Tumor and Cerebrospinal Fluid microRNAs in Primary Central Nervous System Lymphomas
Abstract
:1. Introduction
2. Results
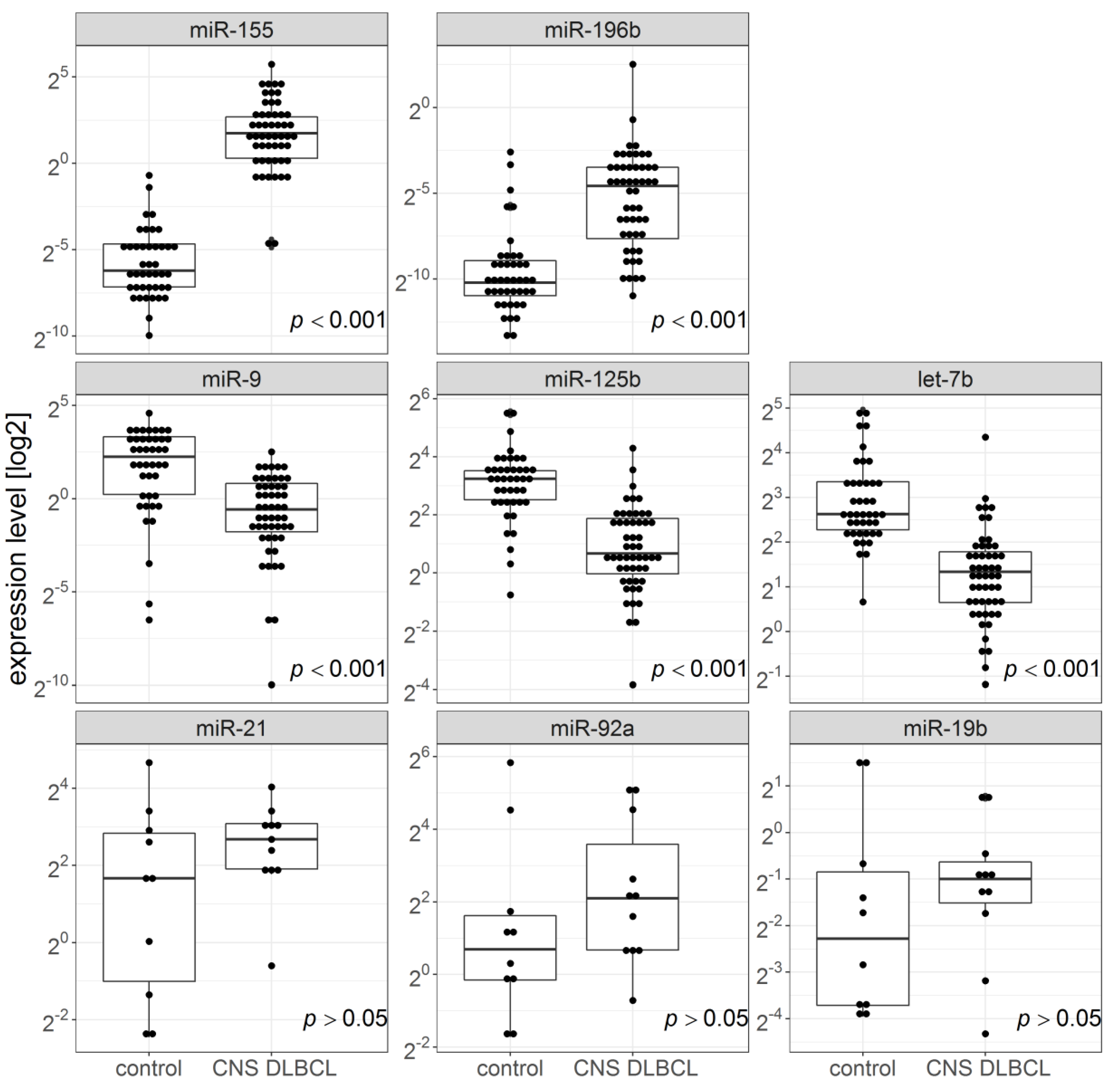
2.1. miR-155, miR-196b, miR-9, miR-125b, and miR-Let-7b Levels in FFPET Brain Biopsies and CSF Samples from Patients with CNS, DLBCL vs. n-ML
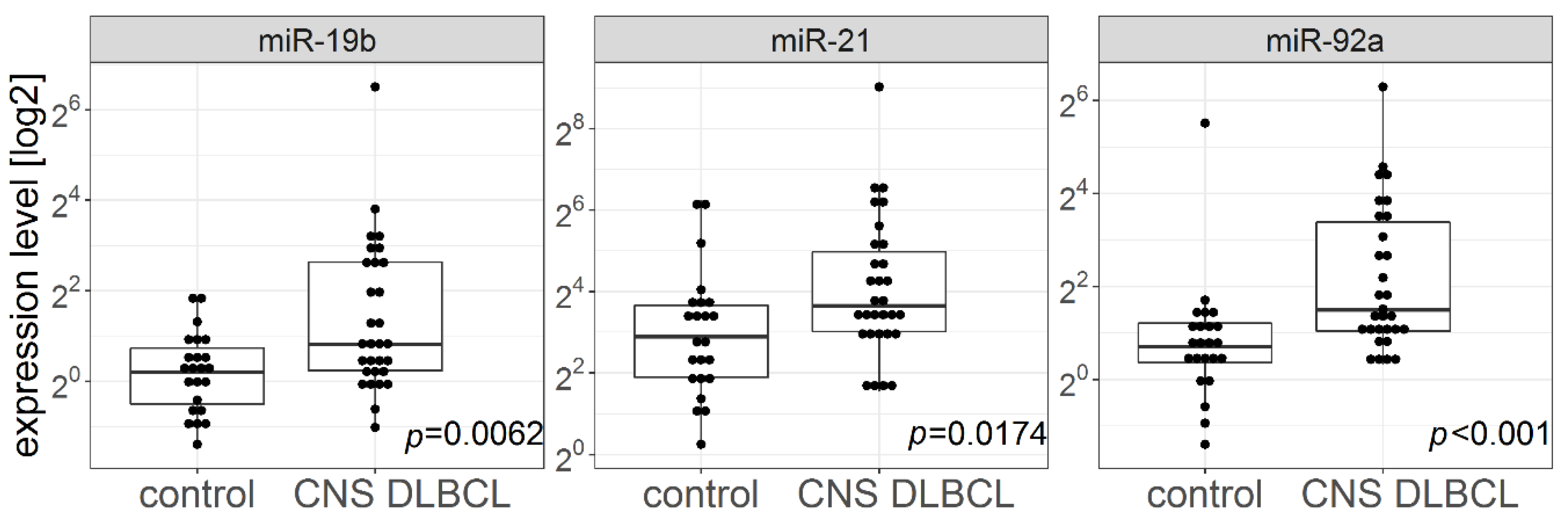
2.2. miR-21, miR-19b, and miR-92a Levels in CSFs and Brain Biopsies from Patients with CNS DLBCL vs. n-ML
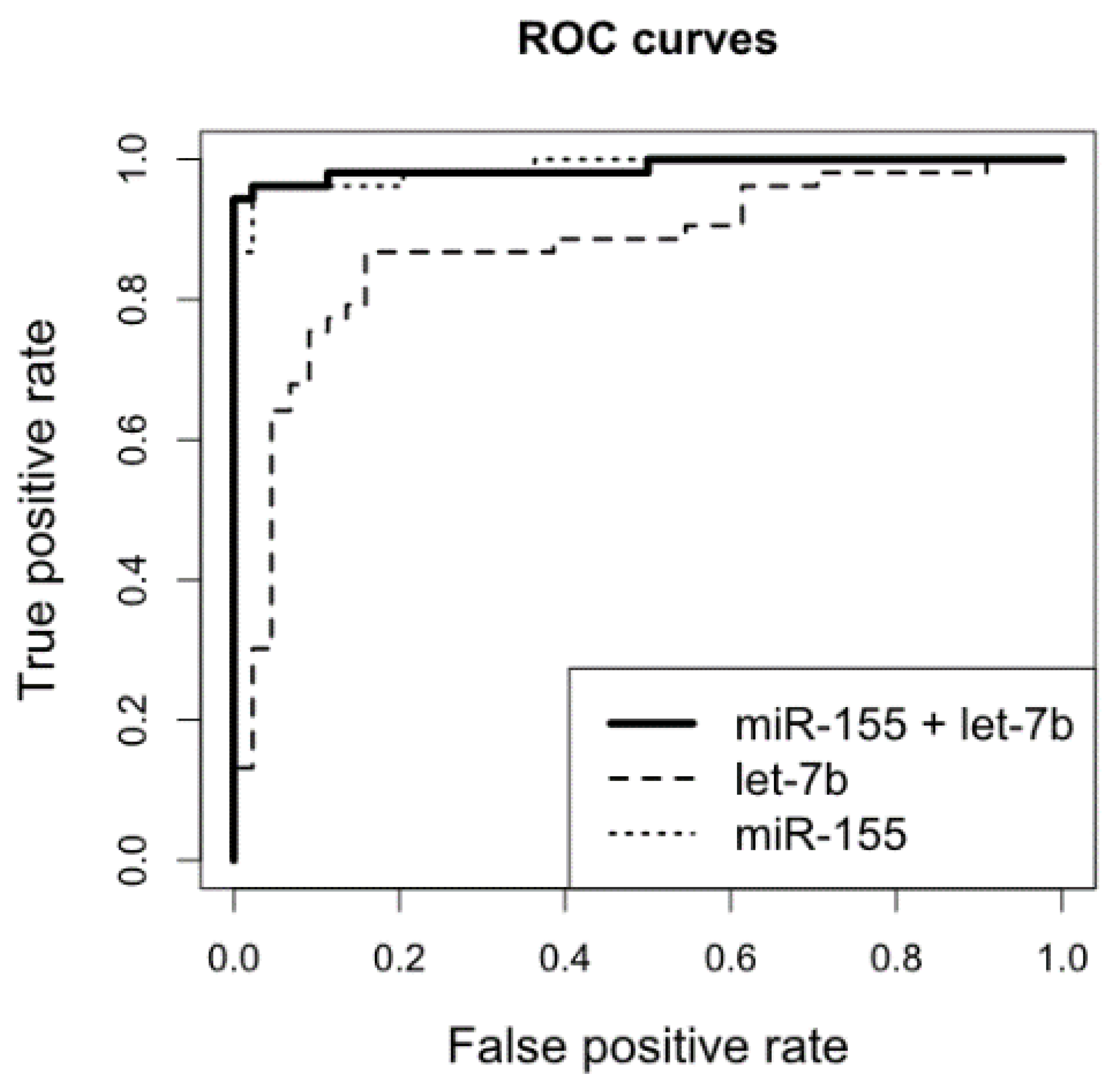
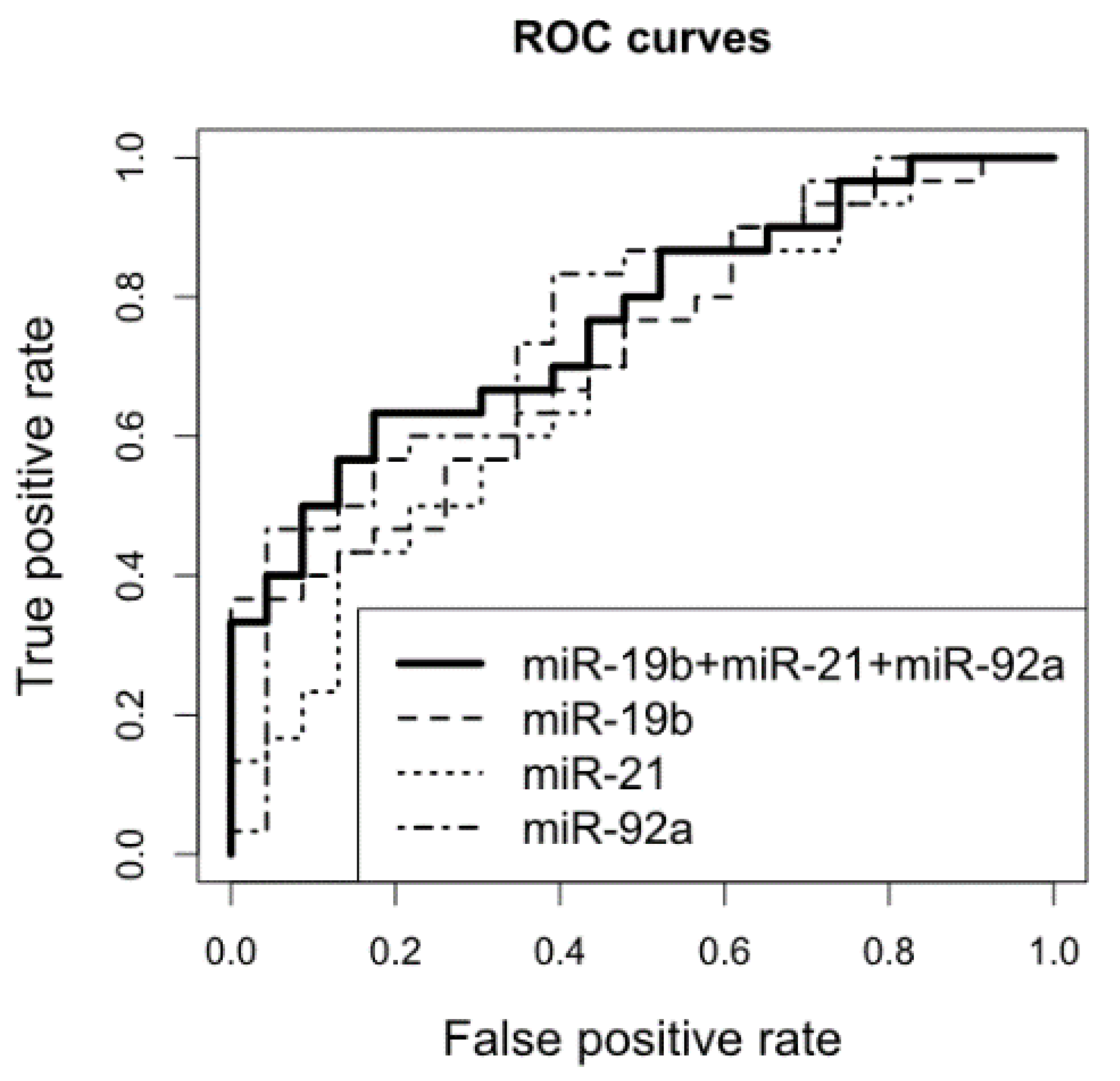
2.3. Survival Analyses
2.4. CNS DLBCL Immunophenotypes and miR Expression
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.1.1. Patients
4.1.2. Sample Collection
4.1.3. Diagnostic Procedures: Immunohistochemical Staining of FFPET Samples
4.1.4. Diagnostic Procedures: Immunophenotyping, Morphology and Proliferation Evaluation of CSF Cells
4.1.5. Final Diagnosis
4.2. RNA Extraction and RT-qPCR
4.3. Statistical Analyses
5. Conclusions
- Tumor miR-155, miR-196b, miR-9, miR-125b, and miR-let-7b expression levels are significantly different in PCNSL and in non-malignant brain lesions.
- A logistic regression model is proposed to discriminate between PCNSL and non-malignant brain lesions.
- We confirm the value of cerebrospinal fluid miR-21, miR-19b, and miR-92a profiles as potential CNS DLBCL markers.
- PCNSL CSFs and the relevant biopsy samples are characterized by specific, different microRNA profiles.
- The examined microRNA profiles do not influence overall survival of PCNSL patients.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Han, C.H.; Batchelor, T.T. Diagnosis and management of primary central nervous system lymphoma. Cancer 2017, 123, 4314–4324. [Google Scholar] [CrossRef] [PubMed]
- Kluin, P.M.; Deckert, M.; Ferry, J.A. Primary diffuse large B-cell lymphoma of the CNS. In WHO Classification of Tumours of the Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; IARC Press: Lyon, France, 2017; Volume 2, pp. 300–302. [Google Scholar]
- Abrey, L.E.; Batchelor, T.T.; Ferreri, A.J.; Gospodarowicz, M.; Pulczynski, E.J.; Zucca, E.; Smith, J.R.; Korfel, A.; Soussain, C.; DeAngelis, L.M.; et al. Report of an International Workshop to Standardize Baseline Evaluation and Response Criteria for Primary CNS Lymphoma. J. Clin. Oncol. 2005, 23, 5034–5043. [Google Scholar] [CrossRef]
- Hoang-Xuan, K.; Bessell, E.; Bromberg, J.; Hottinger, A.F.; Preusser, M.; Ruda, R.; Schlegel, U.; Siegal, T.; Soussain, C.; Abacioglu, U.; et al. European Association for Neuro-Oncology Task Force on Primary, CNS. Lymphoma, Diagnosis and treatment of primary CNS lymphoma in immunocompetent patients: Guidelines from the European Association for Neuro-Oncology. Lancet Oncol. 2015, 16, e322–e332. [Google Scholar] [CrossRef]
- Sasayama, T.; Nakamizo, S.; Nishihara, M.; Kawamura, A.; Tanaka, H.; Mizukawa, K.; Miyake, S.; Taniguchi, M.; Hosoda, K.; Kohmura, E. Cerebrospinal fluid interleukin-10 is a potentially useful biomarker in immunocompetent primary central nervous system lymphoma (PCNSL). Neuro-Oncology 2012, 14, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Rubenstein, J.L.; Wong, V.S.; Kadoch, C.; Gao, H.X.; Barajas, R.; Chen, L.; Josephson, S.A.; Scott, B.; Douglas, V.; Maiti, M.; et al. CXCL13 plus interleukin 10 is highly specific for the diagnosis of CNS lymphoma. Blood 2013, 121, 4740–4748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viaccoz, A.; Ducray, F.; Tholance, Y.; Barcelos, G.K.; Thomas-Maisonneuve, L.; Ghesquières, H.; Meyronet, D.; Quadrio, I.; Cartalat-Carel, S.; Louis-Tisserand, G.; et al. CSF neopterin level as a diagnostic marker in primary central nervous system lymphoma. Neuro-Oncology 2015, 17, 1497–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Westrhenen, A.; Smidt, L.C.A.; Seute, T.; Nierkens, S.; Stork, A.J.; Minnema, M.C.; Snijders, T.J. Diagnostic markers for CNS lymphoma in blood and cerebrospinal fluid: A systematic review. Br. J. Haematol. 2018, 182, 384–403. [Google Scholar] [CrossRef]
- Rao, P.; Benito, E.; Fischer, A. MicroRNAs as biomarkers for CNS disease. Front. Mol. Neurosci. 2013, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Wan, Q.; Li, L.; Jin, H.; Liu, Y.; Wang, Y.; Zhang, G. MicroRNAs as Potential Biomarkers for Diagnosing Cancers of Central Nervous System: A Meta-analysis. Mol. Neurobiol. 2015, 51, 1452–1461. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Radhakrishnan, B.; Anand, A.A.P. Role of miRNA-9 in Brain Development. J. Exp. Neurosci. 2016, 10, JEN.S32843. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.Y.; Hu, H.Y.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Li, N.; Chen, W.; Khaitovich, P. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genom. 2010, 11, 409. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Maghnouj, A.; Zollner, H.; Schmiegel, W.; Hahn, S.; Schroers, R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro-Oncology 2012, 14, 29–33. [Google Scholar] [CrossRef]
- Ivo D’Urso, P.; Fernando D’Urso, O.; Damiano Gianfreda, C.; Mezzolla, V.; Storelli, C.; Marsigliante, S. miR-15b and miR-21 as Circulating Biomarkers for Diagnosis of Glioma. Curr. Genom. 2015, 16, 304–311. [Google Scholar] [CrossRef] [Green Version]
- Kiko, T.; Nakagawa, K.; Tsuduki, T.; Furukawa, K.; Arai, H.; Miyazawa, T. MicroRNAs in Plasma and Cerebrospinal Fluid as Potential Markers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 39, 253–259. [Google Scholar] [CrossRef]
- Yang, Q.; Pan, W.; Qian, L. Identification of the miRNA-mRNA regulatory network in multiple sclerosis. Neurol. Res. 2017, 39, 142–151. [Google Scholar] [CrossRef]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef]
- Kluiver, J.; Poppema, S.; De Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Berg, A.V.D.; Kroesen, B.J.; Kroesen, B. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef]
- Coskun, E.; Von Der Heide, E.K.; Schlee, C.; Kühnl, A.; Gökbuget, N.; Hoelzer, D.; Hofmann, W.K.; Thiel, E.; Baldus, C.D. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk. Res. 2011, 35, 208–213. [Google Scholar] [CrossRef]
- Schotte, D.; Lange-Turenhout, E.A.; Stumpel, D.J.; Stam, R.W.; Buijs-Gladdines, J.G.; Meijerink, J.P.; Pieters, R.; Boer, M.L.D. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica 2010, 95, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Zajdel, M.; Rymkiewicz, G.; Chechlinska, M.; Blachnio, K.; Pieńkowska-Grela, B.; Grygalewicz, B.; Goryca, K.; Cieslikowska, M.; Bystydzienski, Z.; Swoboda, P.; et al. miR expression in MYC-negative DLBCL/BL with partial trisomy 11 is similar to classical Burkitt lymphoma and different from diffuse large B–cell lymphoma. Tumor Boil. 2015, 36, 5377–5388. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Chan, A.; Deckert, M.; Gold, R.; Maghnouj, A.; Zöllner, H.; Reinacher-Schick, A.; Schmiegel, W.; et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 2011, 117, 3140–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohe, Y.; Hayashi, T.; Mishima, K.; Nishikawa, R.; Sasaki, A.; Matsuda, H.; Uchino, A.; Tanahashi, N. Central nervous system lymphoma initially diagnosed as tumefactive multiple sclerosis after brain biopsy. Intern. Med. 2013, 52, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, Y.; Kawasaki, T.; Haraoka, S.; Fujiyama, F.; Kakigi, R.; Abe, M.; Tabuchi, K.; Kuroiwa, T.; Kishikawa, T.; Sugihara, H. Autopsy report of primary CNS B-cell lymphoma indistinguishable from multiple sclerosis: Diagnosis with the immunoglobulin gene rearrangements analysis. J. Neurol. Sci. 1992, 111, 173–179. [Google Scholar] [CrossRef]
- Yang, L.; Niu, F.; Yao, H.; Liao, K.; Chen, X.; Kook, Y.; Ma, R.; Hu, G.; Buch, S. Exosomal miR-9 Released from HIV Tat Stimulated Astrocytes Mediates Microglial Migration. J. Neuroimmune Pharmacol. 2018, 13, 330–344. [Google Scholar] [CrossRef]
- Katz, S.; Cussigh, D.; Urbán, N.; Blomfield, I.; Guillemot, F.; Bally-Cuif, L.; Coolen, M. A Nuclear Role for miR-9 and Argonaute Proteins in Balancing Quiescent and Activated Neural Stem Cell States. Cell Rep. 2016, 17, 1383–1398. [Google Scholar] [CrossRef] [Green Version]
- Dajas-Bailador, F.; Bonev, B.; Garcez, P.; Stanley, P.; Guillemot, F.; Papalopulu, N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012, 15, 697–699. [Google Scholar] [CrossRef]
- Pogue, A.; Cui, J.; Li, Y.; Zhao, Y.; Culicchia, F.; Lukiw, W. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci. Lett. 2010, 476, 18–22. [Google Scholar] [CrossRef]
- Abernathy, D.G.; Kim, W.K.; McCoy, M.J.; Lake, A.M.; Ouwenga, R.; Lee, S.W.; Xing, X.; Li, D.; Lee, H.J.; Heuckeroth, R.O.; et al. MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell 2017, 21, 332–348. [Google Scholar] [CrossRef]
- Topol, A.; Zhu, S.; Hartley, B.J.; English, J.; Hauberg, M.E.; Tran, N.; Rittenhouse, C.A.; Simone, A.; Ruderfer, D.M.; Johnson, J.; et al. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep. 2016, 15, 1024–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camkurt, M.A.; Karababa, F.; Erdal, M.E.; Bayazıt, H.; Kandemir, S.B.; Ay, M.E.; Kandemir, H.; Ay, Ö.I.; Çiçek, E.; Selek, S.; et al. Investigation of Dysregulation of Several MicroRNAs in Peripheral Blood of Schizophrenia Patients. Clin. Psychopharmacol. Neurosci. 2016, 14, 256–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. Micro RNA -125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.M.; Krüger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Derkow, K.; Rossling, R.; Schipke, C.; Kruger, C.; Bauer, J.; Fahling, M.; Stroux, A.; Schott, E.; Ruprecht, K.; Peters, O.; et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer’s disease. PLoS ONE 2018, 13, e0200602. [Google Scholar] [CrossRef] [PubMed]
- Parisi, C.; Napoli, G.; Amadio, S.; Spalloni, A.; Apolloni, S.; Longone, P.; Volonté, C. MicroRNA-125b regulates microglia activation and motor neuron death in ALS. Cell Death Differ. 2016, 23, 531–541. [Google Scholar] [CrossRef]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008, 28, 14341–14346. [Google Scholar] [CrossRef]
- Sørensen, S.S.; Nygaard, A.B.; Carlsen, A.L.; Heegaard, N.H.H.; Bak, M.; Christensen, T. Elevation of brain-enriched miRNAs in cerebrospinal fluid of patients with acute ischemic stroke. Biomark. Res. 2017, 5, 24. [Google Scholar] [CrossRef]
- Long, G.; Wang, F.; Li, H.; Yin, Z.; Sandip, C.; Lou, Y.; Wang, Y.; Chen, C.; Wang, D.W. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013, 13, 178. [Google Scholar] [CrossRef]
- Freiesleben, S.; Hecker, M.; Zettl, U.K.; Fuellen, G.; Taher, L. Analysis of microRNA and Gene Expression Profiles in Multiple Sclerosis: Integrating Interaction Data to Uncover Regulatory Mechanisms. Sci. Rep. 2016, 6, 34512. [Google Scholar] [CrossRef]
- Robertus, J.L.; Harms, G.; Blokzijl, T.; Booman, M.; De Jong, D.; Van Imhoff, G.; Rosati, S.; Schuuring, E.; Kluin, P.; Berg, A.V.D. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod. Pathol. 2009, 22, 547–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, L.; Hummel, M.; Korfel, A.; Lenze, D.; Joehrens, K.; Thiel, E. Differential micro-RNA expression in primary CNS and nodal diffuse large B-cell lymphomas. Neuro-Oncology 2011, 13, 1090–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, W.; Du, J.; Jin, Y.; Zhao, M.; Li, L.; Wang, Y. Prognostic impact of miR-196a/b expression in adult acute myeloid leukaemia: A single-centre, retrospective cohort study. J. Int. Med Res. 2018, 46, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Kawaguchi, A.; Iwadate, Y.; Hondoh, H.; Fukai, J.; Kajiwara, K.; Hayano, A.; Yamanaka, R. MicroRNA signature constituted of miR-30d, miR-93, and miR-181b is a promising prognostic marker in primary central nervous system lymphoma. PLoS ONE 2019, 14, e0210400. [Google Scholar] [CrossRef]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Schmiegel, W.; Hahn, S.; Schroers, R. MicroRNAs in cerebrospinal fluid as biomarker for disease course monitoring in primary central nervous system lymphoma. J. Neuro-Oncol. 2012, 109, 239–244. [Google Scholar] [CrossRef]
- Quintana, E.; Ortega, F.J.; Robles-Cedeño, R.; Villar, M.L.; Buxó, M.; Mercader, J.M.; Alvarez-Cermeño, J.C.; Pueyo, N.; Perkal, H.; Fernández-Real, J.M.; et al. miRNAs in cerebrospinal fluid identify patients with MS and specifically those with lipid-specific oligoclonal IgM bands. Mult. Scler. J. 2017, 23, 1716–1726. [Google Scholar] [CrossRef]
- Drusco, A.; Bottoni, A.; Lagana’, A.; Acunzo, M.; Fassan, M.; Cascione, L.; Antenucci, A.; Kumchala, P.; Vicentini, C.; Gardiman, M.P.; et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 2015, 6, 20829–20839. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncology 2012, 14, 689–700. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.S.; Taylor, J.; Valentine, H.R.; Irlam, J.J.; Eustace, A.; Hoskin, P.J.; Miller, C.J.; West, C.M.L. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br. J. Cancer 2012, 107, 684–694. [Google Scholar] [CrossRef] [Green Version]
- Kakimoto, Y.; Tanaka, M.; Kamiguchi, H.; Ochiai, E.; Osawa, M. MicroRNA Stability in FFPE Tissue Samples: Dependence on GC Content. PLoS ONE 2016, 11, e0163125. [Google Scholar] [CrossRef]
- Shalaby, T.; Grotzer, M.A. Tumor-Associated CSF MicroRNAs for the Prediction and Evaluation of CNS Malignancies. Int. J. Mol. Sci. 2015, 16, 29103–29119. [Google Scholar] [CrossRef] [PubMed]
- Waldera-Lupa, D.M.; Etemad-Parishanzadeh, O.; Brocksieper, M.; Kirchgaessler, N.; Seidel, S.; Kowalski, T.; Montesinos-Rongen, M.; Deckert, M.; Schlegel, U.; Stühler, K. Proteomic changes in cerebrospinal fluid from primary central nervous system lymphoma patients are associated with protein ectodomain shedding. Oncotarget 2017, 8, 110118–110132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, A.M. The role of the blood–CNS barrier in CNS disorders and their treatment. Neurobiol. Dis. 2010, 37, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. J. Neurol. Sci. 2017, 381, 970. [Google Scholar] [CrossRef]
- Cookson, V.J.; Bentley, M.A.; Hogan, B.V.; Horgan, K.; Hayward, B.E.; Hazelwood, L.D.; Hughes, T.A. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell. Oncol. 2012, 35, 301–308. [Google Scholar] [CrossRef]
- Konishi, H.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Tsujiura, M.; Takeshita, H.; Morimura, R.; Nagata, H.; Arita, T.; Kawaguchi, T.; et al. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br. J. Cancer 2012, 106, 740–747. [Google Scholar] [CrossRef] [Green Version]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zajdel, M.; Rymkiewicz, G.; Sromek, M.; Cieslikowska, M.; Swoboda, P.; Kulinczak, M.; Goryca, K.; Bystydzienski, Z.; Blachnio, K.; Ostrowska, B.; et al. Tumor and Cerebrospinal Fluid microRNAs in Primary Central Nervous System Lymphomas. Cancers 2019, 11, 1647. https://doi.org/10.3390/cancers11111647
Zajdel M, Rymkiewicz G, Sromek M, Cieslikowska M, Swoboda P, Kulinczak M, Goryca K, Bystydzienski Z, Blachnio K, Ostrowska B, et al. Tumor and Cerebrospinal Fluid microRNAs in Primary Central Nervous System Lymphomas. Cancers. 2019; 11(11):1647. https://doi.org/10.3390/cancers11111647
Chicago/Turabian StyleZajdel, Michalina, Grzegorz Rymkiewicz, Maria Sromek, Maria Cieslikowska, Pawel Swoboda, Mariusz Kulinczak, Krzysztof Goryca, Zbigniew Bystydzienski, Katarzyna Blachnio, Beata Ostrowska, and et al. 2019. "Tumor and Cerebrospinal Fluid microRNAs in Primary Central Nervous System Lymphomas" Cancers 11, no. 11: 1647. https://doi.org/10.3390/cancers11111647
APA StyleZajdel, M., Rymkiewicz, G., Sromek, M., Cieslikowska, M., Swoboda, P., Kulinczak, M., Goryca, K., Bystydzienski, Z., Blachnio, K., Ostrowska, B., Borysiuk, A., Druzd-Sitek, A., Walewski, J., Chechlinska, M., & Siwicki, J. K. (2019). Tumor and Cerebrospinal Fluid microRNAs in Primary Central Nervous System Lymphomas. Cancers, 11(11), 1647. https://doi.org/10.3390/cancers11111647




