Exosomal microRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study ‖
Abstract
1. Introduction
2. Results
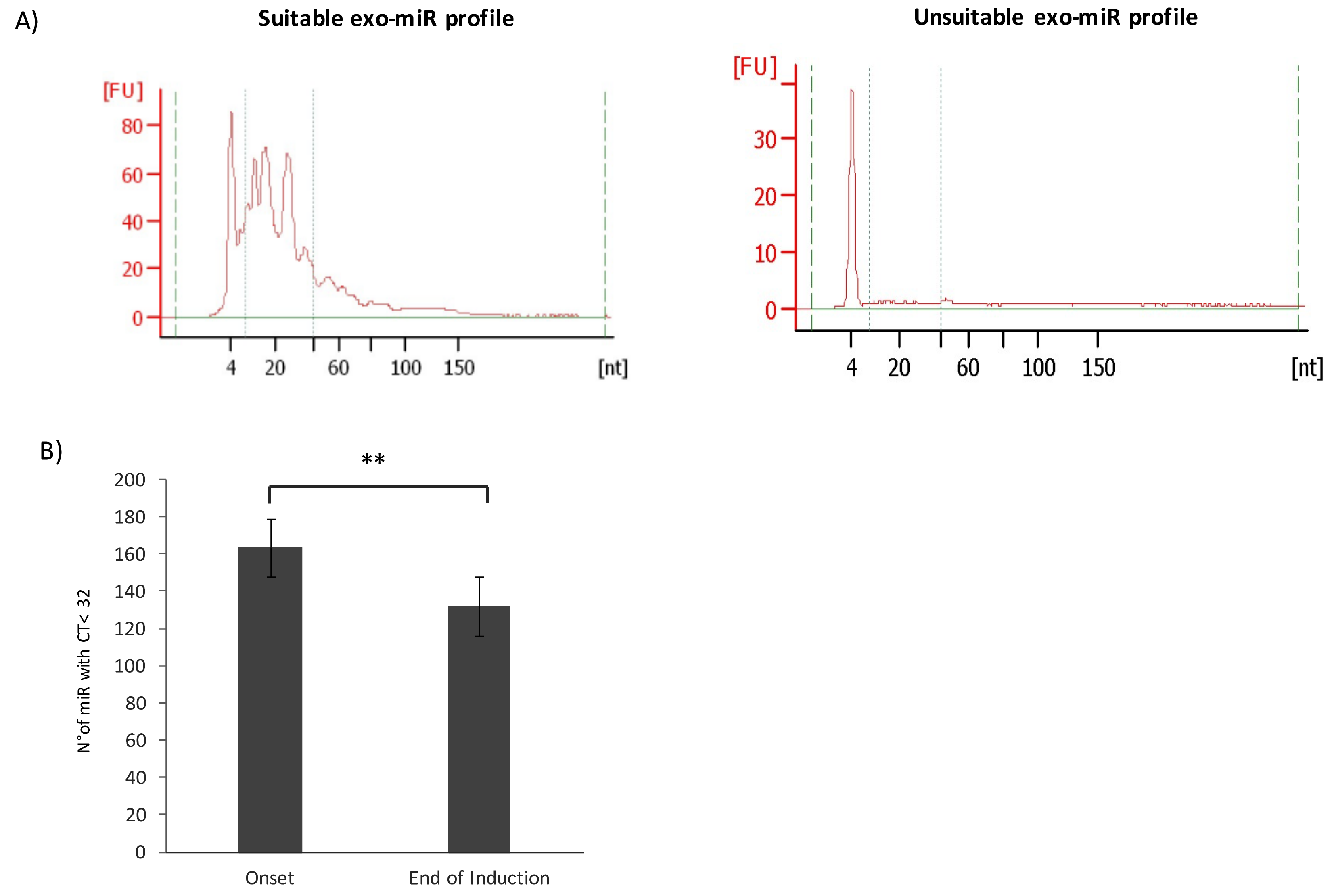
2.1. Characterization of Exosomes Isolated from Plasma of HR-NB Patients
2.2. Differential Plasma exo-miRNAs Expression Profile in HR-NB Patients before and after Induction Chemotherapy
2.3. A Three-miRNAs Signature Differentiates Poor Responders from Good Responders
2.4. The Three-miR Signature Depicts Tumor Growth and Chemoresistance
2.5. Chemoresistance Index Based on exo-miRNAs Modulation after Induction Chemotherapy is Indicative of Event-Free Survival (EFS)
3. Discussion
4. Materials and Methods
4.1. Study Population and Blood Sample Collection
4.2. Exosome Isolation and miRNAs Purification
4.3. Flow Cytometric Analysis
4.4. RTqPCR Analysis
4.5. Bioinformatic Procedures and Statistical Analysis
4.6. Study Approval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Zeka, F.; Decock, A.; Van Goethem, A.; Vanderheyden, K.; Demuynck, F.; Lammens, T.; Helsmoortel, H.H.; Vermeulen, J.; Noguera, R.; Berbegall, A.P.; et al. Circulating microRNA biomarkers for metastatic disease in neuroblastoma patients. JCI Insight. 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yànez-Mò, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Whiteside, T.L. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Proteomic Analysis of Plasma-Derived Exosomes in Defining Their Role as Biomarkers of Disease Progression, Response to Therapy and Outcome. Proteomes 2019, 7, 27. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef]
- Liew, A.W.; Law, N.F.; Yan, H. Missing value imputation for gene expression data: Computational techniques to recover missing data from available information. Brief. Bioinform. 2011, 12, 498–513. [Google Scholar] [CrossRef]
- Chen, C.; Grennan, K.; Badner, J.; Zhang, D.; Gershon, E.; Jin, L.; Liu, C. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS ONE 2011, 6, e17238. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Bagatell, R.; Cohn, S.L.; Pearson, A.D.; Villablanca, J.G.; Berthold, F.; Burchill, S.; Boubaker, A.; McHugh, K.; Nuchtern, J.G.; et al. Revisions of the international neuroblastoma response criteria: A consensus statement from the national cancer isntitute clinical trials planning meeting. J. Clin. Oncol. 2017, 35, 2580–2587. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.R.; Brosens, J.J.; Lam, E.W. Resist or die: FOXO transcription factors determine the cellular response to chemotherapy. Cell Cycle 2008, 7, 3133–3136. [Google Scholar] [CrossRef] [PubMed]
- Startelet, H.; Oligny, L.L.; Vassal, G. AKT pathway in neuroblastoma and its therapeutic implications. Expert Rev. Anticancer Ther. 2008, 8, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Hodkinson, P.S.; Elliott, T.; Wong, W.S.; Rintoul, R.C.; Mackinnon, A.C.; Haslett, C.; Sethi, T. ECM overrides DNA damage-induced cell cycle arrest and apoptosis in small-cell lung cancer cells through β1 integrin-dependent activation of PI3-kinase. Cell Death Differ. 2006, 13, 1776–1788. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Lan, F.; Yan, X.; Xiao, Z.; Wu, Y.; Zhang, Q. Hypoxia exposure induced cisplatin resistance partially via activating p53 and hypoxia inducible factor-1α in non-small cell lung cancer A549 cells. Oncol. Lett. 2018, 16, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.Y.; Li, Y.J.; Wang, P.Y.; Jiao, F.; Zhang, S.; Zhang, W.J. miRNA-regulated expression of oncogenes and tumor suppressor genes in the cisplatin-inhibited growth of K562 cells. Oncol. Rep. 2010, 23, 1693–1700. [Google Scholar] [CrossRef]
- Hummel, R.; Wang, T.; Watson, D.I.; Michael, M.Z.; Van der Hoek, M.; Haier, J.; Hussey, D.J. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncol. Rep. 2011, 26, 1011–1017. [Google Scholar] [CrossRef]
- Ziliak, D.; Gamazon, E.R.; Lacroix, B.; Kyung Im, H.; Wen, Y.; Huang, R.S. Genetic variation that predicts platinum sensitivity reveals the role of miR-193b* in chemotherapeutic susceptibility. Mol. Cancer Ther. 2012, 11, 2054–2061. [Google Scholar] [CrossRef]
- Kim, C.H.; Kim, H.K.; Rettig, R.L.; Kim, J.; Lee, E.T.; Aprelikova, O.; Choi, I.J.; Munroe, D.J.; Green, J.E. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med. Genom. 2011, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yu, S.; Zhang, Y.; Yuan, Y.; Li, X.; Zhong, J.; Feng, J. miR-590-5p regulates gastric cancer cell growth and chemosensitivity through RECK and the AKT/ERK pathway. Onco Targets Ther. 2016, 9, 6009–6019. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qi, H.; Chen, F.; Cao, C. MicroRNA-25 contributes to cisplatin resistance in gastric cancer cells by inhibiting forkhead box O3a. Oncol. Lett. 2017, 14, 6097–6102. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, S.; Li, Y.; Liu, Y.; Zhang, D.; Li, Y.; Zhang, J. MicroRNA-20a contributes to cisplatin-resistance and migration of OVCAR3 ovarian cancer cell line. Oncol. Lett. 2017, 14, 1780–1786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sylvestre, Y.; De Guire, V.; Querido, E.; Mukhopadhyay, U.K.; Bourdeau, V.; Major, F.; Ferbeyre, G.; Chartrand, P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007, 282, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; Zhang, X.D.; Zhu, J.; Guo, X.Y.; Wang, J.F. Low expression of microRNA-146b-5p and microRNA-320d predicts poor outcome of large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone. Hum. Pathol. 2014, 45, 1664–1673. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, Y.; Yang, M.; Jat, P.; Li, K.; Lombardo, Y.; Xiong, D.; Coombes, R.C.; Raguz, S.; Yagüe, E. The miR-106b~25 cluster promotes bypass of doxorubicin-induced senescence and increase in motility and invasion by targeting the E-cadherin transcriptional activator EP300. Cell Death Differ. 2014, 21, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Milazzo, M.; Chieco, P.; Negrini, M.; Calin, G.A.; Grazi, G.L.; Pollutri, D.; Croce, C.M.; Bolondi, L.; Gramantieri, L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010, 70, 5184–5193. [Google Scholar] [CrossRef]
- Cirilo, P.D.R.; de Sousa Andrade, L.N.; Corrêa, B.R.S.; Qiao, M.; Furuya, T.K.; Chammas, R.; Penalva, L.O.F. MicroRNA-195 acts as an anti-proliferative miRNA in human melanoma cells by targeting Prohibitin 1. BMC Cancer 2017, 17, 750. [Google Scholar] [CrossRef]
- Rizzo, S.; Cangemi, A.; Galvano, A.; Fanale, D.; Buscemi, S.; Ciaccio, M.; Russo, A.; Castorina, S.; Bazan, V. Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget 2017, 8, 71924–71932. [Google Scholar] [CrossRef]
- Arrighetti, N.; Cossa, G.; De Cecco, L.; Stucchi, S.; Carenini, N.; Corna, E.; Gandellini, P.; Zaffaroni, N.; Perego, P.; Gatti, L. PKC-alpha modulation by miR-483-3p in platinum-resistant ovarian carcinoma cells. Toxicol. Appl. Pharmacol. 2016, 310, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lamperska, K.M.; Kolenda, T.; Teresiak, A.; Kowalik, A.; Kruszyna-Mochalska, M.; Jackowiak, W.; Bliźniak, R.; Przybyła, W.; Kapałczyńska, M.; Kozlowski, P. Different levels of let-7d expression modulate response of FaDu cells to irradiation and chemotherapeutics. PLoS ONE 2017, 12, e0180265. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yin, J.; Bai, Z.; Zhang, J.; Meng, H.; Cai, J.; Deng, W.; Ma, X.; Zhang, Z. The Profile of Serum microRNAs Predicts Prognosis for Resected Gastric Cancer Patients Receiving Platinum-Based Chemotherapy. Dig. Dis. Sci. 2017, 62, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Yeh, W.L.; Tsai, C.F.; Chen, D.R. Peri-foci adipose-derived stem cells promote chemoresistance in breast cancer. Stem Cell Res. Ther. 2017, 8, 177. [Google Scholar] [CrossRef]
- Boren, T.; Xiong, Y.; Hakam, A.; Wenham, R.; Apte, S.; Chan, G.; Kamath, S.G.; Chen, D.T.; Dressman, H.; Lancaster, J.M. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol. Oncol. 2009, 113, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chen, G.; Sun, D.; Lei, M.; Li, Y.; Zhou, C.; Li, X.; Xue, W.; Wang, H.; Liu, C.; et al. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim. Biophys. Sin. 2017, 49, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Seca, H.; Lima, R.T.; Lopes-Rodrigues, V.; Guimaraes, J.E.; Almeida, G.M.; Vasconcelos, M.H. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr. Drug Targets 2013, 14, 1135–1143. [Google Scholar] [CrossRef]
- Zhou, P.; Ma, L.; Zhou, J.; Jiang, M.; Rao, E.; Zhao, Y.; Guo, F. miR-17-92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cells. Int. J. Oncol. 2016, 48, 1737–1748. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, L.; Yao, Q.; Tao, Z. miR-15b regulates cisplatin resistance and metastasis by targeting PEBP4 in human lung adenocarcinoma cells. Cancer Gene Ther. 2015, 22, 108–114. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, D.; Du, R.; Pan, Y.; Zhao, L.; Sun, S.; Hong, L.; Liu, J.; Fan, D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer. 2008, 123, 372–379. [Google Scholar] [CrossRef]
- Roscigno, G.; Puoti, I.; Giordano, I.; Donnarumma, E.; Russo, V.; Affinito, A.; Adamo, A.; Quintavalle, C.; Todaro, M.; Vivanco, M.D.; et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget 2017, 8, 19507–19521. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; He, N.; Dong, Y.; Jiang, C. MiR-24-BIM-Smac/DIABLO axis controls the sensitivity to doxorubicin treatment in osteosarcoma. Sci. Rep. 2016, 6, 34238. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Dai, X.M.; Li, S.; Qi, G.L.; Cao, G.X.; Zhong, Y.; Yin, P.D.; Yang, X.S. MiR-30c regulates cisplatin-induced apoptosis of renal tubular epithelial cells by targeting Bnip3L and Hspa5. Cell Death Dis. 2017, 8, e2987. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Shen, H.; Cao, Y.; Li, H.; Qin, R.; Chen, Q.; Long, L.; Zhu, X.L.; Xie, C.J.; Xu, W.L. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz. J. Med. Biol. Res. 2014, 47, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef]
- Yu, P.N.; Yan, M.D.; Lai, H.C.; Huang, R.L.; Chou, Y.C.; Lin, W.C.; Yeh, L.T.; Lin, Y.W. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int. J. Cancer 2014, 134, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Kollinerová, S.; Dostál, Z.; Modrianský, M. MicroRNA hsa-miR-29b potentiates etoposide toxicity in HeLa cells via down-regulation of Mcl-1. Toxicol. Vitro 2017, 40, 289–296. [Google Scholar] [CrossRef]
- Li, R.; Wu, S.; Chen, X.; Xu, H.; Teng, P.; Li, W. miR-223/FBW7 axis regulates doxorubicin sensitivity through epithelial mesenchymal transition in non-small cell lung cancer. Am. J. Transl. Res. 2016, 8, 2512–2524. [Google Scholar] [PubMed]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M. Mesenchymal Stem Cell-Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef]
- Zeng, L.P.; Hu, Z.M.; Li, K.; Xia, K. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J. Cell. Mol. Med. 2016, 20, 559–567. [Google Scholar] [CrossRef]
- Yuwen, D.L.; Sheng, B.B.; Liu, J.; Wenyu, W.; Shu, Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2650–2658. [Google Scholar] [PubMed]
- Paik, J.H.; Jang, J.Y.; Jeon, Y.K.; Kim, W.Y.; Kim, T.M.; Heo, D.S.; Kim, C.W. MicroRNA-146a downregulates NFκB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin. Cancer Res. 2011, 17, 4761–4771. [Google Scholar] [CrossRef]
- Wang, T.; Huang, B.; Guo, R.; Ma, J.; Peng, C.; Zu, X.; Tang, H.; Lei, X. A let-7b binding site SNP in the 3′-UTR of the Bcl-xL gene enhances resistance to 5-fluorouracil and doxorubicin in breast cancer cells. Oncol. Lett. 2015, 9, 1907–1911. [Google Scholar] [CrossRef]
- Chen, J.J.; Liu, S.X.; Chen, M.Z.; Zhao, Z.Y. Has-miR-125a and 125b are induced by treatment with cisplatin in nasopharyngeal carcinoma and inhibit apoptosis in a p53-dependent manner by targeting p53 mRNA. Mol. Med. Rep. 2015, 12, 3569–3574. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; An, X.; Li, H.; Ma, L. Effect of miR-155 knockdown on the reversal of doxorubicin resistance in human lung cancer A549/dox cells. Oncol. Lett. 2016, 11, 1161–1166. [Google Scholar] [CrossRef]
- Pang, W.; Tian, X.; Bai, F.; Han, R.; Wang, J.; Shen, H.; Zhang, X.; Liu, Y.; Yan, X.; Jiang, F.; et al. Pim-1 kinase is a target of miR-486-5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol. Cancer 2014, 13, 240. [Google Scholar] [CrossRef]
- Wang, P.; Li, Z.; Liu, H.; Zhou, D.; Fu, A.; Zhang, E. MicroRNA-126 increases chemosensitivity in drug-resistant gastric cancer cells by targeting EZH2. Biochem. Biophys. Res. Commun. 2016, 479, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rajendran, V.; Kulshreshtha, R.; Ghosh, P.C. Enhanced efficacy of anti-miR-191 delivery through stearylamine liposome formulation for the treatment of breast cancer cells. Int. J. Pharm. 2017, 530, 387–400. [Google Scholar] [CrossRef]
- Challagundla, K.B.; Wise, P.M.; Neviani, P.; Chava, H.; Murtadha, M.; Xu, T.; Kennedy, R.; Ivan, C.; Zhang, X.; Vannini, I.; et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- Szajnik, M.; Derbis, M.; Lach, M.; Patalas, P.; Michalak, M.; Drzewiecka, H.; Szpurek, D.; Nowakowski, A.; Spaczynski, M.; Baranowski, W.; et al. Exosomes in plasma of patients with ovarian carcinoma: potential biomarkers of tumor progression and response to therapy. Gynecol. Obstet. 2013, Suppl. 4, 3. [Google Scholar]
- Beckers, A.; Van Peer, G.; Carter, D.R.; Mets, E.; Althoff, K.; Cheung, B.B. MYCN-targeting miRNAs are predominantly downregulated during MYCN-driven neuroblastoma tumor formation. Oncotarget 2015, 6, 5204–5216. [Google Scholar] [CrossRef] [PubMed]
- Li, X.R.; Chu, H.J.; Lv, T.; Wang, L.; Kong, S.F.; Dai, S.Z. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014, 588, 3298–3307. [Google Scholar] [CrossRef] [PubMed]
- Romero-Cordoba, S.L.; Rodriguez-Cuevas, S.; Bautista-Pina, V.; Maffuz-Aziz, A.; D’Ippolito, E.; Cosentino, G.; Baroni, S.; Iorio, M.V.; Hidalgo-Miranda, A. Loss of function of miR-342-3p results in MCT1 over-expression and contributes to oncogenic metabolic reprogramming in triple negative breast cancer. Sci. Rep. 2018, 8, 12252. [Google Scholar] [CrossRef] [PubMed]
- Fardin, P.; Barla, A.; Mosci, S.; Rosasco, L.; Verri, A.; Versteeg, R.; Caron, H.N.; Molenaar, J.J.; Ora, I.; Eva, A.; et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol. Cancer 2010, 9, 185. [Google Scholar] [CrossRef]
- Sun, D.M.; Tang, B.-F.; Li, Z.-X.; Guo, H.-B.; Cheng, J.-L.; Song, P.-P.; Zhao, X. MiR-29c reduces the cisplatin resistance of non-small cell lung cancer cells by negatively regulating the PI3K/Akt pathway. Sci. Rep. 2018, 8, 8007. [Google Scholar] [CrossRef]
- Buechner, J.; Tømte, E.; Haug, B.H.; Henriksen, J.R.; Løkke, C.; Flægstad, T.; Einvik, C. Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br. J. Cancer 2011, 105, 296–303. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef]
- Guo, Y.; Yan, K.; Fang, J.; Qu, Q.; Zhou, M.; Chen, F. Let-7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. J. Exp. Clin. Cancer Res. 2013, 32, 41. [Google Scholar] [CrossRef]
- Marimpietri, D.; Petretto, A.; Raffaghello, L.; Pezzolo, A.; Gagliani, C.; Tacchetti, C.; Mauri, P.; Melioli, G.; Pistoia, V. Proteome profiling of neuroblastoma-derived exosomes reveal the expression of proteins potentially involved in tumor progression. PLoS ONE 2013, 8, e75054. [Google Scholar] [CrossRef]



| Clinical Features | Classification | Patients (n = 52) |
|---|---|---|
| INSS stage | 4 | 47 (91%) |
| 3 | 4 (7%) | |
| 4S | 1 (2%) | |
| MYCN status | AMPL | 22 (42%) |
| NO AMPL | 25 (48%) | |
| NA | 5 (10%) | |
| Induction Response | MR | 6 (12%) |
| PR | 33 (63%) | |
| VGPR | 8 (15%) | |
| NA | 5 (10%) | |
| Relapse | YES | 22 (43%) |
| NO | 30 (57%) | |
| Overall Survival | ALIVE | 40 (77%) |
| DECEASED | 12 (23%) |
| miRNA ID a | Mean ∆CT End | Mean ∆CT Onset | Mean RQ End | Mean RQ Onset | Fold Change b | Adjusted p Value c |
|---|---|---|---|---|---|---|
| hsa-miR-376a | 3.28 | 1.28 | 0.10 | 0.41 | −2.0 | 5.30E−08 |
| hsa-miR-150 | −6.04 | −7.60 | 65.87 | 194.19 | −1.6 | 9.87E−08 |
| hsa-miR-376c | 2.69 | 0.70 | 0.16 | 0.62 | −2.0 | 1.48E−07 |
| hsa-miR-132 | 1.33 | −0.55 | 0.40 | 1.47 | −1.9 | 5.45E−07 |
| hsa-miR-130a | 1.36 | −0.49 | 0.39 | 1.40 | −1.8 | 1.21E−05 |
| hsa-miR-539 | 2.61 | 0.85 | 0.16 | 0.55 | −1.8 | 3.22E−05 |
| hsa-miR-342-3p | −2.59 | −3.70 | 6.03 | 12.96 | −1.1 | 4.81E−05 |
| hsa-miR-192 | 1.40 | 0.06 | 0.38 | 0.96 | −1.3 | 5.95E−05 |
| hsa-miR-340 | 3.02 | 1.78 | 0.12 | 0.29 | −1.2 | 5.95E−05 |
| hsa-miR-320 | −4.01 | −4.97 | 16.10 | 31.28 | −1.0 | 7.29E−05 |
| hsa-let-7g | 0.24 | −1.06 | 0.85 | 2.08 | −1.3 | 9.93E−05 |
| hsa-miR-590-5p | 0.96 | −0.30 | 0.51 | 1.23 | −1.3 | 9.93E−05 |
| hsa-miR-152 | 2.44 | 1.22 | 0.18 | 0.43 | −1.2 | 2.27E−04 |
| hsa-miR-146b | −3.45 | −4.37 | 10.91 | 20.74 | −0.9 | 2.27E−04 |
| hsa-miR-20a | −3.86 | −5.32 | 14.56 | 39.97 | −1.5 | 3.64E−04 |
| hsa-miR-25 | −0.31 | −1.69 | 1.24 | 3.23 | −1.4 | 3.64E−04 |
| hsa-miR-26b | 0.19 | −1.15 | 0.88 | 2.22 | −1.3 | 3.64E−04 |
| hsa-miR-324-3p | 1.73 | 0.62 | 0.30 | 0.65 | −1.1 | 4.52E−04 |
| hsa-miR-199a-3p | −0.01 | −1.41 | 1.01 | 2.65 | −1.4 | 4.95E−04 |
| hsa-miR-106b | −0.59 | −1.75 | 1.50 | 3.36 | −1.2 | 5.83E−04 |
| hsa-miR-19b | −4.65 | −6.15 | 25.11 | 71.00 | −1.5 | 6.23E−04 |
| hsa-miR-374 | −1.50 | −2.61 | 2.82 | 6.11 | −1.1 | 6.54E−04 |
| hsa-miR-29c | 2.42 | 1.03 | 0.19 | 0.49 | −1.4 | 9.50E−04 |
| hsa-miR-20b | −1.84 | −2.92 | 3.57 | 7.55 | −1.1 | 1.03E−03 |
| hsa-miR-26a | −1.83 | −3.01 | 3.57 | 8.07 | −1.2 | 1.09E−03 |
| hsa-miR-140-3p | 3.24 | 2.38 | 0.11 | 0.19 | −0.9 | 1.65E−03 |
| hsa-miR-18a | 1.47 | 0.31 | 0.36 | 0.80 | −1.2 | 1.65E−03 |
| hsa-miR-185 | 1.08 | −0.04 | 0.47 | 1.03 | −1.1 | 1.65E−03 |
| hsa-miR-195 | −0.71 | −1.81 | 1.63 | 3.50 | −1.1 | 1.69E−03 |
| hsa-miR-142-3p | −2.80 | −4.16 | 6.95 | 17.86 | −1.4 | 2.02E−03 |
| hsa-miR-21 | −1.44 | −2.66 | 2.72 | 6.31 | −1.2 | 2.30E−03 |
| hsa-miR-652 | 1.07 | −0.15 | 0.47 | 1.11 | −1.2 | 2.41E−03 |
| hsa-miR-19a | −0.08 | −1.40 | 1.06 | 2.65 | −1.3 | 2.43E−03 |
| hsa-let-7e | −2.27 | −3.31 | 4.81 | 9.94 | −1.0 | 2.55E−03 |
| hsa-miR-106a | −5.59 | −6.46 | 48.08 | 88.20 | −0.9 | 2.55E−03 |
| hsa-miR-660 | 1.40 | 0.29 | 0.38 | 0.82 | −1.1 | 2.99E−03 |
| hsa-miR-598 | 1.94 | 0.64 | 0.26 | 0.64 | −1.3 | 3.56E−03 |
| hsa-let-7d | 0.11 | −0.81 | 0.93 | 1.75 | −0.9 | 3.73E−03 |
| hsa-miR-24 | −4.99 | −5.75 | 31.85 | 53.81 | −0.8 | 5.38E−03 |
| hsa-miR-17 | −5.42 | −6.41 | 42.81 | 85.03 | −1.0 | 5.47E−03 |
| hsa-miR-483-5p | −0.30 | −1.11 | 1.23 | 2.16 | −0.8 | 5.67E−03 |
| hsa-miR-30b | −2.76 | −3.76 | 6.78 | 13.54 | −1.0 | 5.99E−03 |
| hsa-miR-301 | 1.70 | 0.70 | 0.31 | 0.62 | −1.0 | 6.28E−03 |
| hsa-miR-186 | −2.53 | −3.38 | 5.79 | 10.40 | −0.8 | 8.42E−03 |
| hsa-miR-331 | −1.40 | −2.13 | 2.63 | 4.38 | −0.7 | 8.98E−03 |
| hsa-miR-27a | 0.15 | −0.82 | 0.90 | 1.76 | −1.0 | 9.19E−03 |
| hsa-miR-15b | −0.63 | −1.60 | 1.55 | 3.04 | −1.0 | 9.41E−03 |
| hsa-miR-28 | 2.12 | 1.28 | 0.23 | 0.41 | −0.8 | 1.14E−02 |
| hsa-miR-345 | 0.70 | 0.05 | 0.61 | 0.96 | −0.6 | 1.28E−02 |
| hsa-miR-30c | −3.10 | −4.03 | 8.59 | 16.32 | −0.9 | 1.29E−02 |
| hsa-miR-103 | 0.95 | 0.13 | 0.52 | 0.91 | −0.8 | 1.30E−02 |
| hsa-miR-193b | 1.91 | 1.21 | 0.27 | 0.43 | −0.7 | 1.34E−02 |
| hsa-miR-29a | −0.84 | −1.54 | 1.78 | 2.91 | −0.7 | 1.64E−02 |
| hsa-miR-99b | 2.30 | 1.69 | 0.20 | 0.31 | −0.6 | 1.64E−02 |
| hsa-miR-328 | 0.69 | −0.16 | 0.62 | 1.12 | −0.9 | 1.80E−02 |
| hsa-miR-744 | 0.24 | −0.53 | 0.85 | 1.45 | −0.8 | 2.78E−02 |
| hsa-miR-425-5p | 0.38 | −0.23 | 0.77 | 1.18 | −0.6 | 3.23E−02 |
| hsa-miR-92a | −2.30 | −3.03 | 4.94 | 8.15 | −0.7 | 3.23E−02 |
| hsa-miR-16 | −5.99 | −6.86 | 63.78 | 116.19 | −0.9 | 3.25E−02 |
| hsa-miR-142-5p | 2.39 | 1.66 | 0.19 | 0.32 | −0.7 | 3.38E−02 |
| hsa-miR-221 | 0.16 | −0.73 | 0.90 | 1.66 | −0.9 | 4.11E−02 |
| hsa-miR-146a | −5.33 | −5.98 | 40.31 | 62.91 | −0.6 | 4.30E−02 |
| miRNA | Cisplatin | Etoposide | Doxorubicin | Vincristine | Carboplatin | Ref. |
|---|---|---|---|---|---|---|
| hsa-miR-150 | ↑ | [18] | ||||
| hsa-miR-342-3p | ↑ | [19] | ||||
| hsa-miR-320 | ↑ | [20] | ||||
| hsa-let-7g | ↓ | [21] | ||||
| hsa-miR-590-5p | ↑ | [22] | ||||
| hsa-miR-25 | ↑ | [23] | ||||
| hsa-miR-20a | ↑ | ↑ | [24,25] | |||
| hsa-miR-146b | ↑ | ↓ | ↓ | [26] | ||
| hsa-miR-106b | ↑ | [27] | ||||
| hsa-miR-199a-3p | ↓ | [28] | ||||
| hsa-miR-195 | ↓ | ↓ | [29] | |||
| hsa-miR-26a | ↓ | [30] | ||||
| hsa-miR-483-5p | ↑ | [31] | ||||
| hsa-let-7d | ↓ | ↓ | [32] | |||
| hsa-miR-106a | ↑ | ↑ | [33,34] | |||
| hsa-let-7e | ↑ | [35] | ||||
| hsa-miR-21 | ↑ | ↑ | ↑ | [36,37] | ||
| hsa-miR-17 | ↑ | [38] | ||||
| hsa-miR-15b | ↑ | ↓ | [39,40] | |||
| hsa-miR-24 | ↑ | ↓ | ↑ | [41,42] | ||
| hsa-miR-30c | ↑ | ↓ | [43,44] | |||
| hsa-miR-345 | ↓ | [45] | ||||
| hsa-miR-29a | ↓ | ↓ | [46,47] | |||
| hsa-miR-16 | ↓ | [40] | ||||
| hsa-miR-223 | ↑ | ↑ | [48,49] | |||
| hsa-miR-222 | ↑ | ↑ | ↑ | [49,50] | ||
| hsa-miR-146a | ↓ | ↓ | [51,52] | |||
| hsa-let-7b | ↓ | ↓ | [53] | |||
| hsa-miR-125a-5p | ↑ | [54] | ||||
| hsa-miR-155 | ↑ | ↑ | [55] | |||
| hsa-miR-486 | ↑ | [56] | ||||
| hsa-miR-126 | ↓ | [57] | ||||
| hsa-miR-191 | ↑ | ↑ | [58] |
| Drug | Chemoresistance | |
|---|---|---|
| N° Upregulated exo-miRNAs | N° Downregulated exo-miRNAs | |
| Cisplatin | 19 | 7 |
| Etoposide | 1 | 5 |
| Doxorubicin | 10 | 6 |
| Vincristine | 0 | 3 |
| Carboplatin | 2 | 0 |
| Cyclophosphamide | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morini, M.; Cangelosi, D.; Segalerba, D.; Marimpietri, D.; Raggi, F.; Castellano, A.; Fruci, D.; Font de Mora, J.; Cañete, A.; Yáñez, Y.; et al. Exosomal microRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study ‖. Cancers 2019, 11, 1476. https://doi.org/10.3390/cancers11101476
Morini M, Cangelosi D, Segalerba D, Marimpietri D, Raggi F, Castellano A, Fruci D, Font de Mora J, Cañete A, Yáñez Y, et al. Exosomal microRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study ‖. Cancers. 2019; 11(10):1476. https://doi.org/10.3390/cancers11101476
Chicago/Turabian StyleMorini, Martina, Davide Cangelosi, Daniela Segalerba, Danilo Marimpietri, Federica Raggi, Aurora Castellano, Doriana Fruci, Jaime Font de Mora, Adela Cañete, Yania Yáñez, and et al. 2019. "Exosomal microRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study ‖" Cancers 11, no. 10: 1476. https://doi.org/10.3390/cancers11101476
APA StyleMorini, M., Cangelosi, D., Segalerba, D., Marimpietri, D., Raggi, F., Castellano, A., Fruci, D., Font de Mora, J., Cañete, A., Yáñez, Y., Viprey, V., Corrias, M. V., Carlini, B., Pezzolo, A., Schleiermacher, G., Mazzocco, K., Ladenstein, R., Sementa, A. R., Conte, M., ... Varesio, L. (2019). Exosomal microRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study ‖. Cancers, 11(10), 1476. https://doi.org/10.3390/cancers11101476








