Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications
Abstract
1. Introduction
2. The BET Protein Family
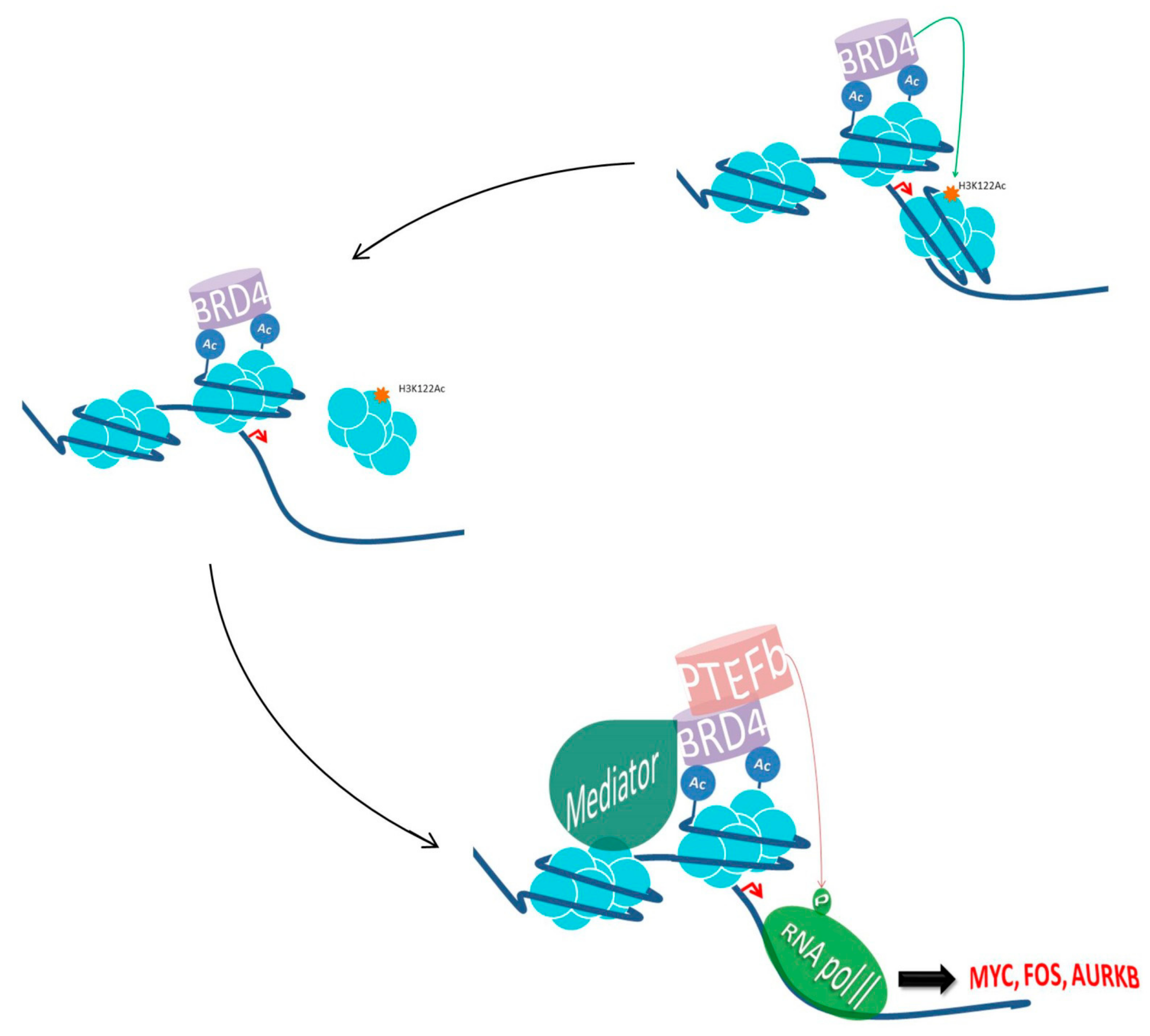
BRD4: Biological Roles and Molecular Mechanisms of Action
3. The Role of BET Proteins in Hematological Cancers
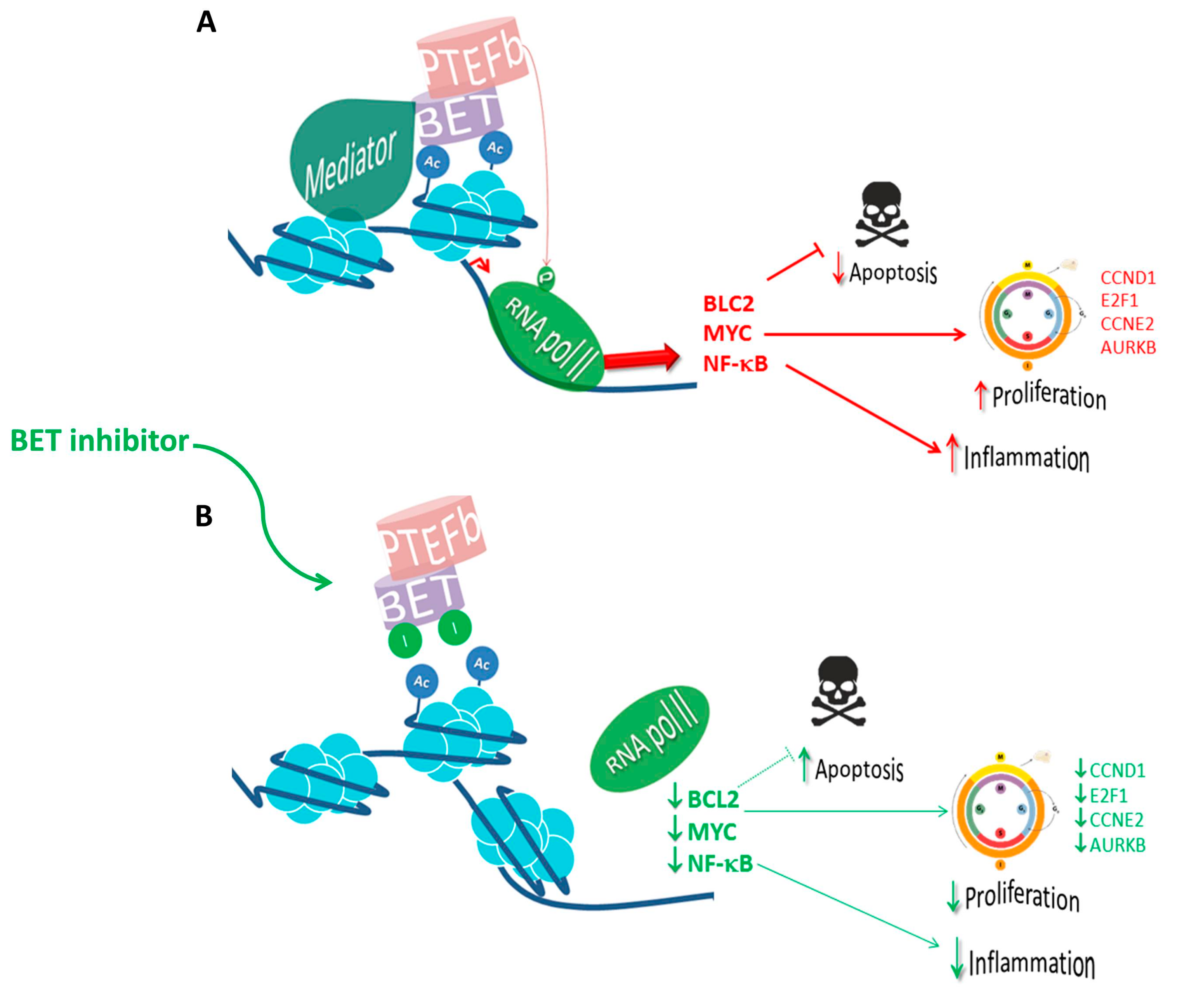
3.1. MYC-Driven Mechanisms of Oncogenesis Regulated by BET Proteins
3.2. BET-Mediated Expression Regulation of Other Oncogenes
4. Characterization and Evaluation of BET Inhibitors
4.1. Preclinical Activity of BET Inhibitors as Monotherapies in Hematological Malignancies
4.2. Evaluation of BET Inhibitors as Part of Combination Therapies in Preclinical Settings
5. Targeting BET in Hematological Cancer Patients
6. Mechanisms of Resistance to BET Inhibitors
7. Future Generations of Bromodomain Inhibitors
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 2001. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Knapp, S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014, 13, 337–356. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Parra, M. Histone deacetylases and cancer. Mol. Oncol. 2012. [Google Scholar] [CrossRef]
- Dhalluin, C.; Carlson, J.E.; Zeng, L.; He, C.; Aggarwal, A.K.; Zhou, M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, L.L.; Wen, X.; Wang, X.Y.; Liu, J.; Cheng, Y.; Huang, J. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014. [Google Scholar] [CrossRef]
- Gillette, T.G.; Hill, J.A. Readers, writers, and erasers: Chromatin as the whiteboard of heart disease. Circ. Res. 2015. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.-P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef]
- Basheer, F.; Huntly, B.J.P. BET bromodomain inhibitors in leukemia. Exp. Hematol. 2015, 43, 718–731. [Google Scholar] [CrossRef]
- Denis, G.V.; Vaziri, C.; Guo, N.; Faller, D. V RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000. [Google Scholar] [CrossRef][Green Version]
- Ostrowski, J.; Florio, S.K.; Denis, G.V.; Suzuki, H.; Bomsztyk, K. Stimulation of p85/RING3 kinase in multiple organs after systemic administration of mitogens into mice. Oncogene 1998. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, H.; Zhang, J.; Shen, W.; Wang, X.; Wu, J.; Wu, J.; Shi, Y. Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails. BMC Struct. Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Umehara, T.; Nakano, K.; Moon, K.J.; Shirouzu, M.; Morita, S.; Uda-Tochio, H.; Hamana, H.; Terada, T.; Adachi, N.; et al. Crystal structure of the human BRD2 bromodomain: Insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 2007. [Google Scholar] [CrossRef] [PubMed]
- Denis, G.V.; McComb, M.E.; Faller, D.V.; Sinha, A.; Romesser, P.B.; Costello, C.E. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 2006. [Google Scholar] [CrossRef] [PubMed]
- LeRoy, G.; Rickards, B.; Flint, S.J. The Double Bromodomain Proteins Brd2 and Brd3 Couple Histone Acetylation to Transcription. Mol. Cell 2008. [Google Scholar] [CrossRef]
- SINHA, A.; FALLER, D.V.; DENIS, G.V. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A 1. Biochem. J. 2005. [Google Scholar] [CrossRef]
- Wu, S.Y.; Chiang, C.M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007. [Google Scholar] [CrossRef]
- Jiang, Y.W.; Veschambre, P.; Erdjument-Bromage, H.; Tempst, P.; Conaway, J.W.; Conaway, R.C.; Kornberg, R.D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 1998. [Google Scholar] [CrossRef]
- Moon, K.J.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005. [Google Scholar] [CrossRef]
- Donner, A.J.; Ebmeier, C.C.; Taatjes, D.J.; Espinosa, J.M. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010. [Google Scholar] [CrossRef]
- Bhagwat, A.S.; Roe, J.S.; Mok, B.Y.L.; Hohmann, A.F.; Shi, J.; Vakoc, C.R. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Ellenberg, J.; Farina, A.; Coleman, A.E.; Maruyama, T.; Sciortino, S.; Lippincott-Schwartz, J.; Ozato, K. A Bromodomain Protein, MCAP, Associates with Mitotic Chromosomes and Affects G2-to-M Transition. Mol. Cell. Biol. 2002. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, K.; Nishiyama, A.; Moon, K.J.; Dey, A.; Ghosh, A.; Tamura, T.; Natsume, H.; Yao, H.; Ozato, K. The bromodomain protein Brd4 stimulates g1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; He, N.; Zhou, Q. Brd4 Recruits P-TEFb to Chromosomes at Late Mitosis To Promote G1 Gene Expression and Cell Cycle Progression. Mol. Cell. Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Rahl, P.B.; Lin, C.Y.; Seila, A.C.; Flynn, R.A.; McCuine, S.; Burge, C.B.; Sharp, P.A.; Young, R.A. C-Myc regulates transcriptional pause release. Cell 2010. [Google Scholar] [CrossRef] [PubMed]
- Schröder, S.; Cho, S.; Zeng, L.; Zhang, Q.; Kaehlcke, K.; Mak, L.; Lau, J.; Bisgrove, D.; Schnölzer, M.; Verdin, E.; et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J. Biol. Chem. 2012. [Google Scholar] [CrossRef]
- Roe, J.S.; Mercan, F.; Rivera, K.; Pappin, D.J.; Vakoc, C.R. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol. Cell 2015. [Google Scholar] [CrossRef]
- Stonestrom, A.J.; Hsu, S.C.; Jahn, K.S.; Huang, P.; Keller, C.A.; Giardine, B.M.; Kadauke, S.; Campbell, A.E.; Evans, P.; Hardison, R.C.; et al. Functions of BET proteins in erythroid gene expression. Blood 2015. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018. [Google Scholar] [CrossRef]
- Chen, L.F.; Fischle, W.; Verdin, E.; Greene, W.C. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 2001. [Google Scholar] [CrossRef]
- Huang, B.; Yang, X.-D.; Zhou, M.-M.; Ozato, K.; Chen, L.-F. Brd4 Coactivates Transcriptional Activation of NF-B via Specific Binding to Acetylated RelA. Mol. Cell. Biol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Q.; Helfer, C.M.; Jiao, J.; You, J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J. Biol. Chem. 2012. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Case-Borden, C.; Gegonne, A.; Hsu, C.H.; Chen, Q.; Meerzaman, D.; Dey, A.; Ozato, K.; Singer, D.S. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Stanlie, A.; Yousif, A.S.; Akiyama, H.; Honjo, T.; Begum, N.A. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol. Cell 2014. [Google Scholar] [CrossRef]
- Li, X.; Baek, G.H.; Ramanand, S.G.; Sharp, A.; Gao, Y.; Yuan, W.; Welti, J.; Rodrigues, D.N.; Dolling, D.; Figueiredo, I.; et al. BRD4 Promotes DNA Repair and Mediates the Formation of TMPRSS2-ERG Gene Rearrangements in Prostate Cancer. Cell Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dulak, A.M.; Hattersley, M.M.; Willis, B.S.; Nikkilä, J.; Wang, A.; Lau, A.; Reimer, C.; Zinda, M.; Fawell, S.E.; et al. BRD4 facilitates replication stress-induced DNA damage response. Oncogene 2018. [Google Scholar] [CrossRef]
- Akıncılar, S.C.; Khattar, E.; Boon, P.L.S.; Unal, B.; Fullwood, M.J.; Tergaonkar, V. Long-range chromatin interactions drive mutant TERT promoter activation. Cancer Discov. 2016. [Google Scholar] [CrossRef]
- Wang, S.; Pike, A.M.; Lee, S.S.; Strong, M.A.; Connelly, C.J.; Greider, C.W. BRD4 inhibitors block telomere elongation. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Zuber, J.; Shi, J.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef]
- Ott, C.J.; Kopp, N.; Bird, L.; Paranal, R.M.; Qi, J.; Bowman, T.; Rodig, S.J.; Kung, A.L.; Bradner, J.E.; Weinstock, D.M. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 2012. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011. [Google Scholar] [CrossRef] [PubMed]
- Mertz, J.A.; Conery, A.R.; Bryant, B.M.; Sandy, P.; Balasubramanian, S.; Mele, D.A.; Bergeron, L.; Sims, R.J. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 2011. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Lovén, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012. [Google Scholar] [CrossRef]
- Frank, S.R.; Parisi, T.; Taubert, S.; Fernandez, P.; Fuchs, M.; Chan, H.M.; Livingston, D.M.; Amati, B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003. [Google Scholar] [CrossRef] [PubMed]
- Rahl, P.B.; Young, R.A. MYC and transcription elongation. Cold Spring Harb. Perspect. Med. 2014. [Google Scholar] [CrossRef]
- Horn, H.; Ziepert, M.; Becher, C.; Barth, T.F.E.; Bernd, H.W.; Feller, A.C.; Klapper, W.; Hummel, M.; Stein, H.; Hansmann, M.L.; et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood 2013. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.A.; Lozanski, G.; Byrd, J.C. Adult Burkitt leukemia and lymphoma. Blood 2004. [Google Scholar] [CrossRef]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Chapuy, B.; McKeown, M.R.; Lin, C.Y.; Monti, S.; Roemer, M.G.M.; Qi, J.; Rahl, P.B.; Sun, H.H.; Yeda, K.T.; Doench, J.G.; et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 2013. [Google Scholar] [CrossRef]
- Pérez-Galán, P.; Dreyling, M.; Wiestner, A. Mantle cell lymphoma: Biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 2011. [Google Scholar] [CrossRef] [PubMed]
- Döhner, K.; Schlenk, R.F.; Habdank, M.; Scholl, C.; Rücker, F.G.; Corbacioglu, A.; Bullinger, L.; Fröhling, S.; Döhner, H. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood 2005. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Gudgin, E.J.; Horton, S.J.; Giotopoulos, G.; Meduri, E.; Robson, S.; Cannizzaro, E.; Osaki, H.; Wiese, M.; Putwain, S.; et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 2014. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Bolli, N.; Shan, J.; Martelli, M.P.; Liso, A.; Pucciarini, A.; Bigerna, B.; Pasqualucci, L.; Mannucci, R.; Rosati, R.; et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood 2006. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhu, L.; Garcia, J.S.; Li, M.X.; Gentles, A.J.; Mitchell, B.S. Brd4 regulates the expression of essential autophagy genes and Keap1 in AML cells. Oncotarget 2018. [Google Scholar] [CrossRef] [PubMed]
- Winters, A.C.; Bernt, K.M. MLL-rearranged leukemias- An update on science and clinical approaches. Front. Pediatr. 2017. [Google Scholar] [CrossRef] [PubMed]
- Thirman, M.J.; Gill, H.J.; Burnett, R.C.; Mbangkollo, D.; McCabe, N.R.; Kobayashi, H.; Ziemin-van der Poel, S.; Kaneko, Y.; Morgan, R.; Sandberg, A.A. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N. Engl. J. Med. 1993. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007. [Google Scholar] [CrossRef]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.-I.; Robson, S.C.; Chung, C.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef]
- Resnick-Silverman, L.; Yan, S.; Mutjaba, S.; Liu, W.-J.; Zeng, L.; Manfredi, J.J.; Zhou, M.-M. Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem. Biol. 2006, 13, 81–90. [Google Scholar] [CrossRef]
- Beesley, A.H.; Stirnweiss, A.; Ferrari, E.; Endersby, R.; Howlett, M.; Failes, T.W.; Arndt, G.M.; Charles, A.K.; Cole, C.H.; Kees, U.R. Comparative drug screening in NUT midline carcinoma. Br. J. Cancer 2014. [Google Scholar] [CrossRef] [PubMed]
- Loosveld, M.; Castellano, R.; Gon, S.; Goubard, A.; Crouzet, T.; Pouyet, L.; Prebet, T.; Vey, N.; Nadel, B.; Collette, Y.; et al. Therapeutic Targeting of c-Myc in T-Cell Acute Lymphoblastic Leukemia (T-ALL). Oncotarget 2014. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- King, B.; Trimarchi, T.; Reavie, L.; Xu, L.; Mullenders, J.; Ntziachristos, P.; Aranda-Orgilles, B.; Perez-Garcia, A.; Shi, J.; Vakoc, C.; et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 2013, 153, 1552–1566. [Google Scholar] [CrossRef] [PubMed]
- Normant, E.; Cummings, R.; Bellon, S.; Bailey, C.; Albrecht, B.; Hewitt, M.; Pardo, E.; Campbell, R.; Jayaram, H.; Poy, F.; et al. Abstract LB-237: In vitro and in vivo characterization of CPI-267203, a potent Inhibitor of bromodomain-containing proteins. In Proceedings of the AACR 103rd Annual Meeting, Chicago, IL, USA, 31 March–4 April 2012; Volume 72, p. LB-237-LB-237. [Google Scholar]
- Moros, A.; Rodríguez, V.; Saborit-Villarroya, I.; Montraveta, A.; Balsas, P.; Sandy, P.; Martínez, A.; Wiestner, A.; Normant, E.; Campo, E.; et al. Synergistic antitumor activity of lenalidomide with the BET bromodomain inhibitor CPI203 in bortezomib-resistant mantle cell lymphoma. Leukemia 2014, 28, 2049–2059. [Google Scholar] [CrossRef]
- Recasens-zorzo, C.; Cardesa-salzmann, T.; Petazzi, P.; Ros-blanco, L.; Esteve-arenys, A.; Clot, G.; Guerrero-hernández, M.; Rodríguez, V.; Soldini, D.; Valera, A.; et al. Pharmacological modulation of CXCR4 cooperates with BET bromodomain inhibition in diffuse large B-cell lymphoma. Haematologica 2018, 104, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Ceribelli, M.; Kelly, P.N.; Shaffer, A.L.; Wright, G.W.; Xiao, W.; Yang, Y.; Mathews Griner, L.A.; Guha, R.; Shinn, P.; Keller, J.M.; et al. Blockade of oncogenic IκB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc. Natl. Acad. Sci. USA 2014, 111, 11365–11370. [Google Scholar] [CrossRef]
- Siegel, M.B.; Liu, S.Q.; Davare, M.A.; Spurgeon, S.E.; Loriaux, M.M.; Druker, B.J.; Scott, E.C.; Tyner, J.W. Small molecule inhibitor screen identifies synergistic activity of the bromodomain inhibitor CPI203 and bortezomib in drug resistant myeloma. Oncotarget 2015, 6, 18921–18932. [Google Scholar] [CrossRef]
- Esteve-Arenys, A.; Valero, J.G.; Chamorro-Jorganes, A.; Gonzalez, D.; Rodriguez, V.; Dlouhy, I.; Salaverria, I.; Campo, E.; Colomer, D.; Martinez, A.; et al. The BET bromodomain inhibitor CPI203 overcomes resistance to ABT-199 (venetoclax) by downregulation of BFL-1/A1 in in vitro and in vivo models of MYC+/BCL2+ double hit lymphoma. Oncogene 2018. [Google Scholar] [CrossRef]
- Albrecht, B.K.; Gehling, V.S.; Hewitt, M.C.; Vaswani, R.G.; Côté, A.; Leblanc, Y.; Nasveschuk, C.G.; Bellon, S.; Bergeron, L.; Campbell, R.; et al. Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials. J. Med. Chem. 2016, 59, 1330–1339. [Google Scholar] [CrossRef]
- Siu, K.T.; Ramachandran, J.; Yee, A.J.; Eda, H.; Santo, L.; Panaroni, C.; Mertz, J.A.; Sims, R.J., III; Cooper, M.R.; Raje, N. Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia 2016, 31, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Zeybek, B.; Lopez, S.; Santin, A.D. BET inhibitors: Betting on improved outcomes in uterine serous carcinoma. Oncotarget 2018, 9, 35470–35471. [Google Scholar] [CrossRef] [PubMed]
- Valero, J.G.; Chamorro-Jorganes, A.; Walker, K.; Guerrero-Hernandez, M.; Rodriguez, V.; Recasens-Zorzo, C.; Esteve-Arenys, A.; Lee-Verges, E.; Matas-Cespedes, A.; Serrat, N.; et al. The BET Inhibitor GS-626510 Converges with Idelalisib through the Inhibition of Tumor-Macrophage Crosstalk and Cytokine Production in Diffuse Large B-Cell Lymphoma. Blood 2017, 130, 4108. [Google Scholar]
- Coudé, M.-M.; Braun, T.; Berrou, J.; Dupont, M.; Bertrand, S.; Masse, A.; Raffoux, E.; Itzykson, R.; Delord, M.; Riveiro, M.E.; et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 2015. [Google Scholar] [CrossRef] [PubMed]
- Boi, M.; Gaudio, E.; Bonetti, P.; Kwee, I.; Bernasconi, E.; Tarantelli, C.; Rinaldi, A.; Testoni, M.; Cascione, L.; Ponzoni, M.; et al. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin. Cancer Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Shah, B.; Fiskus, W.; Qi, J.; Rajapakshe, K.; Coarfa, C.; Li, L.; Devaraj, S.G.T.; Sharma, S.; Zhang, L.; et al. Synergistic activity of BET protein antagonist-based combinations in mantle cell lymphoma cells sensitive or resistant to ibrutinib. Blood 2015, 126, 1565–1574. [Google Scholar] [CrossRef]
- Picaud, S.; Da Costa, D.; Thanasopoulou, A.; Filippakopoulos, P.; Fish, P.V.; Philpott, M.; Fedorov, O.; Brennan, P.; Bunnage, M.E.; Owen, D.R.; et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lasorsa, E.; Smonksey, M.; Kirk, J.S.; Rosario, S.; Hernandez-Ilizaliturri, F.J.; Ellis, L. Mitochondrial protection impairs BET bromodomain inhibitor-mediated cell death and provides rationale for combination therapeutic strategies. Cell Death Dis. 2015. [Google Scholar] [CrossRef][Green Version]
- Bui, M.H.; Lin, X.; Albert, D.H.; Li, L.; Lam, L.T.; Faivre, E.J.; Warder, S.E.; Huang, X.; Wilcox, D.; Donawho, C.K.; et al. Preclinical Characterization of BET Family Bromodomain Inhibitor ABBV-075 Suggests Combination Therapeutic Strategies. Cancer Res. 2017, 77, 2976–2989. [Google Scholar] [CrossRef] [PubMed]
- Picaud, S.; Leonards, K.; Lambert, J.-P.; Dovey, O.; Wells, C.; Fedorov, O.; Monteiro, O.; Fujisawa, T.; Wang, C.-Y.; Lingard, H.; et al. Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia. Sci. Adv. 2016. [Google Scholar] [CrossRef]
- Stubbs, M.C.; Burn, T.C.; Sparks, R.; Maduskuie, T.; Diamond, S.; Rupar, M.; Wen, X.; Volgina, A.; Zolotarjova, N.; Waeltz, P.; et al. The Novel Bromodomain and Extraterminal Domain Inhibitor INCB054329 Induces Vulnerabilities in Myeloma Cells That Inform Rational Combination Strategies. Clin. Cancer Res. 2019, 25, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Gaudio, E.; Tarantelli, C.; Ponzoni, M.; Odore, E.; Rezai, K.; Bernasconi, E.; Cascione, L.; Rinaldi, A.; Stathis, A.; Riveiro, E.; et al. Bromodomain inhibitor OTX015 (MK-8628) combined with targeted agents shows strong in vivo antitumor activity in lymphoma. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Harris, A.W.; Pinkert, C.A.; Corcoran, L.M.; Alexander, W.S.; Cory, S.; Palmiter, R.D.; Brinster, R.L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985. [Google Scholar] [CrossRef] [PubMed]
- Tomska, K.; Kurilov, R.; Lee, K.S.; Hüllein, J.; Lukas, M.; Sellner, L.; Walther, T.; Wagner, L.; Oleś, M.; Brors, B.; et al. Drug-based perturbation screen uncovers synergistic drug combinations in Burkitt lymphoma. Sci. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Lewis, J.M.; Cyrenne, B.M.; Monico, P.F.; Mirza, F.N.; Carlson, K.R.; Foss, F.M.; Girardi, M. BET inhibition in advanced cutaneous T cell lymphoma is synergistically potentiated by BCL2 inhibition or HDAC inhibition. Oncotarget 2018. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009. [Google Scholar] [CrossRef]
- Wyce, A.; Ganji, G.; Smitheman, K.N.; Chung, C.-W.; Korenchuk, S.; Bai, Y.; Barbash, O.; Le, B.C.; Craggs, P.D.; McCabe, M.T.; et al. BET Inhibition Silences Expression of MYCN and BCL2 and Induces Cytotoxicity in Neuroblastoma Tumor Models. PLoS ONE 2013. [Google Scholar] [CrossRef]
- Wang, H.; Hong, B.; Li, X.; Deng, K.; Li, H.; Lui, V.W.Y.; Lin, W. JQ1 synergizes with the Bcl-2 inhibitor ABT-263 against MYCN-amplified small cell lung cancer. Oncotarget 2017. [Google Scholar] [CrossRef][Green Version]
- Hogg, S.J.; Newbold, A.; Vervoort, S.J.; Cluse, L.A.; Martin, B.P.; Gregory, G.P.; Lefebure, M.; Vidacs, E.; Tothill, R.W.; Bradner, J.E.; et al. BET Inhibition Induces Apoptosis in Aggressive B-Cell Lymphoma via Epigenetic Regulation of BCL-2 Family Members. Mol. Cancer Ther. 2016, 15, 2030–2041. [Google Scholar] [CrossRef]
- Zhao, L.; Okhovat, J.P.; Hong, E.K.; Kim, Y.H.; Wood, G.S. Preclinical Studies Support Combined Inhibition of BET Family Proteins and Histone Deacetylases as Epigenetic Therapy for Cutaneous T-Cell Lymphoma. Neoplasia (United States) 2019, 21, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Mill, C.P.; Cai, T.; Fiskus, W.; Borthakur, G.; Kornblau, S.M.; Kadia, T.M.; DiNardo, C.D.; Nowak, A.J.; Saenz, D.T.; Saenz, D.N.; et al. Mechanisms Underlying Superior Efficacy of Co-Targeting BET Proteins and Anti-Apoptotic BCL2 or MCL1 Protein Against AML Blast Progenitor Cells. Blood 2018, 132, 1351. [Google Scholar]
- Bhadury, J.; Nilsson, L.M.; Muralidharan, S.V.; Green, L.C.; Li, Z.; Gesner, E.M.; Hansen, H.C.; Keller, U.B.; McLure, K.G.; Nilsson, J.A. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc. Natl. Acad. Sci. USA 2014. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, R.; Matta, H.; Tolani, B.; Triche, T., Jr.; Chaudhary, P.M. Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene 2015, 35, 1797–1810. [Google Scholar] [CrossRef] [PubMed]
- Doroshow, D.B.; Eder, J.P.; LoRusso, P.M. BET inhibitors: A novel epigenetic approach. Ann. Oncol. 2017. [Google Scholar] [CrossRef]
- Berthon, C.; Raffoux, E.; Thomas, X.; Vey, N.; Gomez-Roca, C.; Yee, K.; Taussig, D.C.; Rezai, K.; Roumier, C.; Herait, P.; et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016, 3, e186–e195. [Google Scholar] [CrossRef]
- Amorim, S.; Stathis, A.; Gleeson, M.; Iyengar, S.; Magarotto, V.; Leleu, X.; Morschhauser, F.; Karlin, L.; Broussais, F.; Rezai, K.; et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016, 3, e196–e204. [Google Scholar] [CrossRef]
- Kremyanskaya, M.; Hoffman, R.; Mascarenhas, J.; Verstovsek, S.; Mertz, J.; Garner, F.; Senderowicz, A. A Phase 2 Study of Cpi-0610, a Bromodomain and Extraterminal (BET) Inhibitor, in Patients with Myelofibrosis (MF). Blood 2018, 132, 5481. [Google Scholar]
- Abramson, J.S.; Blum, K.A.; Flinn, I.W.; Gutierrez, M.; Goy, A.; Maris, M.; Cooper, M.; O’Meara, M.; Borger, D.; Jennifer Mertz, J.; et al. BET inhibitor CPI-0610 is well tolerated and induces responses in diffuse large B-cell lymphoma and follicular lymphoma: Preliminary analysis of an ongoing phase 1 study. Blood 2015, 126, 1491. [Google Scholar]
- Fong, C.Y.; Gilan, O.; Lam, E.Y.N.; Rubin, A.F.; Ftouni, S.; Tyler, D.; Stanley, K.; Sinha, D.; Yeh, P.; Morison, J.; et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 2015. [Google Scholar] [CrossRef]
- Rathert, P.; Roth, M.; Neumann, T.; Muerdter, F.; Roe, J.S.; Muhar, M.; Deswal, S.; Cerny-Reiterer, S.; Peter, B.; Jude, J.; et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Lin, C.Y.; He, H.H.; Witwicki, R.M.; Tabassum, D.P.; Roberts, J.M.; Janiszewska, M.; Huh, S.J.; Liang, Y.; Ryan, J.; et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Fu, L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine 2018, 36, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Bondeson, D.P.; Crews, C.M. Targeted Protein Degradation by Small Molecules. Annu. Rev. Pharmacol. Toxicol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zengerle, M.; Chan, K.H.; Ciulli, A. Selective Small Molecule Induced Degradation of the BET Bromodomain Protein BRD4. ACS Chem. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Fiskus, W.; Qian, Y.; Rajapakshe, K.; Raina, K.; Coleman, K.G.; Crew, A.P.; Shen, A.; Saenz, D.T.; Mill, C.P.; et al. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia 2018, 32, 343–352. [Google Scholar] [CrossRef]
- Winter, G.E.; Buckley, D.L.; Paulk, J.; Roberts, J.M.; Souza, A.; Dhe-Paganon, S.; Bradner, J.E. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348, 1376–1381. [Google Scholar] [CrossRef]
- Winter, G.E.; Mayer, A.; Buckley, D.L.; Erb, M.A.; Roderick, J.E.; Vittori, S.; Reyes, J.M.; di Iulio, J.; Souza, A.; Ott, C.J.; et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol. Cell 2017, 67, 5–18.e19. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Lewis, B.A.; Cherman, N.; Hewitt, M.C.; Albrecht, B.K.; Robey, P.G.; Ozato, K.; Sims, R.J.; Singer, D.S. BRD4 is an atypical kinase that phosphorylates Serine2 of the RNA Polymerase II carboxy-terminal domain. Proc. Natl. Acad. Sci. USA 2012, 109, 6927–6932. [Google Scholar] [CrossRef]
- Ember, S.W.J.; Zhu, J.-Y.; Olesen, S.H.; Martin, M.P.; Becker, A.; Berndt, N.; Georg, G.I.; Schönbrunn, E. Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors. ACS Chem. Biol. 2014, 9, 1160–1171. [Google Scholar] [CrossRef]
| BET Inhibitor | Target | Drug Combination | Disease | Study Phase (Number) | Status |
|---|---|---|---|---|---|
| GSK525762/C | BRD2/3/4 | - | MM | I (NCT01587703) | Active, not recruiting |
| OTX015/MK-8628 | Entinostat, Molibresib | Lymphomas | I (NCT03925428) | Not yet recruiting | |
| - | AML, MDS, DLBCL | I (NCT02698189) | Active, not recruiting | ||
| CC-90010 | BDR2 | - | R/R-NHL | I (NCT03220347) | Recruiting |
| CPI0610 | Ruxolitinib | AML, MDS | II (NCT02158858) | Recruiting | |
| ABBV-744 | BRD4 | - | AML | I (NCT03360006) | Recruiting |
| BI 894999 | - | DLBCL | I (NCT02516553) | Recruiting | |
| BMS-986158 | - | Lymphoma | I (NCT03936465) | Recruiting | |
| RO6870810 | Daratumumab | MM | I (NCT03068351) | Active, not recruiting | |
| Venetoclax, Rituximab | R/R-DLBCL, High-grade B-cell lymphoma | I (NCT03255096) | Active, not recruiting |
| BETi | Drug Combination | Target Disease and Criteria of Inclusion | Clinical Trial (Number of Patients, Age) | Response | Adverse Effects |
|---|---|---|---|---|---|
| ABBV-075 | Venetoclax | AML, MM | NCT02391480 (n = 128, ≥18 years old) | No data available | No data available |
| FT-1101 | Azacitidine | R/R AML (FLT3-ITD or FLT3-TKD mutated) MDS (eligible to receive azacitidine), R/R NHL (primary mediastinal, DLBCL and B-cell lymphoma) | NCT02543879 (n = 94, ≥18 years old) | No data available | No data available |
| OTX015/MK-8628 | - | AML Ph + ALL MM (exposed to at least one alkylating agent/corticosteroid/IMiD and bortezomib) DLBCL (≥1 nonirradiated tumor mass ≥750 mm3). (failure of all standard therapies and life expectancy ≥3 months) | NCT01713582 (n = 141, ≥18 years old) | DOR in 6,6% of DLBCL patients and clinical activity without ORC in 13,3% patients | Serious adverse effects: Thrombocytopenia, febrile neutropenia, leukocytosis, sinus bradycardia, gastrointestinal events (grade 1–2), fatigue, asthenia (grade 1–2) and infections |
| Azacitidine | AML | NCT02303782 † | No data available | No data available | |
| CPI0610 | - | MM (progressed after at least one line of standard therapy) | NCT02157636 (n = 30, ≥18 years old) | No data available | No data available |
| - | Lymphoma (progressed after/prior treatment and without available/effective standard therapy) | NCT01949883 (n = 64, ≥18 years old) | No data available | No data available | |
| PLX51107 | - | R/R AML or NHL (life expectancy ≥3 months) | NCT02683395 †† (n = 50, ≥18 years old) | No data available | No data available |
| BAY1238097 | - | MM (refractory to standard treatment or with no standard therapy available, life expectancy ≥3 months) | NCT02369029 †† (n = 8, ≥18 years old) | No data available | No data available |
| GS-5829 | Exemestane, Fulvestrant | DLBCL, PTCL (refractory to or intolerant of standard therapy or no standard therapy available) | NCT02392611 (n = 33, ≥18 years old) | No data available | No data available |
| INCB054329 | - | DLBCL, AML, MM, ALL, BL (progressed following at least 1 line of therapy without further approved therapy available) | NCT02431260 †† (n = 69, ≥18 years old) | PK variations among individuals | Serious adverse effects Anemia, neutropenia, thrombocytopenia, gastrointestinal disorders, fatigue, hyperbilirubinemia, pneumonia, multi-organ failure, hepatic vein thrombosis, infections increased alanine and aspartate aminotransferases, hyperbilinubinemia, respiratory, thoracic, and mediastinal disorders |
| RO6870810 | - | R/R AML and MDS (allogeneic stem cell transplant not treated with immunosuppressors for ≥2 weeks, life expectancy ≥2 months) | NCT02308761 (n = 26, ≥18 years old) | No data available | No data available |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes-Garau, D.; Ribeiro, M.L.; Roué, G. Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications. Cancers 2019, 11, 1483. https://doi.org/10.3390/cancers11101483
Reyes-Garau D, Ribeiro ML, Roué G. Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications. Cancers. 2019; 11(10):1483. https://doi.org/10.3390/cancers11101483
Chicago/Turabian StyleReyes-Garau, Diana, Marcelo L. Ribeiro, and Gaël Roué. 2019. "Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications" Cancers 11, no. 10: 1483. https://doi.org/10.3390/cancers11101483
APA StyleReyes-Garau, D., Ribeiro, M. L., & Roué, G. (2019). Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications. Cancers, 11(10), 1483. https://doi.org/10.3390/cancers11101483






