; Rohan, T.E.; Pieri, E.; Chen, X.; Adler, E.; Cox, D.; Maleki, S.; Bresnick, A.; et al. Invasive breast carcinoma cells from patients exhibit MenaINV- and macrophage-dependent transendothelial migration. Sci. Signal 2014, 7, ra112. [Google Scholar] [CrossRef]
- Yang, J.; Liao, D.; Chen, C.; Liu, Y.; Chuang, T.H.; Xiang, R.; Markowitz, D.; Reisfeld, R.A.; Luo, Y. Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells 2013, 31, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Spicer, J.D.; McDonald, B.; Cools-Lartigue, J.J.; Chow, S.C.; Giannias, B.; Kubes, P.; Ferri, L.E. Neutrophils Promote Liver Metastasis via Mac-1–Mediated Interactions with Circulating Tumor Cells. Cancer Res. 2012, 72, 3919–3927. [Google Scholar] [CrossRef] [PubMed]
- Wculek, S.K.; Malanchi, I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 2015, 528, 413–417. [Google Scholar] [CrossRef]
- Psaila, B.; Lyden, D. The metastatic niche: Adapting the foreign soil. Nat. Rev. Cancer 2009, 9, 285–293. [Google Scholar] [CrossRef]
- Deng, J.; Liu, Y.; Lee, H.; Herrmann, A.; Zhang, W.; Zhang, C.; Shen, S.; Priceman, S.J.; Kujawski, M.; Pal, S.K.; et al. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell 2012, 21, 642–654. [Google Scholar] [CrossRef]
- Kanai, T.; Seki, S.; Jenks, J.A.; Kohli, A.; Kawli, T.; Martin, D.P.; Snyder, M.; Bacchetta, R.; Nadeau, K.C. Identification of STAT5A and STAT5B target genes in human T cells. PLoS ONE 2014, 9, e86790. [Google Scholar] [CrossRef]
- Grebien, F.; Kerenyi, M.A.; Kovacic, B.; Kolbe, T.; Becker, V.; Dolznig, H.; Pfeffer, K.; Klingmuller, U.; Muller, M.; Beug, H.; et al. Stat5 activation enables erythropoiesis in the absence of EpoR and Jak2. Blood 2008, 111, 4511–4522. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.M.; McLaughlin, S.K.; Na, R.; Liu, J.; Cui, Y.; Martin, C.; Kimura, A.; Robinson, G.W.; Andrews, N.C.; Hennighausen, L. Hematopoietic-specific Stat5-null mice display microcytic hypochromic anemia associated with reduced transferrin receptor gene expression. Blood 2008, 112, 2071–2080. [Google Scholar] [CrossRef] [Green Version]
- Kerenyi, M.A.; Grebien, F.; Gehart, H.; Schifrer, M.; Artaker, M.; Kovacic, B.; Beug, H.; Moriggl, R.; Mullner, E.W. Stat5 regulates cellular iron uptake of erythroid cells via IRP-2 and TfR-1. Blood 2008, 112, 3878–3888. [Google Scholar] [CrossRef] [Green Version]
- Barash, I. Stat5 in the mammary gland: Controlling normal development and cancer. J. Cell Physiol. 2006, 209, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Cella, N.; Groner, B.; Hynes, N.E. Characterization of Stat5a and Stat5b homodimers and heterodimers and their association with the glucocortiocoid receptor in mammary cells. Mol. Cell Biol. 1998, 18, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Aoki, Y.; Yoshida, M.; Arai, K.; Watanabe, S. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J. Immunol. 2002, 168, 4567–4575. [Google Scholar] [CrossRef] [PubMed]
- Meinke, A.; Barahmand-Pour, F.; Wohrl, S.; Stoiber, D.; Decker, T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol. Cell Biol. 1996, 16, 6937–6944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarino, A.V.; Sciumè, G.; Davis, F.P.; Iwata, S.; Zitti, B.; Robinson, G.W.; Hennighausen, L.; Kanno, Y.; O’Shea, J.J. Subset- and tissue-defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J. Exp. Med. 2017, 214, 2999–3014. [Google Scholar] [CrossRef]
- Maurer, B.; Farlik, M.; Sexl, V. It is a differentiation game: STAT5 in a new role. Cell Death Differ. 2017, 24, 953–954. [Google Scholar] [CrossRef] [Green Version]
- Heltemes-Harris, L.M.; Farrar, M.A. The role of STAT5 in lymphocyte development and transformation. Curr. Opin. Immunol. 2012, 24, 146–152. [Google Scholar] [CrossRef] [Green Version]
- Hoelbl, A.; Kovacic, B.; Kerenyi, M.A.; Simma, O.; Warsch, W.; Cui, Y.; Beug, H.; Hennighausen, L.; Moriggl, R.; Sexl, V. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood 2006, 107, 4898–4906. [Google Scholar] [CrossRef] [Green Version]
- Minieri, V.; De Dominici, M.; Porazzi, P.; Mariani, S.A.; Spinelli, O.; Rambaldi, A.; Peterson, L.F.; Porcu, P.; Nevalainen, M.T.; Calabretta, B. Targeting STAT5 or STAT5-Regulated Pathways Suppresses Leukemogenesis of Ph+ Acute Lymphoblastic Leukemia. Cancer Res. 2018, 78, 5793–5807. [Google Scholar] [CrossRef]
- Gleixner, K.V.; Schneeweiss, M.; Eisenwort, G.; Berger, D.; Herrmann, H.; Blatt, K.; Greiner, G.; Byrgazov, K.; Hoermann, G.; Konopleva, M.; et al. Combined targeting of STAT3 and STAT5: A novel approach to overcome drug resistance in chronic myeloid leukemia. Haematologica 2017, 102, 1519–1529. [Google Scholar] [CrossRef]
- Kaltenecker, D.; Themanns, M.; Mueller, K.M.; Spirk, K.; Golob-Schwarzl, N.; Friedbichler, K.; Kenner, L.; Haybaeck, J.; Moriggl, R. STAT5 deficiency in hepatocytes reduces diethylnitrosamine-induced liver tumorigenesis in mice. Cytokine 2018. [Google Scholar] [CrossRef] [PubMed]
- Boutillon, F.; Pigat, N.; Sala, L.S.; Reyes-Gomez, E.; Moriggl, R.; Guidotti, J.E.; Goffin, V. STAT5a/b Deficiency Delays, but does not Prevent, Prolactin-Driven Prostate Tumorigenesis in Mice. Cancers 2019, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.K.; Man, K.; Poon, R.T.P.; Lo, C.M.; Yuen, A.P.; Ng, I.O.; Ng, K.T.; Leonard, W.; Fan, S.T. Signal Transducers and Activators of Transcription 5b Activation Enhances Hepatocellular Carcinoma Aggressiveness through Induction of Epithelial-Mesenchymal Transition. Cancer Res. 2006, 66, 9948–9956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, A.N.; Bu, W.; Hein, S.; Garcia, S.; Camacho, L.; Xue, L.; Qin, L.; Nagi, C.; Hilsenbeck, S.G.; Kapali, J.; et al. Hyperprolactinemia-inducing antipsychotics increase breast cancer risk by activating JAK-STAT5 in precancerous lesions. Breast Cancer Res. 2018, 20, 42. [Google Scholar] [CrossRef] [Green Version]
- Vafaizadeh, V.; Klemmt, P.; Brendel, C.; Weber, K.; Doebele, C.; Britt, K.; Grez, M.; Fehse, B.; Desrivieres, S.; Groner, B. Mammary epithelial reconstitution with gene-modified stem cells assigns roles to Stat5 in luminal alveolar cell fate decisions, differentiation, involution, and mammary tumor formation. Stem Cells 2010, 28, 928–938. [Google Scholar] [CrossRef]
- Bessette, K.; Lang, M.L.; Fava, R.A.; Grundy, M.; Heinen, J.; Horne, L.; Spolski, R.; Al-Shami, A.; Morse, H.C., 3rd; Leonard, W.J.; et al. A Stat5b transgene is capable of inducing CD8+ lymphoblastic lymphoma in the absence of normal TCR/MHC signaling. Blood 2008, 111, 344–350. [Google Scholar] [CrossRef] [Green Version]
- Xiong, H.; Su, W.Y.; Liang, Q.C.; Zhang, Z.G.; Chen, H.M.; Du, W.; Chen, Y.X.; Fang, J.Y. Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Lab. Investig. 2009, 89, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Koppikar, P.; Lui, V.W.; Man, D.; Xi, S.; Chai, R.L.; Nelson, E.; Tobey, A.B.; Grandis, J.R. Constitutive activation of signal transducer and activator of transcription 5 contributes to tumor growth, epithelial-mesenchymal transition, and resistance to epidermal growth factor receptor targeting. Clin. Cancer Res. 2008, 14, 7682–7690. [Google Scholar] [CrossRef]
- Talati, P.G.; Gu, L.; Ellsworth, E.M.; Girondo, M.A.; Trerotola, M.; Hoang, D.T.; Leiby, B.; Dagvadorj, A.; McCue, P.A.; Lallas, C.D.; et al. Jak2-Stat5a/b Signaling Induces Epithelial-to-Mesenchymal Transition and Stem-Like Cell Properties in Prostate Cancer. Am. J. Pathol. 2015, 185, 2505–2522. [Google Scholar] [CrossRef] [Green Version]
- Creamer, B.A.; Sakamoto, K.; Schmidt, J.W.; Triplett, A.A.; Moriggl, R.; Wagner, K.U. Stat5 promotes survival of mammary epithelial cells through transcriptional activation of a distinct promoter in Akt1. Mol. Cell Biol. 2010, 30, 2957–2970. [Google Scholar] [CrossRef]
- Schmidt, J.W.; Wehde, B.L.; Sakamoto, K.; Triplett, A.A.; Anderson, S.M.; Tsichlis, P.N.; Leone, G.; Wagner, K.U. Stat5 regulates the phosphatidylinositol 3-kinase/Akt1 pathway during mammary gland development and tumorigenesis. Mol. Cell Biol. 2014, 34, 1363–1377. [Google Scholar] [CrossRef] [PubMed]
- Tsuruyama, T.; Hiratsuka, T.; Jin, G.; Imai, Y.; Takeuchi, H.; Maruyama, Y.; Kanaya, K.; Ozeki, M.; Takakuwa, T.; Haga, H.; et al. Murine leukemia retrovirus integration induces the formation of transcription factor complexes on palindromic sequences in the signal transducer and activator of transcription factor 5a gene during the development of pre-B lymphomagenesis. Am. J. Pathol. 2011, 178, 1374–1386. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Q.; Zhang, L.; Zhong, Y.; Fan, G.; Zhang, Z.; Wang, R.; Jin, M.; Qiu, Y.; Kong, D. Stellettin B induces apoptosis in human chronic myeloid leukemia cells via targeting PI3K and Stat5. Oncotarget 2017, 8, 28906–28921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagvadorj, A.; Collins, S.; Jomain, J.B.; Abdulghani, J.; Karras, J.; Zellweger, T.; Li, H.; Nurmi, M.; Alanen, K.; Mirtti, T.; et al. Autocrine prolactin promotes prostate cancer cell growth via Janus kinase-2-signal transducer and activator of transcription-5a/b signaling pathway. Endocrinology 2007, 148, 3089–3101. [Google Scholar] [CrossRef] [PubMed]
- Latha, K.; Li, M.; Chumbalkar, V.; Gururaj, A.; Hwang, Y.; Dakeng, S.; Sawaya, R.; Aldape, K.; Cavenee, W.K.; Bogler, O.; et al. Nuclear EGFRvIII-STAT5b complex contributes to glioblastoma cell survival by direct activation of the Bcl-XL promoter. Int. J. Cancer 2013, 132, 509–520. [Google Scholar] [CrossRef]
- Chueh, F.-Y.; Leong, K.-F.; Yu, C.-L. Mitochondrial translocation of signal transducer and activator of transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated cells. Biochem. Biophys. Res. Commun. 2010, 402, 778–783. [Google Scholar] [CrossRef] [Green Version]
- Walker, S.R.; Xiang, M.; Frank, D.A. Distinct roles of STAT3 and STAT5 in the pathogenesis and targeted therapy of breast cancer. Mol. Cell Endocrinol. 2014, 382, 616–621. [Google Scholar] [CrossRef]
- Sultan, A.S.; Xie, J.; LeBaron, M.J.; Ealley, E.L.; Nevalainen, M.T.; Rui, H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene 2005, 24, 746–760. [Google Scholar] [CrossRef]
- Mallette, F.A.; Gaumont-Leclerc, M.F.; Ferbeyre, G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007, 21, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Nishikawa, H.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol. 2014, 27, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Knol, A.C.; Nguyen, J.M.; Quereux, G.; Brocard, A.; Khammari, A.; Dreno, B. Prognostic value of tumor-infiltrating Foxp3+ T-cell subpopulations in metastatic melanoma. Exp. Derm. 2011, 20, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Sasada, T.; Kimura, M.; Yoshida, Y.; Kanai, M.; Takabayashi, A. CD4+CD25+ regulatory T cells in patients with gastrointestinal malignancies: Possible involvement of regulatory T cells in disease progression. Cancer 2003, 98, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Li, J.; Tian, Y.; Mao, J.; Liu, X.; Feng, S.; Li, J.; Bian, Q.; Ji, H.; Zhang, L. Restoring Th17/Treg balance via modulation of STAT3 and STAT5 activation contributes to the amelioration of chronic obstructive pulmonary disease by Bufei Yishen formula. J. Ethnopharmacol. 2018, 217, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Burchill, M.A.; Yang, J.; Vogtenhuber, C.; Blazar, B.R.; Farrar, M.A. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007, 178, 280–290. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Paul, W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 2010, 327, 1098–1102. [Google Scholar] [CrossRef]
- Li, X.; Kostareli, E.; Suffner, J.; Garbi, N.; Hammerling, G.J. Efficient Treg depletion induces T-cell infiltration and rejection of large tumors. Eur. J. Immunol. 2010, 40, 3325–3335. [Google Scholar] [CrossRef]
- Fisher, S.A.; Aston, W.J.; Chee, J.; Khong, A.; Cleaver, A.L.; Solin, J.N.; Ma, S.; Lesterhuis, W.J.; Dick, I.; Holt, R.A.; et al. Transient Treg depletion enhances therapeutic anti-cancer vaccination. Immun. Inflamm. Dis. 2017, 5, 16–28. [Google Scholar] [CrossRef]
- Schmidt, A.; Zhang, X.M.; Joshi, R.N.; Iqbal, S.; Wahlund, C.; Gabrielsson, S.; Harris, R.A.; Tegner, J. Human macrophages induce CD4(+)Foxp3(+) regulatory T cells via binding and re-release of TGF-beta. Immunol. Cell Biol. 2016, 94, 747–762. [Google Scholar] [CrossRef]
- Goswami, K.K.; Ghosh, T.; Ghosh, S.; Sarkar, M.; Bose, A.; Baral, R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017, 316, 1–10. [Google Scholar] [CrossRef]
- Müller-Hermelink, N.; Braumüller, H.; Pichler, B.; Wieder, T.; Mailhammer, R.; Schaak, K.; Ghoreschi, K.; Yazdi, A.; Haubner, R.; Sander, C.A.; et al. TNFR1 Signaling and IFN-γ Signaling Determine whether T Cells Induce Tumor Dormancy or Promote Multistage Carcinogenesis. Cancer Cell 2008, 13, 507–518. [Google Scholar] [CrossRef]
- Edwards, J.P.; Thornton, A.M.; Shevach, E.M. Release of active TGF-beta1 from the latent TGF-beta1/GARP complex on T regulatory cells is mediated by integrin beta8. J. Immunol. 2014, 193, 2843–2849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schafer, C.C.; Hough, K.P.; Tousif, S.; Duncan, S.R.; Kearney, J.F.; Ponnazhagan, S.; Hsu, H.C.; Deshane, J.S. Myeloid-Derived Suppressor Cells Impair B Cell Responses in Lung Cancer through IL-7 and STAT5. J. Immunol. 2018, 201, 278–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurieva, R.I.; Podd, A.; Chen, Y.; Alekseev, A.M.; Yu, M.; Qi, X.; Huang, H.; Wen, R.; Wang, J.; Li, H.S.; et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J. Biol. Chem. 2012, 287, 11234–11239. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.D.; O’Sullivan, D.; Klein Geltink, R.I.; Curtis, J.D.; Chang, C.H.; Sanin, D.E.; Qiu, J.; Kretz, O.; Braas, D.; van der Windt, G.J.; et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 2016, 166, 63–76. [Google Scholar] [CrossRef] [Green Version]
- Berod, L.; Friedrich, C.; Nandan, A.; Freitag, J.; Hagemann, S.; Harmrolfs, K.; Sandouk, A.; Hesse, C.; Castro, C.N.; Bahre, H.; et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014, 20, 1327–1333. [Google Scholar] [CrossRef]
- Michalek, R.D.; Gerriets, V.A.; Jacobs, S.R.; Macintyre, A.N.; MacIver, N.J.; Mason, E.F.; Sullivan, S.A.; Nichols, A.G.; Rathmell, J.C. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011, 186, 3299–3303. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.X.; Li, P.; Liu, D.; Jin, H.T.; He, J.; Ata Ur Rasheed, M.; Rochman, Y.; Wang, L.; Cui, K.; Liu, C.; et al. Critical Role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 2012, 36, 586–599. [Google Scholar] [CrossRef]
- Owen, D.L.; Farrar, M.A. STAT5 and CD4 (+) T Cell Immunity. F1000Research 2017, 6, 32. [Google Scholar] [CrossRef]
- Grange, M.; Buferne, M.; Verdeil, G.; Leserman, L.; Schmitt-Verhulst, A.M.; Auphan-Anezin, N. Activated STAT5 promotes long-lived cytotoxic CD8+ T cells that induce regression of autochthonous melanoma. Cancer Res. 2012, 72, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Gotthardt, D.; Putz, E.M.; Grundschober, E.; Prchal-Murphy, M.; Straka, E.; Kudweis, P.; Heller, G.; Bago-Horvath, Z.; Witalisz-Siepracka, A.; Cumaraswamy, A.A.; et al. STAT5 Is a Key Regulator in NK Cells and Acts as a Molecular Switch from Tumor Surveillance to Tumor Promotion. Cancer Discov. 2016, 6, 414–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, D.; Sopper, S.; Pircher, A.; Gastl, G.; Wolf, A.M. Treg(s) in Cancer: Friends or Foe? J. Cell Physiol. 2015, 230, 2598–2605. [Google Scholar] [CrossRef] [PubMed]
- Turkson, J.; Kim, J.S.; Zhang, S.; Yuan, J.; Huang, M.; Glenn, M.; Haura, E.; Sebti, S.; Hamilton, A.D.; Jove, R. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol. Cancer 2004, 3, 261–269. [Google Scholar]
- Fontaine, F.; Overman, J.; Francois, M. Pharmacological manipulation of transcription factor protein-protein interactions: Opportunities and obstacles. Cell Regen. 2015, 4, 2. [Google Scholar] [CrossRef]
- Siddiquee, K.; Zhang, S.; Guida, W.C.; Blaskovich, M.A.; Greedy, B.; Lawrence, H.R.; Yip, M.L.; Jove, R.; McLaughlin, M.M.; Lawrence, N.J.; et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. USA 2007, 104, 7391–7396. [Google Scholar] [CrossRef] [Green Version]
- Siddiquee, K.A.; Gunning, P.T.; Glenn, M.; Katt, W.P.; Zhang, S.; Schrock, C.; Sebti, S.M.; Jove, R.; Hamilton, A.D.; Turkson, J. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem. Biol. 2007, 2, 787–798. [Google Scholar] [CrossRef]
- Sato, K.; Tsuchiya, M.; Saldanha, J.; Koishihara, Y.; Ohsugi, Y.; Kishimoto, T.; Bendig, M.M. Reshaping a human antibody to inhibit the interleukin 6-dependent tumor cell growth. Cancer Res. 1993, 53, 851–856. [Google Scholar]
- Mihara, M.; Kasutani, K.; Okazaki, M.; Nakamura, A.; Kawai, S.; Sugimoto, M.; Matsumoto, Y.; Ohsugi, Y. Tocilizumab inhibits signal transduction mediated by both mIL-6R and sIL-6R, but not by the receptors of other members of IL-6 cytokine family. Int. Immunopharmacol. 2005, 5, 1731–1740. [Google Scholar] [CrossRef]
- Nishimoto, N.; Sasai, M.; Shima, Y.; Nakagawa, M.; Matsumoto, T.; Shirai, T.; Kishimoto, T.; Yoshizaki, K. Improvement in Castleman’s disease by humanized anti-interleukin-6 receptor antibody therapy. Blood 2000, 95, 56–61. [Google Scholar] [CrossRef]
- Nishimoto, N.; Yoshizaki, K.; Miyasaka, N.; Yamamoto, K.; Kawai, S.; Takeuchi, T.; Hashimoto, J.; Azuma, J.; Kishimoto, T. Treatment of rheumatoid arthritis with humanized anti-interleukin-6 receptor antibody: A multicenter, double-blind, placebo-controlled trial. Arthritis Rheum. 2004, 50, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Intravenous Tocilizumab: A Review of Its Use in Adults with Rheumatoid Arthritis. BioDrugs 2013, 28, 75–106. [Google Scholar] [CrossRef] [PubMed]
- Le, R.Q.; Li, L.; Yuan, W.; Shord, S.S.; Nie, L.; Habtemariam, B.A.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist 2018, 23, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Dambacher, J.; Beigel, F.; Seiderer, J.; Haller, D.; Goke, B.; Auernhammer, C.J.; Brand, S. Interleukin 31 mediates MAP kinase and STAT1/3 activation in intestinal epithelial cells and its expression is upregulated in inflammatory bowel disease. Gut 2007, 56, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Smith, M.A.; Doshi, P.; Sasser, K.; Fulp, W.; Altiok, S.; Haura, E.B. Antitumor efficacy of the anti-interleukin-6 (IL-6) antibody siltuximab in mouse xenograft models of lung cancer. J. Thorac. Oncol. 2014, 9, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Chen, B. Siltuximab (CNTO 328): A promising option for human malignancies. Drug Des. Devel. 2015, 9, 3455–3458. [Google Scholar] [CrossRef] [PubMed]
- Fizazi, K.; De Bono, J.S.; Flechon, A.; Heidenreich, A.; Voog, E.; Davis, N.B.; Qi, M.; Bandekar, R.; Vermeulen, J.T.; Cornfeld, M.; et al. Randomised phase II study of siltuximab (CNTO 328), an anti-IL-6 monoclonal antibody, in combination with mitoxantrone/prednisone versus mitoxantrone/prednisone alone in metastatic castration-resistant prostate cancer. Eur. J. Cancer 2012, 48, 85–93. [Google Scholar] [CrossRef]
- Takata, F.; Dohgu, S.; Matsumoto, J.; Machida, T.; Sakaguchi, S.; Kimura, I.; Yamauchi, A.; Kataoka, Y. Oncostatin M-induced blood-brain barrier impairment is due to prolonged activation of STAT3 signaling in vitro. J. Cell Biochem. 2018, 119, 9055–9063. [Google Scholar] [CrossRef]
- Jorcyk, C.L.; Holzer, R.G.; Ryan, R.E. Oncostatin M induces cell detachment and enhances the metastatic capacity of T-47D human breast carcinoma cells. Cytokine 2006, 33, 323–336. [Google Scholar] [CrossRef]
- Sterbova, S.; Karlsson, T.; Persson, E. Oncostatin M induces tumorigenic properties in non-transformed human prostate epithelial cells, in part through activation of signal transducer and activator of transcription 3 (STAT3). Biochem. Biophys. Res. Commun. 2018, 498, 769–774. [Google Scholar] [CrossRef]
- Richards, C.D. The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm. 2013, 2013, 512103. [Google Scholar] [CrossRef] [PubMed]
- Hintzen, C.; Evers, C.; Lippok, B.E.; Volkmer, R.; Heinrich, P.C.; Radtke, S.; Hermanns, H.M. Box 2 region of the oncostatin M receptor determines specificity for recruitment of Janus kinases and STAT5 activation. J. Biol. Chem. 2008, 283, 19465–19477. [Google Scholar] [CrossRef]
- Hermanns, H.M.; Radtke, S.; Schaper, F.; Heinrich, P.C.; Behrmann, I. Non-redundant signal transduction of interleukin-6-type cytokines. The adapter protein Shc is specifically recruited to rhe oncostatin M receptor. J. Biol. Chem. 2000, 275, 40742–40748. [Google Scholar] [CrossRef] [PubMed]
- Choy, E.H.; Bendit, M.; McAleer, D.; Liu, F.; Feeney, M.; Brett, S.; Zamuner, S.; Campanile, A.; Toso, J. Safety, tolerability, pharmacokinetics and pharmacodynamics of an anti- oncostatin M monoclonal antibody in rheumatoid arthritis: Results from phase II randomized, placebo-controlled trials. Arthritis Res. 2013, 15, R132. [Google Scholar] [CrossRef] [PubMed]
- Poling, J.; Gajawada, P.; Richter, M.; Lorchner, H.; Polyakova, V.; Kostin, S.; Shin, J.; Boettger, T.; Walther, T.; Rees, W.; et al. Therapeutic targeting of the oncostatin M receptor-beta prevents inflammatory heart failure. Basic Res. Cardiol. 2014, 109, 396. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Du, Y.; Li, K.; Zhang, W.; Feng, X.; Hao, J.; Li, H.; Liu, S. Anti-OSM Antibody Inhibits Tubulointerstitial Lesion in a Murine Model of Lupus Nephritis. Mediat. Inflamm. 2017, 2017, 3038514. [Google Scholar] [CrossRef] [PubMed]
- Kucia-Tran, J.A.; Tulkki, V.; Scarpini, C.G.; Smith, S.; Wallberg, M.; Paez-Ribes, M.; Araujo, A.M.; Botthoff, J.; Feeney, M.; Hughes, K.; et al. Anti-oncostatin M antibody inhibits the pro-malignant effects of oncostatin M receptor overexpression in squamous cell carcinoma. J. Pathol. 2018, 244, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.J.; Turner, R.C.; Naser, Z.J.; McCurdy, C.R.; O’Callaghan, J.P.; Huber, J.D.; Matsumoto, R.R. SN79, a sigma receptor antagonist, attenuates methamphetamine-induced astrogliosis through a blockade of OSMR/gp130 signaling and STAT3 phosphorylation. Exp. Neurol. 2014, 254, 180–189. [Google Scholar] [CrossRef] [Green Version]
- Bottos, A.; Gotthardt, D.; Gill, J.W.; Gattelli, A.; Frei, A.; Tzankov, A.; Sexl, V.; Wodnar-Filipowicz, A.; Hynes, N.E. Decreased NK-cell tumour immunosurveillance consequent to JAK inhibition enhances metastasis in breast cancer models. Nat. Commun. 2016, 7, 12258. [Google Scholar] [CrossRef] [Green Version]
- Doherty, M.R.; Jackson, M.W. Using interferon-beta to combat cancer stem cell properties in triple negative breast cancer. Oncoscience 2018, 5, 169–171. [Google Scholar] [CrossRef]
- Doherty, M.R.; Parvani, J.G.; Tamagno, I.; Junk, D.J.; Bryson, B.L.; Cheon, H.J.; Stark, G.R.; Jackson, M.W. The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer. Breast Cancer Res. 2019, 21, 54. [Google Scholar] [CrossRef] [PubMed]
- Sistigu, A.; Yamazaki, T.; Vacchelli, E.; Chaba, K.; Enot, D.P.; Adam, J.; Vitale, I.; Goubar, A.; Baracco, E.E.; Remedios, C.; et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 2014, 20, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Fu, M.L.; Weichselbaum, R.R.; Gajewski, T.F.; Guo, Y.; Fu, Y.X. Targeting the tumor microenvironment with interferon-beta bridges innate and adaptive immune responses. Cancer Cell 2014, 25, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Marini, F.; Konopleva, M.; Schober, W.; Shi, Y.; Burks, J.; Clise-Dwyer, K.; Wang, R.Y.; Zhang, W.; Yuan, X.; et al. Mesenchymal Stem Cells Overexpressing IFN-beta Inhibit Breast Cancer Growth and Metastases through Stat3 Signaling in a Syngeneic Tumor Model. Cancer Microenviron. 2010, 3, 83–95. [Google Scholar] [CrossRef]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef]
- Bidwell, B.N.; Slaney, C.Y.; Withana, N.P.; Forster, S.; Cao, Y.; Loi, S.; Andrews, D.; Mikeska, T.; Mangan, N.E.; Samarajiwa, S.A.; et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat. Med. 2012, 18, 1224–1231. [Google Scholar] [CrossRef]
- Liu, S.; Lachapelle, J.; Leung, S.; Gao, D.; Foulkes, W.D.; Nielsen, T.O. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012, 14, R48. [Google Scholar] [CrossRef]
- Xie, C.; Liu, C.; Wu, B.; Lin, Y.; Ma, T.; Xiong, H.; Wang, Q.; Li, Z.; Ma, C.; Tu, Z. Effects of IRF1 and IFN-beta interaction on the M1 polarization of macrophages and its antitumor function. Int. J. Mol. Med. 2016, 38, 148–160. [Google Scholar] [CrossRef]
- Moreira-Teixeira, L.; Sousa, J.; McNab, F.W.; Torrado, E.; Cardoso, F.; Machado, H.; Castro, F.; Cardoso, V.; Gaifem, J.; Wu, X.; et al. Type I IFN Inhibits Alternative Macrophage Activation during Mycobacterium tuberculosis Infection and Leads to Enhanced Protection in the Absence of IFN-gamma Signaling. J. Immunol. 2016, 197, 4714–4726. [Google Scholar] [CrossRef]
- Pylaeva, E.; Lang, S.; Jablonska, J. The Essential Role of Type I Interferons in Differentiation and Activation of Tumor-Associated Neutrophils. Front. Immunol. 2016, 7, 629. [Google Scholar] [CrossRef] [Green Version]
- Brockwell, N.K.; Owen, K.L.; Zanker, D.; Spurling, A.; Rautela, J.; Duivenvoorden, H.M.; Baschuk, N.; Caramia, F.; Loi, S.; Darcy, P.K.; et al. Neoadjuvant Interferons: Critical for Effective PD-1-Based Immunotherapy in TNBC. Cancer Immunol. Res. 2017, 5, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Essers, M.A.; Offner, S.; Blanco-Bose, W.E.; Waibler, Z.; Kalinke, U.; Duchosal, M.A.; Trumpp, A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009, 458, 904–908. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Jin, S.; Yang, W.; Zhou, G.; Zhao, M.; Fang, S.; Zhang, Z.; Hu, J. Interferon-alpha enhances the antitumour activity of EGFR-targeted therapies by upregulating RIG-I in head and neck squamous cell carcinoma. Br. J. Cancer 2018, 118, 509–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheon, H.; Holvey-Bates, E.G.; Schoggins, J.W.; Forster, S.; Hertzog, P.; Imanaka, N.; Rice, C.M.; Jackson, M.W.; Junk, D.J.; Stark, G.R. IFNbeta-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013, 32, 2751–2763. [Google Scholar] [CrossRef]
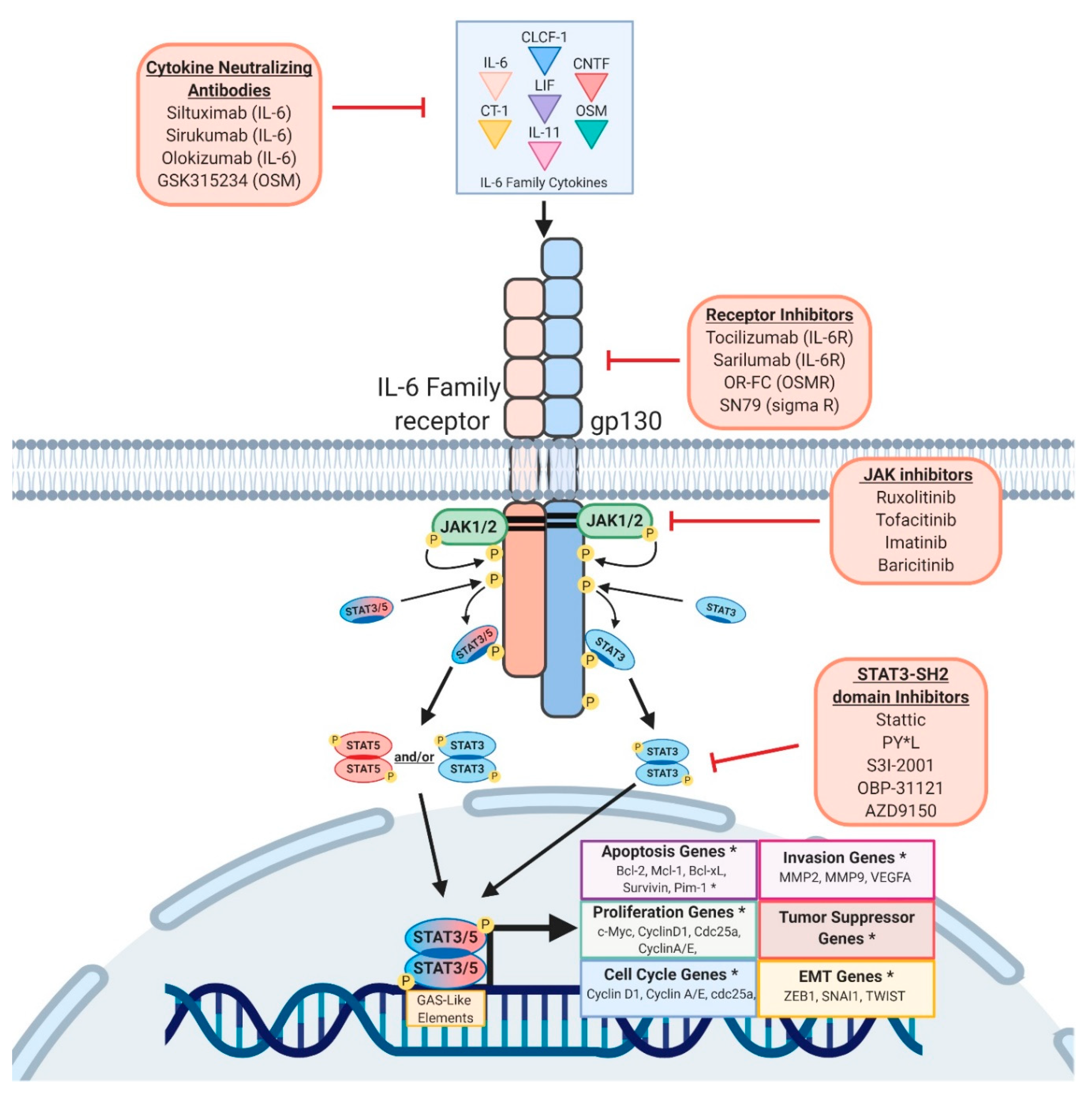
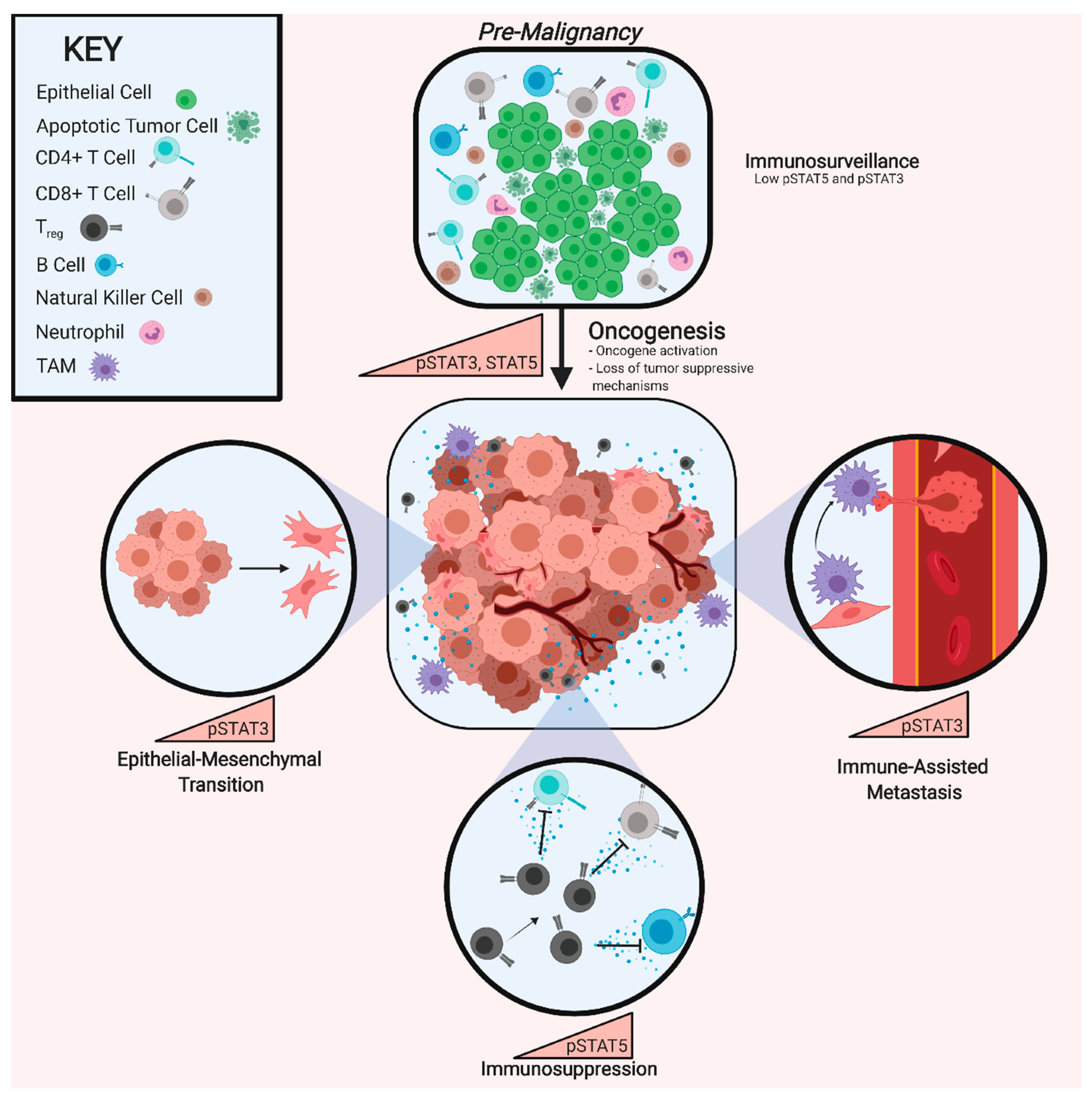
| STAT-Family Protein | pYSTAT3/5-Activating Cytokines and Growth Factors | Normal Immune Function | Pro-Tumorigenic Genes Activated | Pro-Tumorigenic Effects | Therapies and Drugs in Development |
|---|---|---|---|---|---|
| STAT3 | Growth Factors: EGFs, FGFs, HGFs, Leptin (REF), PDGFs Lymphoid Cytokines: IL-4, IL-13 Myeloid Cytokines: G-CSF, IL-10 Ubiquitous Cytokines: CNTF, LIF, IL-6, OSM | Development of Mature Neutrophils [58] Neutrophil Mobilization from Bone Marrow [59] | Apoptosis: BCL2, BCL-XL, MCL1, PIM1 Cell Cycle: Cyclin A, Cyclin D1, CDC25A EMT: ZEB1, SNAI1, TWIST, CDH2, Vimentin Migration/Invasion: MMP2, MMP9, VEGFA Proliferation: cMYC, NOTCH4 Stem Cell: SOX2, NANOG, OCT4 Tumor Suppressor: p53 | Transformation Activation of Anti-Apoptotic Proteins EMT M2 Polarization N2 Polarization Enhanced Metastasis Therapy Resistance | Imatinib (JAK1/2) Ruxolitinib (JAK1/2) Tofacitinib (JAK1/2) Siltuximab (IL-6) Tocilizumab (IL-6R) GSK315234 (OSM) OS-FC (OSMR) PY*L (SH2 Domain) S3I-2001 (SH2 Domain) SN79 (Sigma Receptor) IFNβ |
| STAT5 | Growth Factors: EGFs, FGFs, FLT3L, Leptin [60], PDGFs, Prolactin, SCF Lymphoid Cytokines: IL-2, IL-4, IL-7 [61], IL-9 [62], IL-13, IL-15, Thymic Stromal Lymphopoietin [63] Myeloid Cytokines: Erythropoietin [64], G-CSF, GM-CSF, IL-3, IL-5, IL-10, Thrombopoietin [65] Ubiquitous Cytokines: IL-21 [66], IL-31 [67], OSM | Differentiation, Survival, and Lineage Expansion of NK and NKT Cells Dendritic Cell Function [68] T Cell Differentiation and Expansion Effector Memory CD8+ T Cell-Mediated Cancer Cell Clearance [69] Myeloid Cell Differentiation and Survival [70] | Apoptosis: BCL-2, BCL-XL, MCL1, PIM1, Survivin Cell Cycle: Cyclin D1, Cyclin E Immune Function: PRDM1, BCL-6 Migration/Invasion: MMP2, MMP9, VEGFA Proliferation: AKT1, cMYC, PI3K Tumor Suppressor: p53 | Transformation Activation of Anti-Apoptotic Proteins Treg Population Expansion M2 Polarization [71] Megakaryopoeisis [72] Erythropoeisis [73] | Imatinib (JAK1/2) Ruxolitinib (JAK1/2) Tofacitinib (JAK1/2) Siltuximab (IL-6) Tocilizumab (IL-6R) GSK315234 (OSM) OS-FC (OSMR) AC-4-130 (SH2 Domain) [74] BP-1-107 (SH2 Domain) [75] BP-1-108 (SH2 Domain) [75] Pomstafib-2 (SH2 Domain) [76] SF-1-087 (SH2 Domain) [75] SF-1-088 (SH2 Domain) [75] IFNβ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polak, K.L.; Chernosky, N.M.; Smigiel, J.M.; Tamagno, I.; Jackson, M.W. Balancing STAT Activity as a Therapeutic Strategy. Cancers 2019, 11, 1716. https://doi.org/10.3390/cancers11111716
Polak KL, Chernosky NM, Smigiel JM, Tamagno I, Jackson MW. Balancing STAT Activity as a Therapeutic Strategy. Cancers. 2019; 11(11):1716. https://doi.org/10.3390/cancers11111716
Chicago/Turabian StylePolak, Kelsey L., Noah M. Chernosky, Jacob M. Smigiel, Ilaria Tamagno, and Mark W. Jackson. 2019. "Balancing STAT Activity as a Therapeutic Strategy" Cancers 11, no. 11: 1716. https://doi.org/10.3390/cancers11111716
APA StylePolak, K. L., Chernosky, N. M., Smigiel, J. M., Tamagno, I., & Jackson, M. W. (2019). Balancing STAT Activity as a Therapeutic Strategy. Cancers, 11(11), 1716. https://doi.org/10.3390/cancers11111716





