Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance
Abstract
:1. Introduction
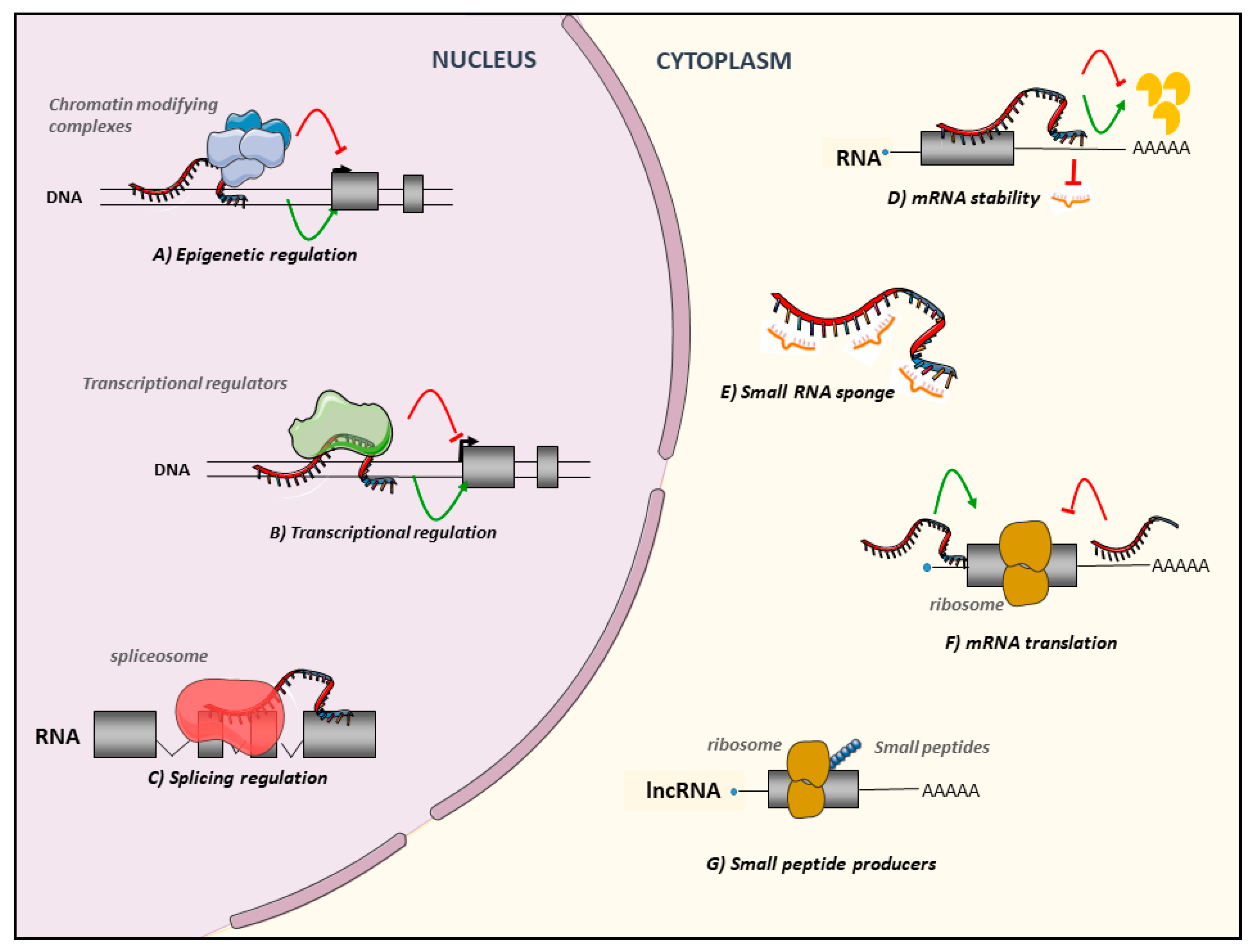
2. Background on lncRNAs
3. Regulatory Roles of lncRNAs in AML
4. LncRNAs as Biomarker Candidates among Acute Myeloid Leukemia
4.1. Specific lncRNA Signatures among AML Subtypes
4.2. Prognostic lncRNA Biomarker Candidates
4.2.1. Good Prognostic lncRNA Biomarkers
4.2.2. Poor Prognostic lncRNA Biomarkers
4.2.3. LncRNAs Involved in Resistance to Treatments
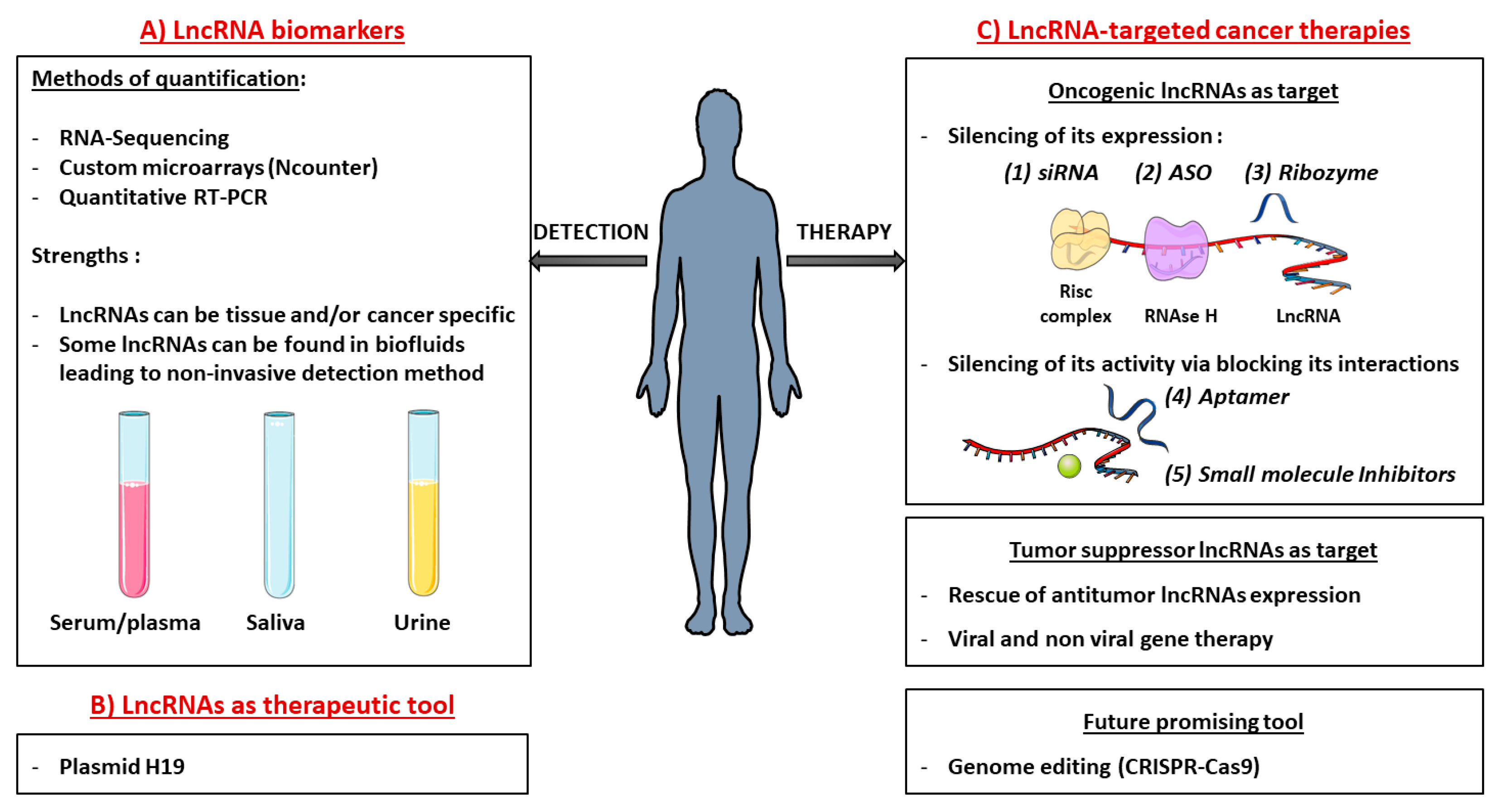
5. From Fundamental to Clinical Research: Incorporation of lncRNAs
5.1. Detection and Quantification of lncRNAs In Clinic
5.2. Development of lncRNA-targeted Cancer Therapies
5.2.1. Targeting Oncogenic lncRNAs
5.2.2. Targeting Tumor Suppressor lncRNAs
5.3. LncRNAs as Tools
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Döhner, H.; Weisdorf, D.J.; Bloomfield, B.C. Clinical presentation of Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef]
- Aziz, H.; Ping, C.Y.; Alias, H.; Ab Mutalib, N.S.; Jamal, R. Gene mutations as emerging biomarkers and therapeutic targets for relapsed acute myeloid leukemia. Front. Pharmacol. 2017, 8, 897. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, G.; Hoadley, K.; Triche, T.J.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. Molecular biology: The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Ulitsky, I.; Shkumatava, A.; Jan, C.H.; Sive, H.; Bartel, D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011, 147, 1537–1550. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Kitagawa, M.; Kitagawa, K.; Kotake, Y.; Niida, H.; Ohhata, T. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 2013, 70, 4785–4794. [Google Scholar] [CrossRef] [Green Version]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struhl, K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007, 14, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Merkel, A.; Gonzalez, D.; Lagarde, J.; et al. The GENCODE v7 catalogue of human long non-coding RNAs: Analysis of their structure, evolution and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Washietl, S.; Kellis, M.; Garber, M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014, 24, 616–628. [Google Scholar] [CrossRef] [Green Version]
- Cabili, M.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [Green Version]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007, 23, 614–622. [Google Scholar] [CrossRef]
- Yang, L.; Duff, M.O.; Graveley, B.R.; Carmichael, G.G.; Chen, L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011, 12, R16. [Google Scholar] [CrossRef]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long Noncoding RNAs with snoRNA Ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Chujo, T.; Hirose, T. Nuclear bodies built on architectural long noncoding RNAs: Unifying principles of their construction and function. Mol. Cells 2017, 40, 889–896. [Google Scholar] [CrossRef]
- Yang, C.; Yang, H.; Liu, J.; Zhu, L.; Yu, S.; Zhang, X.; Gao, L. Focus on exosomes: Novel pathogenic components of leukemia. Am. J. Cancer Res. 2019, 9, 1815–1829. [Google Scholar]
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet. 2010, 19, R152–R161. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- Blanco, M.R.; Guttman, M. Re-evaluating the foundations of lnc RNA–Polycomb function. EMBO J. 2017, 36, 964–966. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef] [Green Version]
- Melé, M.; Rinn, J.L. “Cat’s Cradling” the 3D Genome by the Act of LncRNA Transcription. Mol. Cell 2016, 62, 657–664. [Google Scholar] [CrossRef] [PubMed]
- He, R.Z.; Luo, D.X.; Mo, Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019, 6, 6–15. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome profiling provides evidence that large non-coding RNAs do not encode proteins. Cell 2014, 154, 240–251. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Choi, S.-W.; Kim, H.-W.; Nam, J.-W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2018. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.S.; Jackson, S.E.; Wills, M.R.; Weissman, J.S. Ribosome Profiling Reveals Pervasive Translation Outside of Annotated Protein-Coding Genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosmarin, A.G.; Yang, Z.; Resendes, K.K. Transcriptional regulation in myelopoiesis: Hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp. Hematol. 2005, 33, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Dominguez, J.R.; Lodish, H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 2017, 130, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Delás, M.J.; Sabin, L.R.; Dolzhenko, E.; Knott, S.R.; Munera Maravilla, E.; Jackson, B.T.; Wild, S.A.; Kovacevic, T.; Stork, E.M.; Zhou, M.; et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. Elife 2017, 6, e25607. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- De Kok, J.B.; Verhaegh, G.W.; Roelofs, R.W.; Hessels, D.; Kiemeney, L.A.; Aalders, T.W.; Swinkels, D.W.; Schalken, J.A. DD3PCA3, a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002, 62, 2695–2698. [Google Scholar]
- Day, J.R.; Jost, M.; Reynolds, M.A.; Groskopf, J.; Rittenhouse, H. PCA3: From basic molecular science to the clinical lab. Cancer Lett. 2011, 301, 1–6. [Google Scholar] [CrossRef]
- Groskopf, J.; Aubin, S.M.J.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef]
- Lee, G.L.; Dobi, A.; Srivastava, S. Prostate cancer: Diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 2011, 8, 123–124. [Google Scholar] [CrossRef]
- Chandra Gupta, S.; Nandan Tripathi, Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int. J. Cancer 2017, 140, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Long Non Coding RNA HOTAIR and Midkine as Biomarkers in Thyroid Cancer. ClinicalTrials.gov Identifier: NCT03469544. Available online: ClinicalTrials.gov (accessed on 19 March 2018).
- Pasmant, E.; Sabbagh, A.; Vidaud, M.; Bièche, I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011, 25, 444–448. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Margeron, R.; Reinberg, D. The Polycomb Complex PRC2 and its Mark in Life Raphaël. Nature 2013, 469, 343–349. [Google Scholar] [CrossRef]
- Sun, L.Y.; Li, X.J.; Sun, Y.M.; Huang, W.; Fang, K.; Han, C.; Chen, Z.H.; Luo, X.Q.; Chen, Y.Q.; Wang, W.T. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Mol. Cancer 2018, 17, 127. [Google Scholar] [CrossRef]
- Wang, Y.; Dang, Y.; Liu, J.; Ouyang, X. The function of homeobox genes and lncRNAs in cancer. Oncol. Lett. 2016, 12, 1635–1641. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.L.; Licht, J.D. HOX deregulation in acute myeloid leukemia. J. Clin. Investig. 2007, 117, 865–868. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; David, J.; Tsai, M.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta-Rev. Cancer 2015, 1856, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Xing, C.Y.; Hu, X.Q.; Xie, F.Y.; Yu, Z.J.; Li, H.Y.; Bin-Zhou; Wu, J.B.; Tang, L.Y.; Gao, S.M. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015, 589, 1981–1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Zhou, B.; Li, H.; Huang, X.; Wu, Y.; Xing, C.; Yu, X.; Ji, Y. Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp. Hematol. 2018, 67, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Huang, Y.; Su, R.; Yu, Y.Y. Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 2019, 19, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lian, Z.; Padden, C.; Gerstein, M.B.; Rozowsky, J.; Snyder, M.; Gingeras, T.R.; Kapranov, P.; Weissman, S.M.; Newburger, P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009, 113, 2526–2534. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Q.D.; Dostie, J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017, 45, 1091–1104. [Google Scholar] [CrossRef]
- Zhang, X.; Weissman, S.M.; Newburger, P.E. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014, 11, 777–787. [Google Scholar] [CrossRef] [Green Version]
- De Braekeleer, E.; Douet-Guilbert, N.; Morel, F.; Le Bris, M.J.; Férec, C.; De Braekeleer, M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011, 7, 77–91. [Google Scholar] [CrossRef]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef]
- Wang, H.; Li, W.; Guo, R.; Sun, J.; Cui, J.; Wang, G.; Hoffman, A.R.; Hu, J.F. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int. J. Cancer 2014, 135, 2783–2794. [Google Scholar] [CrossRef]
- Sun, J.; Li, W.; Sun, Y.; Yu, D.; Wen, X.; Wang, H.; Cui, J.; Wang, G.; Hoffman, A.R.; Hu, J.F. A novel antisense long noncoding RNA within the IGF1Rgene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 2014, 42, 9588–9601. [Google Scholar] [CrossRef]
- Pashaiefar, H.; Izadifard, M.; Yaghmaie, M.; Montazeri, M.; Gheisari, E.; Ahmadvand, M.; Momeny, M.; Ghaffari, S.H.; Kasaeian, A.; Alimoghaddam, K.; et al. Low Expression of Long Noncoding RNA IRAIN Is Associated with Poor Prognosis in Non-M3 Acute Myeloid Leukemia Patients. Genet. Test. Mol. Biomark. 2018, 22, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.M.; Legnini, I.; Salvatori, B.; Masciarelli, S.; Marchioni, M.; Fazi, F.; Morlando, M.; Bozzoni, I.; Fatica, A. C/EBPα-p30 protein induces expression of the oncogenic long non-coding RNA UCA1 in acute myeloid leukemia. Oncotarget 2015, 6, 18534–18544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Xu, X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J. Cell Biochem. 2018, 119, 6296–6308. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.D.; Zheng, Y.Q.; Wang, L.P.; Zhao, H.T.; Yang, S. Long noncoding RNA UCA1 promotes cell proliferation, migration and invasion of human leukemia cells via sponging miR-126. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.X.; Wang, W.; Cao, L.; Li, Z.; Wang, X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol. Cells 2016, 39, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, Y.; Shen, Y.; Lou, S.; Deng, J. Comprehensive analysis the potential biomarkers for the high-risk of childhood acute myeloid leukemia based on a competing endogenous RNA network. Blood Cells Mol. Dis. 2019, 79, 102352. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Lu, P.; Guan, J.; Zhou, Y.; Zou, L.; Yi, X.; Cheng, H. LncRNA KCNQ1OT1 controls cell proliferation, differentiation and apoptosis by sponging miR-326 to regulate c-Myc expression in acute myeloid leukemia. Neoplasma 2019, 181215N972. [Google Scholar] [CrossRef]
- Yin, X.; Huang, S.; Xu, A.; Fan, F.; Chen, L.; Sun, C.; Hu, Y. Identification of distinctive long noncoding RNA competitive interactions and a six-methylated-gene prognostic signature in acute myeloid leukemia with -5/del(5q) or -7/del(7q). J. Cell. Biochem. 2019. [Google Scholar] [CrossRef]
- Bao, X.L.; Zhang, L.; Song, W.P. LncRNA SNHG1 overexpression regulates the proliferation of acute myeloid leukemia cells through miR-488-5p/NUP205 axis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5896–5903. [Google Scholar] [CrossRef]
- Zhao, T.T.; Liu, X. LncRNA-H19 inhibits apoptosis of acute myeloid leukemia cells via targeting miR-29a-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 224–231. [Google Scholar] [CrossRef]
- Tian, Y.J.; Wang, Y.H.; Xiao, A.J.; Li, P.L.; Guo, J.; Wang, T.J.; Zhao, D.J. Long noncoding RNA SBF2-AS1 act as a ceRNA to modulate cell proliferation via binding with miR-188-5p in acute myeloid leukemia. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1730–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, N.; Chen, L.; Wang, C.; Zhao, H. MALAT1 knockdown inhibits proliferation and enhances cytarabine chemosensitivity by upregulating miR-96 in acute myeloid leukemia cells. Biomed. Pharmacother. 2019, 112, 108720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tao, W. Long Noncoding RNA LINC00152 Facilitates the Leukemogenesis of Acute Myeloid Leukemia by Promoting CDK9 Through miR-193a. DNA Cell Biol. 2019, 38, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Song, W.; Wang, J. TUG1 confers Adriamycin resistance in acute myeloid leukemia by epigenetically suppressing miR-34a expression via EZH2. Biomed. Pharmacother. 2019, 109, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Ma, P.; Ma, J.; Wang, W.; Han, H.; Chen, L.; Li, X.; Wu, F.; Sun, H. Knockdown of ZFAS1 suppresses the progression of acute myeloid leukemia by regulating microRNA-150/Sp1 and microRNA-150/Myb pathways. Eur. J. Pharmacol. 2019, 844, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Fang, Z.; Yu, M.; Zhang, L.; Xiao, R.; Li, X.; Pan, G.; Liu, J. Knockdown of long noncoding RNA HOXA-AS2 suppresses chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 Axis. Cell. Physiol. Biochem. 2018, 51, 886–896. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, X.; Tian, X. High expression of long intergenic non-coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA-340-5p. Eur. J. Pharmacol. 2019, 859, 172535. [Google Scholar] [CrossRef]
- Zhao, T.F.; Jia, H.Z.; Zhang, Z.Z.; Zhao, X.S.; Zou, Y.F.; Zhang, W.; Wan, J.; Chen, X.F. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol. Med. Rep. 2017, 16, 3687–3693. [Google Scholar] [CrossRef]
- El Hajj, J.; Nguyen, E.; Liu, Q.; Bouyer, C.; Adriaenssens, E.; Hilal, G.; Ségal-Bendirdjian, E. Telomerase regulation by the long non-coding RNA H19 in human acute promyelocytic leukemia cells. Mol. Cancer 2018, 17, 85. [Google Scholar] [CrossRef]
- Chen, Z.H.; Wang, W.T.; Huang, W.; Fang, K.; Sun, Y.M.; Liu, S.R.; Luo, X.Q.; Chen, Y.Q. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017, 24, 212–224. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.B.; Kobayashi, N.; Todo, Y.; Watari, H. Long non-coding RNA NEAT1: A novel target for diagnosis and therapy in human tumors. Front. Genet. 2018, 9, 471. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Xu, Y.; Xu, L.; Yu, X.; Cheng, J.; Yang, L.; Chen, S.; Li, Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 2014, 14, 693. [Google Scholar] [CrossRef] [PubMed]
- Fallik, N.; Bar-Lavan, Y.; Greenshpan, Y.; Goldstein, O.; Grosch, M.; Drukker, M.; Gazit, R. Neat1 in Hematopoietic Stem Cells. Oncotarget 2017, 8, 109575–109586. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, S.; Zhao, Y.; Du, F.; Wang, W.; Lv, P.; Qi, L. Long noncoding RNA NEAT1 modulates cell proliferation and apoptosis by regulating miR-23a-3p/SMC1A in acute myeloid leukemia. J. Cell. Physiol. 2019, 234, 6161–6172. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, J.; Zhang, L.; Liu, S.; Ma, Y.; Wen, X.; Hao, J.; Li, Z.; Ni, Y.; Li, X.; et al. Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development. Cell Stem Cell 2019, 24, 285–298. [Google Scholar] [CrossRef]
- Venkatraman, A.; He, X.C.; Thorvaldsen, J.L.; Sugimura, R.; Perry, J.M.; Tao, F.; Zhao, M.; Christenson, M.K.; Sanchez, R.; Yu, J.Y.; et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 2013, 500, 345–349. [Google Scholar] [CrossRef]
- Zhang, T.J.; Zhou, J.D.; Zhang, W.; Lin, J.; Ma, J.C.; Wen, X.M.; Yuan, Q.; Li, X.X.; Xu, Z.J.; Qian, J. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin. Epigenet. 2018, 10, 47. [Google Scholar] [CrossRef]
- Wu, S.; Zheng, C.; Chen, S.; Cai, X.; Shi, Y.; Lin, B.; Chen, Y. Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol. Lett. 2015, 10, 2410–2414. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Y.; Huang, S.H.; Zhou, H.R.; Chen, C.J.; Tian, L.H.; Shen, J.Z. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol. Rep. 2016, 36, 3113–3122. [Google Scholar] [CrossRef] [Green Version]
- Hao, S.; Shao, Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int. J. Clin. Exp. Pathol. 2015, 8, 7223. [Google Scholar]
- Wang, X.; Zhang, L.; Zhao, F.; Xu, R.; Jiang, J.; Zhang, C.; Liu, H.; Huang, H. Long non-coding RNA taurine-upregulated gene 1 correlates with poor prognosis, induces cell proliferation, and represses cell apoptosis via targeting aurora kinase A in adult acute myeloid leukemia. Ann. Hematol. 2018, 97, 1375–1389. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Bao, H.; Li, H. Correlation of long non-coding RNA taurine-upregulated gene 1 with disease conditions and prognosis, as well as its effect on cell activities in acute myeloid leukemia. Cancer Biomark. 2018, 23, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Yu, H.; Zou, X.; Ni, X.; Wei, J. Long non-coding RNA taurine-upregulated gene 1 correlates with unfavorable prognosis in patients with refractory or relapsed acute myeloid leukemia treated by purine analogue based chemotherapy regimens. Cancer Biomark. 2018, 23, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhou, J.D.; Zhang, T.J.; Ma, J.C.; Xiao, G.F.; Chen, Q.; Deng, Z.Q.; Lin, J.; Qian, J.; Yao, D.M. Overexpression of lncRNA PANDAR predicts adverse prognosis in acute myeloid leukemia. Cancer Manag. Res. 2018, 10, 4999–5007. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, C.-K. Long noncoding RNA SNHG5 is up-regulated and serves as a potential prognostic biomarker in acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3342–3347. [Google Scholar] [CrossRef] [PubMed]
- El-Khazragy, N.; Elayat, W.; Matbouly, S.; Seliman, S.; Sami, A.; Safwat, G.; Diab, A. The prognostic significance of the long non-coding RNAs “CCAT1, PVT1” in t (8;21) associated Acute Myeloid Leukemia. Gene 2019, 707, 172–177. [Google Scholar] [CrossRef]
- Zeng, C.; Yu, X.; Lai, J.; Yang, L.; Chen, S.; Li, Y. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J. Hematol. Oncol. 2015, 8, 126. [Google Scholar] [CrossRef]
- Díaz-Beyá, M.; Brunet, S.; Nomdedéu, J.; Pratcorona, M.; Cordeiro, A.; Gallardo, D.; Escoda, L.; Tormo, M.; Heras, I.; Ribera, J.M.; et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015, 6, 31613–31627. [Google Scholar] [CrossRef]
- Hirano, T.; Yoshikawa, R.; Harada, H.; Harada, Y.; Ishida, A.; Yamazaki, T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol. Cancer 2015, 14, 90. [Google Scholar] [CrossRef]
- Radtke, I.; Mullighan, C.G.; Ishii, M.; Su, X.; Cheng, J.; Ma, J.; Ganti, R.; Cai, Z.; Goorha, S.; Pounds, S.B.; et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2009, 106, 12944–12949. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Wang, P.; Mo, W.; Zhang, Y.; Zhou, W.; Deng, T.; Zhou, M.; Chen, X.; Wang, S.; Wang, C. lncRNA-CCDC26, as a novel biomarker, predicts prognosis in acute myeloid leukemia. Oncol. Lett. 2019, 18, 2203–2211. [Google Scholar] [CrossRef]
- De Clara, E.; Gourvest, M.; Ma, H.; Vergez, F.; Tosolini, M.; Dejean, S.; Demur, C.; Delabesse, E.; Recher, C.; Touriol, C.; et al. Long non-coding RNA expression profile in cytogenetically normal acute myeloid leukemia identifies a distinct signature and a new biomarker in NPM1-mutated patients. Haematologica 2017, 102, 1718–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bill, M.; Papaioannou, D.; Karunasiri, M.; Kohlschmidt, J.; Pepe, F.; Walker, C.J.; Walker, A.E.; Brannan, Z.; Pathmanathan, A.; Zhang, X.; et al. Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia 2019, 33, 2169–2182. [Google Scholar] [CrossRef] [PubMed]
- Benetatos, L.; Hatzimichael, E.; Dasoula, A.; Dranitsaris, G.; Tsiara, S.; Syrrou, M.; Georgiou, I.; Bourantas, K.L. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 2010, 34, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Duan, M.; Lin, L.; Wu, C.; Fu, X.; Wang, H.; Guo, L.; Chen, W.; Huang, L.; Liu, D.; et al. TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget 2017, 8, 18337–18347. [Google Scholar] [CrossRef] [Green Version]
- Sellers, Z.P.; Bolkun, L.; Kloczko, J.; Wojtaszewska, M.L.; Lewandowski, K.; Moniuszko, M.; Ratajczak, M.Z.; Schneider, G. Increased methylation upstream of the MEG3 promotor is observed in acute myeloid leukemia patients with better overall survival. Clin. Epigenet. 2019, 11, 50. [Google Scholar] [CrossRef]
- Merkerova, M.; Remesova, H.; Krejcik, Z.; Loudova, N.; Hrustincova, A.; Szikszai, K.; Cermak, J.; Jonasova, A.; Belickova, M. Relationship between Altered miRNA Expression and DNA Methylation of the DLK1-DIO3 Region in Azacitidine-Treated Patients with Myelodysplastic Syndromes and Acute Myeloid Leukemia with Myelodysplasia-Related Changes. Cells 2018, 7, 138. [Google Scholar] [CrossRef]
- Fernando, T.R.; Contreras, J.R.; Zampini, M.; Rodriguez-Malave, N.I.; Alberti, M.O.; Anguiano, J.; Tran, T.M.; Palanichamy, J.K.; Gajeton, J.; Ung, N.M.; et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol. Cancer 2017, 16, 126. [Google Scholar] [CrossRef]
- Garzon, R.; Volinia, S.; Papaioannou, D.; Nicolet, D.; Kohlschmidt, J.; Yan, P.S.; Mrózek, K.; Bucci, D.; Carroll, A.J.; Baer, M.R.; et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2014, 111, 18679–18684. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Beya, M.; Navarro, A.; Cordeiro, A.; Pratcorona, M.; Castellano, J.; Torrente, M.A.; Nomdedeu, M.; Risueño, R.; Rozman, M.; Monzo, M.; et al. Exploring the Expression Profile of Long Non-Coding RNA (lncRNA) in Different Acute Myeloid Leukemia (AML) Subtypes: T (8;16) (p11; p13)/MYST3-Crebbp AML Harbors a Distinctive Lncrna Signature. Blood 2015, 126, 1397. [Google Scholar] [CrossRef]
- Zhang, J.; Griffith, M.; Miller, C.A.; Griffith, O.L.; Spencer, D.H.; Walker, J.R.; Magrini, V.; McGrath, S.D.; Ly, A.; Helton, N.M.; et al. Comprehensive discovery of noncoding RNAs in acute myeloid leukemia cell transcriptomes. Exp. Hematol. 2017, 55, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, A.; Emmrich, S.; Schmidt, F.; Beck, D.; Ng, M.; Reimer, C.; Adams, F.F.; Grasedieck, S.; Witte, D.; Käbler, S.; et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat. Commun. 2017, 8, 218. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Yao, C.Y.; Tien, F.M.; Tang, J.L.; Kuo, Y.Y.; Chiu, Y.C.; Lin, C.C.; Tseng, M.H.; Peng, Y.L.; Liu, M.C.; et al. Incorporation of long non-coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients. EBioMedicine 2019, 40, 240–250. [Google Scholar] [CrossRef] [Green Version]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [Green Version]
- Balik, V.; Srovnal, J.; Sulla, I.; Kalita, O.; Foltanova, T.; Vaverka, M.; Hrabalek, L.; Hajduch, M. MEG3: A novel long noncoding potentially tumour-suppressing RNA in meningiomas. J. Neurooncol. 2013, 112, 1–8. [Google Scholar] [CrossRef]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; Zhou, Y.; Klibanski, A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 2010, 151, 939–947. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Mehta, K.R.; Danila, D.C.; Scolavino, S.; Johnson, S.R.; Klibanski, A. A Pituitary-Derived MEG3 Isoform Functions as a Growth Suppressor in Tumor Cells. J. Clin. Endocrinol. Metab. 2003, 88, 5119–5126. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.; Fan, H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed. Pharmacother. 2015, 72, 113–118. [Google Scholar] [CrossRef]
- Lu, M.; Liu, Z.; Li, B.; Wang, G.; Li, D.; Zhu, Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J. Cancer Res. Clin. Oncol. 2017, 143, 71–81. [Google Scholar] [CrossRef]
- Zhan, Y.; Lin, J.; Liu, Y.; Chen, M.; Chen, X.; Zhuang, C.; Liu, L.; Xu, W.; Chen, Z.; He, A.; et al. Up-regulation of long non-coding RNA PANDAR is associated with poor prognosis and promotes tumorigenesis in bladder cancer. J. Exp. Clin. Cancer Res. 2016, 35, 83. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Zhong, Y.; Wu, J.; Xiao, H.; Zhang, X.; Liao, X.; Li, J.; Mao, X.; Liu, Y.; Zhang, F. Long non-coding PANDAR as a novel biomarker in human cancer: A systematic review. Cell Prolif. 2018, 51, e12422. [Google Scholar] [CrossRef] [PubMed]
- Le Beau, M.M.; Davis, E.M.; Patel, B.; Phan, V.T.; Sohal, J.; Kogan, S.C. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice identify cooperating events and genetic pathways to acute promyelocytic leukemia. Blood 2003, 102, 1072–1074. [Google Scholar] [CrossRef] [PubMed]
- Metzeler, K.H.; Hummel, M.; Bloomfield, C.D.; Spiekermann, K.; Braess, J.; Sauerland, M.C.; Heinecke, A.; Radmacher, M.; Marcucci, G.; Whitman, S.P.; et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008, 112, 4193–4201. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Q.; Tang, S.; Li, M.; Feng, A.; Qin, L.; Liu, Z.; Wang, X. The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology 2017, 22, 208–216. [Google Scholar] [CrossRef]
- Shang, C.; Guo, Y.; Zhang, H.; Xue, Y.X. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother. Pharmacol. 2016, 77, 507–513. [Google Scholar] [CrossRef]
- Yin, W.; Rossin, A.; Clifford JL, G.H. Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. Oncogene 2006, 25, 3735–3744. [Google Scholar] [CrossRef] [Green Version]
- Kapranov, P.; St Laurent, G.; Raz, T.; Ozsolak, F.; Reynolds, C.P.; Sorensen, P.H.; Reaman, G.; Milos, P.; Arceci, R.J.; Thompson, J.F.; et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is “dark matter” un-annotated RNA. BMC Biol. 2011, 9, 1. [Google Scholar] [CrossRef]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019, 58, 100–108. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Jiang, N.; Pan, J.; Fang, S.; Zhou, C.; Han, Y.; Chen, J.; Meng, X.; Jin, X.; Gong, Z. Liquid biopsy: Circulating exosomal long noncoding RNAs in cancer. Clin. Chim. Acta 2019, 495, 331–337. [Google Scholar] [CrossRef]
- Papaioannou, D.; Nicolet, D.; Ozer, H.G.; Mrózek, K.; Volinia, S.; Fadda, P.; Carroll, A.J.; Kohlschmidt, J.; Kolitz, J.E.; Wang, E.S.; et al. Prognostic and Biologic Relevance of Clinically Applicable Long Noncoding RNA Profiling in Older Patients with Cytogenetically Normal Acute Myeloid Leukemia. Mol. Cancer Ther. 2019, 18, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Bernard, P.S.; Parker, J.S.; Mullins, M.; Cheung, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Liu, M.C.; Pitcher, B.N.; Mardis, E.R.; Davies, S.R.; Friedman, P.N.; Snider, J.E.; Vickery, T.L.; Reed, J.P.; Deschryver, K.; Singh, B.; et al. PAM50 gene signatures and breast cancer prognosis with adjuvant anthracycline-and taxane-based chemotherapy: Correlative analysis of C9741 (alliance). NPJ Breast Cancer 2016, 2, 15023. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef]
- Coelho, T.; Adams, D.; Silva, A.; Lozeron, P.; Hawkins, P.N.; Mant, T.; Perez, J.; Chiesa, J.; Warrington, S.; Tranter, E.; et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013, 369, 819–829. [Google Scholar] [CrossRef]
- Gonzalez, V.; Palumaa, K.; Turman, K.; Munoz, F.J.; Jordan, J.; Garcia, J.; Ussa, F.; Anton, A.; Gutierrez, E. Phase 2 of bamosiran (SYL040012), a novel RNAi based compound for the treatment of increased intraocular pressure associated to glaucoma. Investig. Ophthalmol. Vis. Sci. 2014, 55, 564. [Google Scholar]
- Nguyen, Q.D.; Schachar, R.A.; Nduaka, C.I.; Sperling, M.; Basile, A.S.; Klamerus, K.J.; Chi-Burris, K.; Yan, E.; Paggiarino, D.A.; Rosenblatt, I.; et al. Phase 1 dose-escalation study of a siRNA targeting the RTP801 gene in age-related macular degeneration patients. Eye 2012, 26, 1099–1105. [Google Scholar] [CrossRef] [Green Version]
- Titze-de-Almeida, R.; David, C.; Titze-de-Almeida, S.S. The Race of 10 Synthetic RNAi-Based Drugs to the Pharmaceutical Market. Pharm. Res. 2017, 34, 1339–1363. [Google Scholar] [CrossRef]
- Huang, J.; Ke, P.; Guo, L.; Wang, W.; Tan, H.; Liang, Y.; Yao, S. Lentivirus-mediated RNA interference targeting the long noncoding RNA HOTAIR inhibits proliferation and invasion of endometrial carcinoma cells in vitro and in vivo. Int. J. Gynecol. Cancer 2014, 24, 635–642. [Google Scholar] [CrossRef]
- Wu, X.; Tudoran, O.M.; Calin, G.A.; Ivan, M. The Many Faces of Long Noncoding RNAs in Cancer. Antioxid. Redox Signal. 2018, 29, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Ideue, T.; Hino, K.; Kitao, S.; Yokoi, T.; Hirose, T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. Rna 2009, 15, 1578–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansoori, B.; Shotorbani, S.S.; Baradaran, B. RNA interference and its role in cancer therapy. Adv. Pharm. Bull. 2014, 4, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Pahle, J.; Walther, W. Vectors and strategies for nonviral cancer gene therapy. Expert Opin. Biol. Ther. 2016, 16, 443–461. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Fedor, M.J.; Williamson, J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005, 6, 399–412. [Google Scholar] [CrossRef]
- Pavco, P.A.; Bouhana, K.S.; Gallegos, A.M.; Agrawal, A.; Blanchard, K.S.; Grimm, S.L.; Jensen, K.L.; Andrews, L.E.; Wincott, F.E.; Pitot, P.A.; et al. Antitumor and antimetastatic activity of ribozymes targeting the messenger RNA of vascular endothelial growth factor receptors. Clin. Cancer Res. 2000, 6, 2094–2103. [Google Scholar]
- Vitiello, M.; Tuccoli, A.; Poliseno, L. Long non-coding RNAs in cancer: Implications for personalized therapy. Cell. Oncol. 2015, 38, 17–28. [Google Scholar] [CrossRef]
- Fatemi, R.P.; Velmeshev, D.; Faghihi, M.A. De-repressing LncRNA-targeted genes to upregulate gene expression: Focus on small molecule therapeutics. Mol. Ther.-Nucleic Acids 2014, 3, e196. [Google Scholar] [CrossRef]
- Pedram Fatemi, R.; Salah-Uddin, S.; Modarresi, F.; Khoury, N.; Wahlestedt, C.; Faghihi, M.A. Screening for small-molecule modulators of long noncoding RNA-protein interactions using alphascreen. J. Biomol. Screen. 2015, 20, 1132–1141. [Google Scholar] [CrossRef]
- Yang, H.; Shang, D.; Xu, Y.; Zhang, C.; Feng, L.; Sun, Z.; Shi, X.; Zhang, Y.; Han, J.; Su, F.; et al. The LncRNA Connectivity Map: Using LncRNA Signatures to Connect Small Molecules, LncRNAs, and Diseases. Sci. Rep. 2017, 7, 6655. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.K.; Hurley, B.; Kerrigan, R.; Shu, L.; Chin, D.N.; Shen, Y.; O’Brien, G.; Sung, M.J.; Hou, Y.; Axford, J.; et al. Discovery of Small Molecule Splicing Modulators of Survival Motor Neuron-2 (SMN2) for the Treatment of Spinal Muscular Atrophy (SMA). J. Med. Chem. 2018, 61, 11021–11036. [Google Scholar] [CrossRef] [PubMed]
- Connelly, C.M.; Moon, M.H.; Schneekloth, J.S. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem. Biol. 2016, 23, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Nayerossadat, N.; Ali, P.; Maedeh, T. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1, 27. [Google Scholar] [CrossRef]
- Sakuma, T.; Yamamoto, T. Acceleration of cancer science with genome editing and related technologies. Cancer Sci. 2018, 109, 3679–3685. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, W.; Shao, Z. Association between long non-coding RNA polymorphisms and cancer risk: A meta-analysis. Biosci. Rep. 2018, 38, BSR20180365. [Google Scholar] [CrossRef]
- Gao, P.; Wei, G.H. Genomic insight into the role of lncRNA in cancer susceptibility. Int. J. Mol. Sci. 2017, 18, 1239. [Google Scholar] [CrossRef]
- Smaldone, M.C.; Davies, B.J. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr. Opin. Mol. Ther. 2010, 12, 607–616. [Google Scholar]
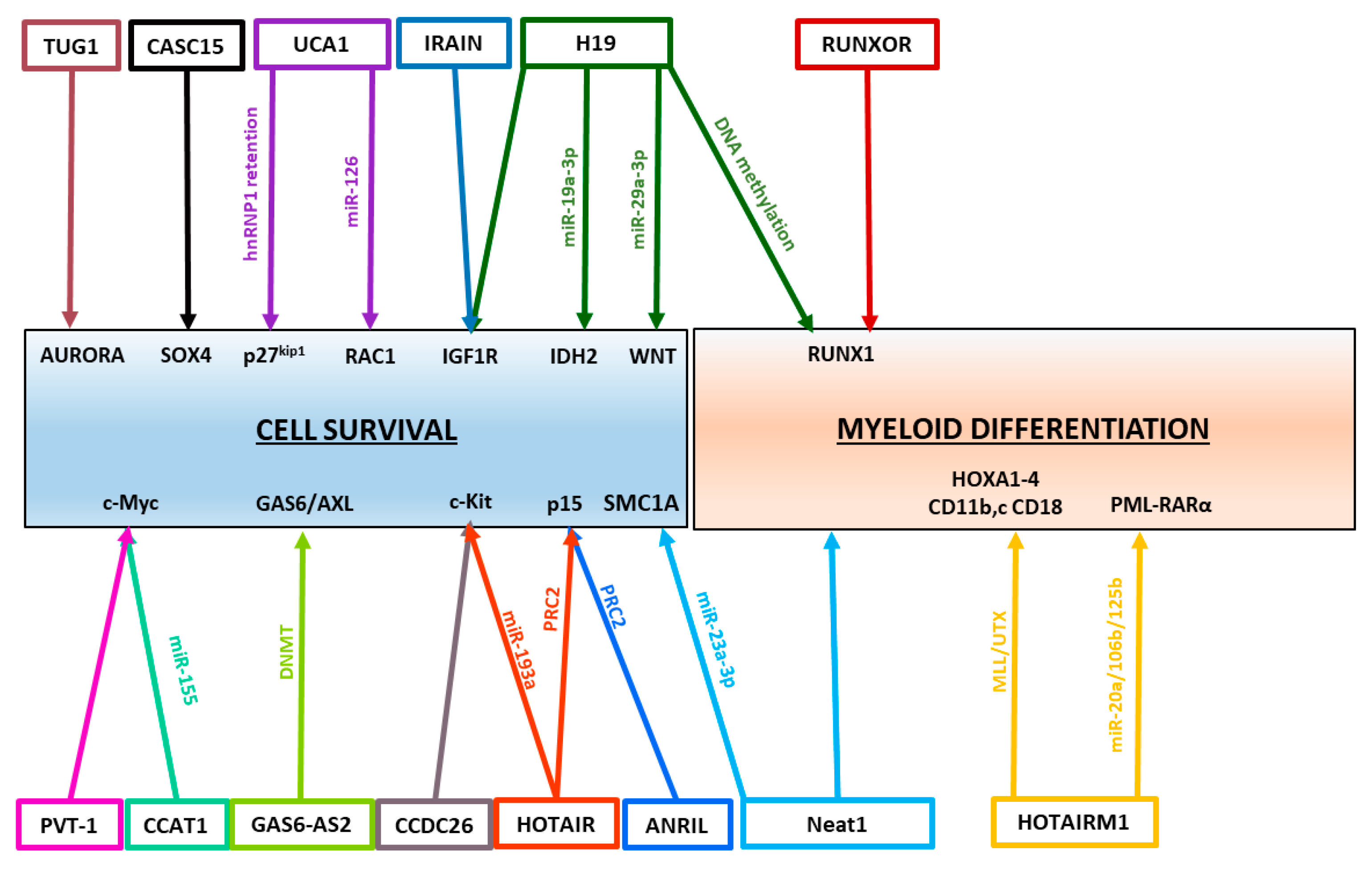
| LncRNA | Clinical Significance | Mechanisms of Actions | Functions in Leukemogenesis | Ref |
|---|---|---|---|---|
| H19 | —Upregulated in AML —Correlated with WBC count/karyotypic classifications/FLT3-ITD and DNMT3a mutations/chemotherapy response/OS; high at diagnosis/relapse | —Maternal imprinting of IGF2 gene —miR-19a-3p and 29a-3p sponges —Promoter methylation of hematopoietic transcription factors (RUNX1/SP1) | —Sustain Adult HSC quiescence, leukemic cell proliferation —Limit apoptosis | [96,97] [98] |
| HOTAIR | —Upregulated in AML —Correlated with OS/DFS/drug resistance —Downregulated after treatment | —Repress p15 expression (PRC2) —Regulate c-KIT expression by sponging miR-193a | —Sustain Cell growth —Inhibit apoptosis | [62,63,64] [99,100,101] |
| TUG1 | —Upregulated in AML with monosomal karyotype/FLT3-ITD mutation/ poor-risk patients —Correlated with high WBC count/shorter OS/lower rate of CR —Implicated in Adriamycin resistance | Target Aurora Kinase | —Promote cell proliferation —Inhibit Apoptosis —Doxorubicin resistance | [85] [102,103,104] |
| UCA1 | —Upregulated in CN-AML with dominant negative C/EBPα —Upregulated in Adriamycin resistant pediatric AML cases | —Inhibit p27kip1 translation by titrating hnRNPI factor —miR-125a and miR-126 sponges | —Sustain AML cell proliferation, migration, invasion —Inhibit Apoptosis —Doxorubicin Resistance | [73,74,75,76] |
| PANDAR | —Upregulated in AML —Associated with higher AML blasts/older patients/poor karyotypes/lower OS and CR | [105] | ||
| RUNXOR | —Upregulated in t(8;21) AML and after ARA-C treatment | —Intrachromosomal loop between RUNX1 promoter and enhancers —Interacts with PRC2 and RUNX1 protein to regulate RUNX1 expression | [70] | |
| SNHG5 | —Upregulated in AML patients —Associated with advanced FAB classification and unfavorable cytogenetics, Shorter OS | —miR-205-5P sponge | [106] | |
| ANRIL | —Upregulated in AML at diagnosis and downregulated after CR | —Silencing of p15INK4B by scaffolding PRC2 —Regulate expression of AdiR1 | —AML cell maintenance —Implicated in Glucose metabolism | [57] |
| PVT-1 | —Upregulated in APL and t(8;21) AML —Associated with high-risk criteria/shorter OS and DFS | —miR-200 sponge: c-MYC regulation | —Sustain proliferation of promyelocytes | [107] [108] |
| CCAT1 | —Upregulated in AML patients (mostly in M4-M5 subtypes) | —miR-155 sponge: c-MYC regulation | —Repress monocytic differentiation —Promote cell growth | [107] |
| HOTAIRM1 | —High expression associated with shorter OS and DFS/higher incidence of relapse in IR-AML, mostly in NPM1 mutated patients | —Activate expression of proximal HOXA1-4, HOXA4, CD11b, c, and CD18 genes —miR-20a/106b and miR-125b sponges | —Regulate myeloid differentiation, cell cycle, and autophagy pathways | [65,66,67] [109] |
| CCDC26 | —Upregulated at diagnosis and relapse —Associated with age, anemia, risk stratification, remission, shorter OS | Repress c-Kit expression | —Sustain AML cell proliferation —Resistance to anticancer drugs (ATRA treatment) | [110,111,112] |
| XLOC_ 109948 | —Low expression indicates a good prognosis, especially for NPM1-mutated AML patients | Apoptosis/ drug resistance | [113] | |
| DANCR | —Knock-down in LSCs resulted in decreased stem-cell renewal and quiescence | [114] |
| LncRNA | Clinical Significance | Mechanisms of Actions | Functions in Hematopoiesis | Ref |
|---|---|---|---|---|
| IRAIN | Downregulated in AML cell lines; Lower in patients with high-risk AML —Associated with higher WBC counts/shorter OS and DFS/Refractory response to chemotherapies/relapse | —Intrachromosomal enhancer/promoter loop of IGF1R gene | —Inhibit tumor cell migration | [71,72] |
| NEAT1 | —Downregulated in AML patients with PML-RARα translocation | —miR-23a-3p sponge | —Regulate myeloid differentiation —Inhibit AML cell proliferation, induce cell-cycle arrest and apoptosis | [93,94,95] |
| MEG3 | —Hypermethylation of MEG3 promoter in AML —Downregulated in AML —Associated with longer OS and DFS | —Positive regulation of p53 expression | —Regulate cell cycle, apoptosis | [115,116,117,118] |
| CASC15 | —Upregulated in RUNX1-rearranged AML —Highest level found in AML with the t(8;21) translocation —Associated with a good prognosis | —CASC15 activates expression of SOX4 gene, by regulating the activity of the YY1 transcription factor | —Increase apoptosis and myeloid cells number | [119] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gourvest, M.; Brousset, P.; Bousquet, M. Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance. Cancers 2019, 11, 1638. https://doi.org/10.3390/cancers11111638
Gourvest M, Brousset P, Bousquet M. Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance. Cancers. 2019; 11(11):1638. https://doi.org/10.3390/cancers11111638
Chicago/Turabian StyleGourvest, Morgane, Pierre Brousset, and Marina Bousquet. 2019. "Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance" Cancers 11, no. 11: 1638. https://doi.org/10.3390/cancers11111638
APA StyleGourvest, M., Brousset, P., & Bousquet, M. (2019). Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance. Cancers, 11(11), 1638. https://doi.org/10.3390/cancers11111638





