The Capacity of Long-Term In Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome
Abstract
1. Introduction
2. Results
2.1. Long-Term In Vitro Proliferation of AML Cells in the Absence of Supportive Non-Leukemic Cells Is Detected Only for a Subset of Patients
2.2. The Capacity of Cytokine-Supported Long-Term In Vitro Proliferation Is Not Associated with Patient Age, Morphological Differentiation, CD34 Expression, Karyotype, or Molecular Genetic Abnormalities
2.3. A high Frequency of AML Cells Showing Long-Term Cytokine-Supported Proliferation Is Associated with Reduced Survival for Patients Receiving Intensive Anti-Leukemic Therapy
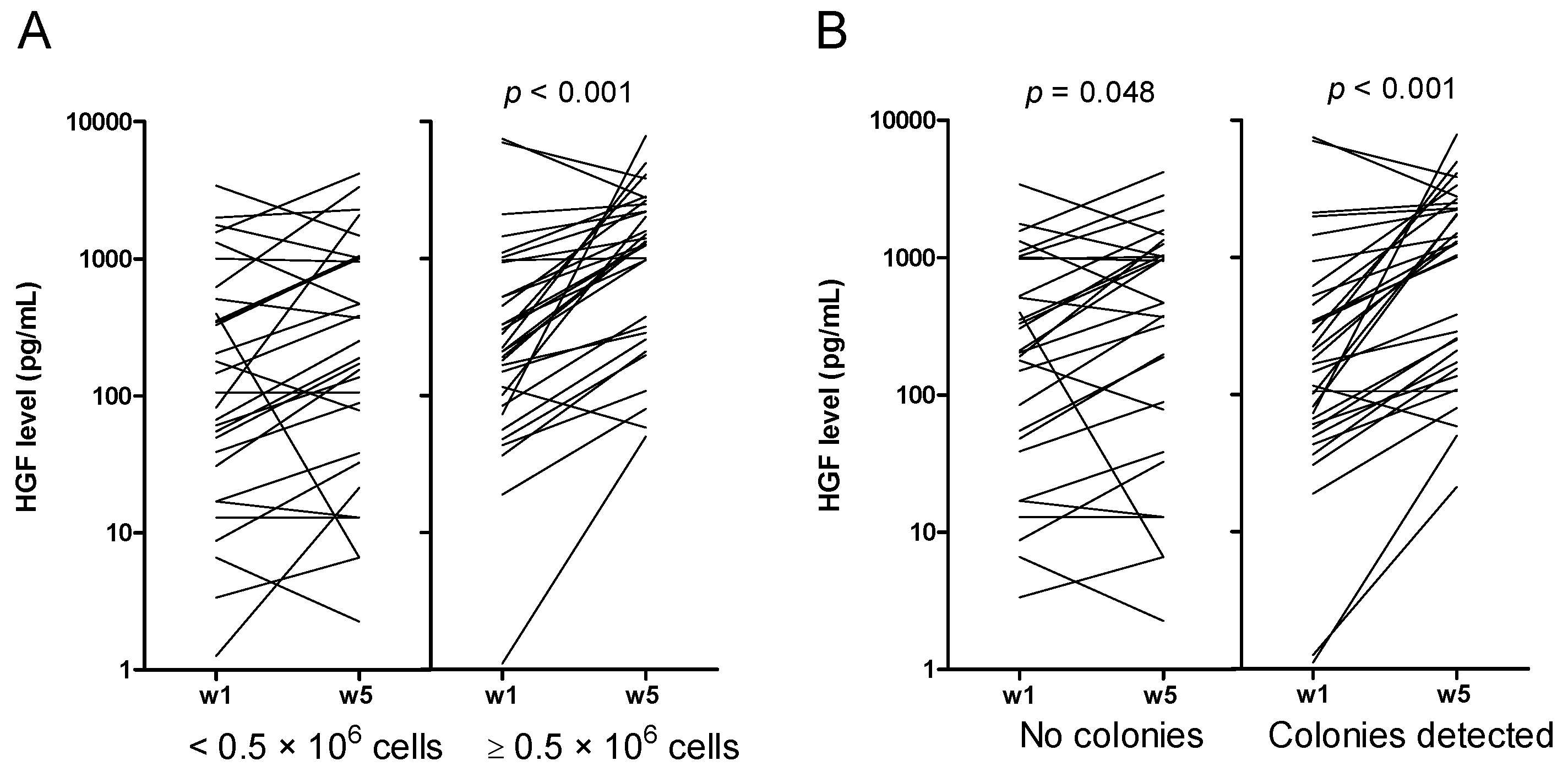
2.4. Increased Levels of HGF and IL-1RA during Long-Term Suspension Culture Are Associated with Maintenance of Clonogenic Cells during Culture
2.5. High Constitutive Cytokine Release Is Associated with an Altered AML Cell Phenotype during the Initial Five Weeks of Suspension Culture
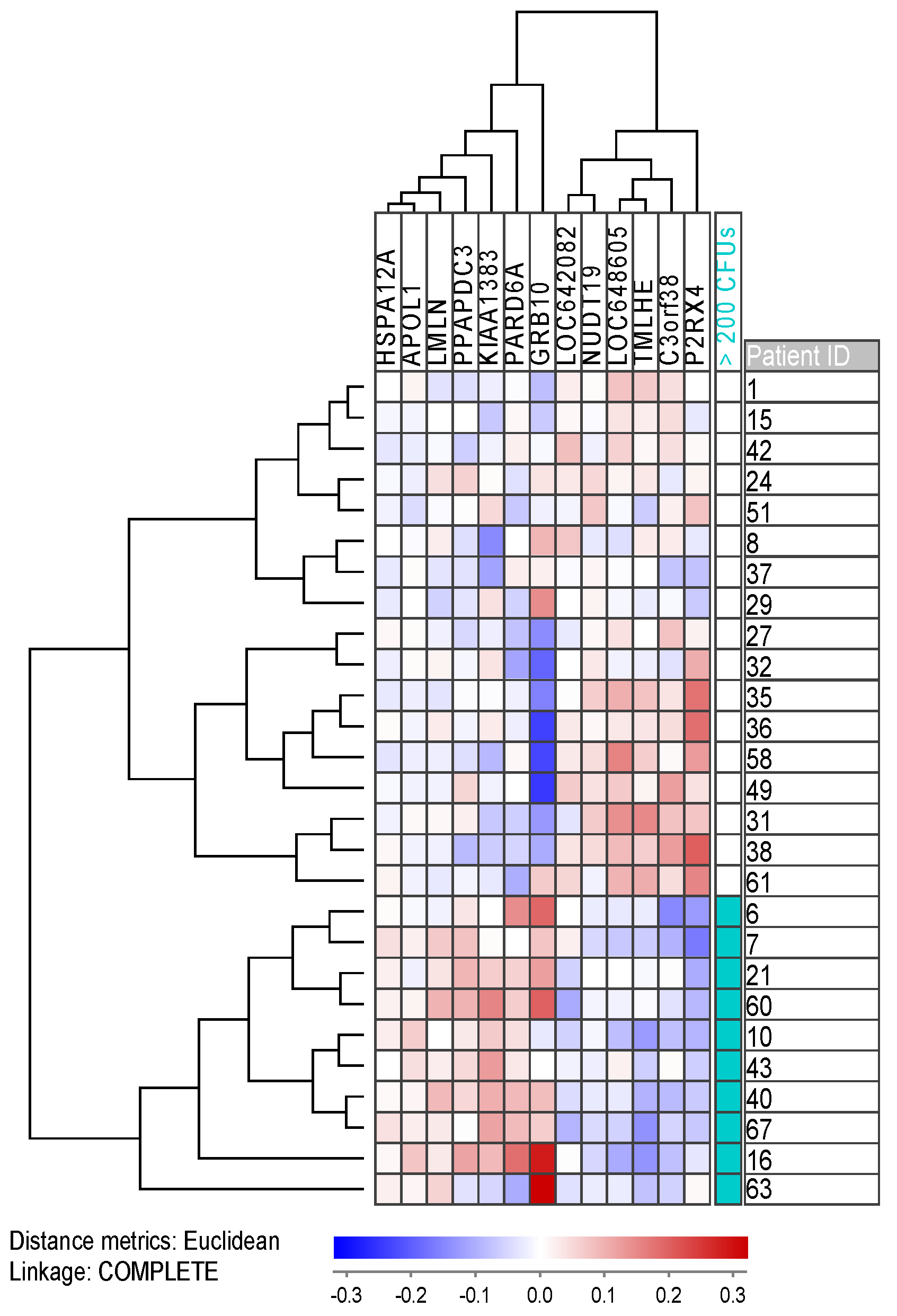
2.6. Gene Expression and Mutation Profiling Identifies Differences between Patient Groups with High and Low Numbers of Clonogenic Cells
2.7. Gene Expression Profiling Identifies Differentially Expressed Genes for Both Cultures Which Present with an Altered AML Phenotype and Cultures with Erythroid Colonies
2.8. Proteomics Analysis Identifies Proteins Involved in Transcription to Be More Abundant for Patients with a High Number of Clonogenic Cells
3. Discussion
4. Materials and Methods
4.1. Patients and Preparation of AML Cells
4.2. Preparation of Enriched AML Cells
4.3. Analyses of AML Cell Viability and Short-Term Proliferation
4.3.1. Analysis of AML Cell Viability
4.3.2. Proliferation Assay
4.4. Long-Term Suspension Cultures
4.5. Colony Formation Assay
4.6. Analysis of Soluble Mediator Levels in Cell Culture Supernatants
4.7. RNA Preparation and Analysis of Global Gene Expression
4.8. Proteomic and Phosphoproteomic Analyses
4.9. Statistical and Bioinformatical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caceres-Cortes, R.J. Blastic leukaemias (AML): A biologist’s view. Cell Biochem. Biophys. 2013, 66, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Burnett, A.; Wetzler, M.; Löwenberg, B. Therapeutic advances in acute myeloid leukemia. J. Clin. Oncol. 2011, 29, 487–494. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposal for the recognition of minimally differentiated acute myeloid leukaemia (AML-MO). Br. J. Haematol. 1991, 78, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Neame, P.B.; Soamboonsrup, P.; Browman, G.P.; Meyer, R.M.; Benger, A.; Wilson, W.E.; Walker, I.R.; Saeed, N.; McBride, J.A. Classifying acute leukemia by immunophenotyping: A combined FAB-immunologic classification of AML. Blood 1986, 68, 1355–1362. [Google Scholar]
- Sidney, L.E.; Branch, M.J.; Dunphy, S.E.; Dua, H.S.; Hopkinson, A. Concise review: Evidence for CD34 as a common marker for diverse progenitors. Stem Cells 2014, 32, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Gjertsen, B.T.; Foss, B.; Huang, T.S. New strategies in the treatment of acute myelogenous leukemia (AML): In vitro culture of AML cells—The present use in experimental studies and the possible importance for future therapeutic approaches. Stem Cells 2001, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ryningen, A.; Stapnes, C.; Bruserud, Ø. Clonogenic acute myelogenous leukemia cells are heterogeneous with regard to regulation of differentiation and effect of epigenetic pharmacological targeting. Leuk Res. 2007, 31, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, H.J.; Blair, A.; Zapf, R.W. Characterization of a hierarchy in human acute myeloid leukemia progenitor cells. Blood 1996, 87, 4754–4761. [Google Scholar] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef]
- Eppert, K.; Takenaka, K.; Lechman, E.R.; Waldron, L.; Nilsson, B.; van Galen, P.; Metzeler, K.H.; Poeppl, A.; Ling, V.; Beyene, J.; et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011, 17, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Prada-Arismendy, J.; Arroyave, J.C.; Röthlisberger, S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. 2017, 31, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, K.J.; Reikvam, H.; Bruserud, Ø. Identification of a subset of patients with acute myeloid leukemia characterized by long-term in vitro proliferation and altered cell cycle regulation of the leukemic cells. Expert Opin. Targets 2014, 18, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Brenner, A.K.; Tvedt, T.H.; Nepstad, I.; Rye, K.P.; Hagen, K.M.; Reikvam, H.; Bruserud, Ø. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin. Targets 2017, 21, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Ryningen, A.; Olsnes, A.M.; Stordrange, L.; Øyan, A.M.; Kalland, K.H.; Gjertsen, B.T. Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica 2007, 92, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Casas, S.; Ollila, J.; Aventin, A.; Vihinen, M.; Sierra, J.; Knuutila, S. Changes in apoptosis-related pathways in acute myelocytic leukemia. Cancer Genet. Cytogenet 2003, 146, 89–101. [Google Scholar] [CrossRef]
- Kazi, J.U.; Ronnstrand, L. FLT3 signals via the adapter protein Grb10 and overexpression of Grb10 leads to aberrant cell proliferation in acute myeloid leukemia. Mol. Oncol. 2013, 7, 402–418. [Google Scholar] [CrossRef]
- Bodner, S.M.; Naeve, C.W.; Rakestraw, K.M.; Jones, B.G.; Valentine, V.A.; Valentine, M.B.; Luthardt, F.W.; Willman, C.L.; Raimondi, S.C.; Downing, J.R.; et al. Cloning and chromosomal localization of the gene encoding human cyclin D-binding Myb-like protein (hDMP1). Gene 1999, 229, 223–228. [Google Scholar] [CrossRef]
- Caligiuri, M.A.; Briesewitz, R.; Yu, J.; Wang, L.; Wei, M.; Arnoczky, K.J.; Marburger, T.B.; Wen, J.; Perrotti, D.; Bloomfield, C.D.; et al. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood 2007, 110, 1022–1024. [Google Scholar] [CrossRef]
- Cui, X.Y.; Skretting, G.; Jing, Y.; Sun, H.; Sandset, P.M.; Sun, L. Hypoxia influences stem cell-like properties in multidrug resistant K562 leukemic cells. Blood Cells Mol. Dis. 2013, 51, 177–184. [Google Scholar] [CrossRef]
- Han, Q.; Lu, J.; Wang, J.; Ye, J.; Jiang, X.; Chen, H.; Liu, C.; Chen, L.; Lin, T.; Chen, S.; et al. H2AFY is a novel fusion partner of MECOM in acute myeloid leukemia. Cancer Genet. 2018, 222–223, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shao, T.; Zhang, H.; Zhang, N.; Shi, X.; Liu, X.; Yao, Y.; Xu, L.; Zhu, S.; Cao, J.; et al. MiR-425 expression profiling in acute myeloid leukemia might guide the treatment choice between allogeneic transplantation and chemotherapy. J. Transl. Med. 2018, 16, 267. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.; Wang, Z.; Coyaud, E.; Voisin, V.; Gronda, M.; Jitkova, Y.; Mattson, R.; Hurren, R.; Babovic, S.; Maclean, N.; et al. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 2015, 27, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Larkin, K.; Byrd, J.C. Antagonizing ClpP: A new power play in targeted therapy for AML. Cancer Cell 2015, 27, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Davison, J.; Du, L.; Storer, B.; Stirewalt, D.L.; Heimfeld, S.; Estey, E.; Appelbaum, F.R.; Fang, M. Identification of differentially methylated markers among cytogenetic risk groups of acute myeloid leukemia. Epigenetics 2015, 10, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Du, Y.; Beckford, J.; Alachkar, H. Upregulation of the EMT marker vimentin is associated with poor clinical outcome in acute myeloid leukemia. J. Transl. Med. 2018, 16, 170. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Lu, F.; Li, P.; Ye, J.; Ji, M.; Ma, D.; Ji, C. SMG1 acts as a novel potential tumor suppressor with epigenetic inactivation in acute myeloid leukemia. Int. J. Mol. Sci. 2014, 15, 17065–17076. [Google Scholar] [CrossRef] [PubMed]
- Germano, G.; Morello, G.; Aveic, S.; Pinazza, M.; Minuzzo, S.; Frasson, C.; Persano, L.; Bonvini, P.; Viola, G.; Bresolin, S.; et al. ZNF521 sustains the differentiation block in MLL-rearranged acute myeloid leukemia. Oncotarget 2017, 8, 26129–26141. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Deng, L.; Song, Y.; Redell, M. DOT1L inhibition sensitizes MLL-rearranged AML to chemotherapy. PLoS ONE 2014, 9, e98270. [Google Scholar] [CrossRef]
- Shafer, D.; Grant, S. Update on rational targeted therapy in AML. Blood Rev. 2016, 30, 275–283. [Google Scholar] [CrossRef]
- Haimovici, A.; Brigger, D.; Torbett, B.E.; Fey, M.F.; Tschan, M.P. Induction of the autophagy-associated gene MAP1S via PU.1 supports APL differentiation. Leuk Res. 2014, 38, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Noura, M.; Tokushige, C.; Maeda, S.; Kiyose, H.; Kashiwazaki, G.; Taniguchi, J.; Bando, T.; Yoshida, K.; Ozaki, T.; et al. Autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid leukemia cells. Sci. Rep. 2017, 7, 16604. [Google Scholar] [CrossRef]
- Niapour, M.; Farr, C.; Minden, M.; Berger, S.A. Elevated calpain activity in acute myelogenous leukemia correlates with decreased calpastatin expression. Blood Cancer J. 2012, 2, e51. [Google Scholar] [CrossRef] [PubMed]
- Olsson, L.; Zettermark, S.; Biloglav, A.; Castor, A.; Behrendtz, M.; Forestier, E.; Paulsson, K.; Johansson, B. The genetic landscape of paediatric de novo acute myeloid leukaemia as defined by single nucleotide polymorphism array and exon sequencing of 100 candidate genes. Br. J. Haematol. 2016, 174, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, Y.; Wu, H. A novel scoring system for acute myeloid leukemia risk assessment based on the expression levels of six genes. Int. J. Mol. Med. 2018, 42, 1495–1507. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.S.Y.; Zhou, J.; Chooi, J.Y.; Chan, Z.L.; Toh, S.H.M.; Tan, T.Z.; Wee, S.; Gunaratne, J.; Zeng, Q.; Chng, W.J. Non-canonical activation of beta-catenin by PRL-3 phosphatase in acute myeloid leukemia. Oncogene 2018. [Google Scholar] [CrossRef] [PubMed]
- Van Rhenen, A.; Feller, N.; Kelder, A.; Westra, A.H.; Rombouts, E.; Zweegman, S.; van der Pol, M.A.; Waisfisz, Q.; Ossenkoppele, G.J.; Schuurhuis, G.J. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin. Cancer Res. 2005, 11, 6520–6527. [Google Scholar] [CrossRef] [PubMed]
- Blair, A.; Hogge, D.E.; Sutherland, H.J. Most acute myeloid leukemia progenitor cells with long-term proliferative ability in vitro and in vivo have the phenotype CD34(+)/CD71(−)/HLA-DR. Blood 1998, 92, 4325–4335. [Google Scholar]
- Terpstra, W.; Prins, A.; Ploemacher, R.E.; Wognum, B.W.; Wagemaker, G.; Löwenberg, B.; Wielenga, J.J. Long-term leukemia-initiating capacity of a CD34-subpopulation of acute myeloid leukemia. Blood 1996, 87, 2187–2194. [Google Scholar]
- Schuurhuis, G.J.; Meel, M.H.; Wouters, F.; Min, L.A.; Terwijn, M.; de Jonge, N.A.; Kelder, A.; Snel, A.N.; Zweegman, S.; Ossenkoppele, G.J.; et al. Normal hematopoietic stem cells within the AML bone marrow have a distinct and higher ALDH activity level than co-existing leukemic stem cells. PLoS ONE 2013, 8, e78897. [Google Scholar] [CrossRef]
- Migliaccio, G.; Migliaccio, A.R.; Visser, J.W. Synergism between erythropoietin and interleukin-3 in the induction of hematopoietic stem cell proliferation and erythroid burst colony formation. Blood 1988, 72, 944–951. [Google Scholar] [PubMed]
- Öhler, L.; Geissler, K. Semi-solid colony growth in acute myeloid leukemia and its relation to cytogenetic risk groups. Leuk Lymphoma 2002, 43, 1743–1747. [Google Scholar] [CrossRef] [PubMed]
- Goodman, J.W.; Hall, E.A.; Miller, K.L.; Shinpock, S.G. Interleukin 3 promotes erythroid burst formation in “serum-free” cultures without detectable erythropoietin. Proc. Natl. Acad. Sci. USA 1985, 82, 3291–3295. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Frostad, S.; Foss, B. In vitro culture of acute myelogenous leukemia blasts: A comparison of four different culture media. J. Hematother. 1999, 8, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Gjertsen, B.T.; von Volkman, H.L. In vitro culture of human acute myelogenous leukemia (AML) cells in serum-free media: Studies of native AML blasts and AML cell lines. J. Hematother. Stem Cell Res. 2000, 9, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.; Delwel, R.; Touw, I.; Mahmoud, L.; Löwenberg, B. Human AML colony growth in serum-free culture. Leuk Res. 1988, 12, 157–165. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Hovland, R.; Wergeland, L.; Huang, T.S.; Gjertsen, B.T. Flt3-mediated signaling in human acute myelogenous leukemia (AML) blasts: A functional characterization of Flt3-ligand effects in AML cell populations with and without genetic Flt3 abnormalities. Haematologica 2003, 88, 416–428. [Google Scholar]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Gjertsen, B.T.; Øyan, A.M.; Marzolf, B.; Hovland, R.; Gausdal, G.; Døskeland, S.O.; Dimitrov, K.; Golden, A.; Kalland, K.H.; Hood, L.; Bruserud, Ø. Analysis of acute myelogenous leukemia: Preparation of samples for genomic and proteomic analyses. J. Hematother. Stem Cell Res. 2002, 11, 469–481. [Google Scholar] [CrossRef]
- Reuss-Borst, M.A.; Klein, G.; Waller, H.D.; Müller, C.A. Differential expression of adhesion molecules in acute leukemia. Leukemia 1995, 9, 869–874. [Google Scholar]
- Nepstad, I.; Hatfield, K.J.; Aasebø, E.; Hernandez-Valladares, M.; Brenner, A.K.; Bartaula-Brevik, S.; Berven, F.; Selheim, F.; Skavland, J.; Gjertsen, B.T.; et al. Two acute myeloid leukemia patient subsets are identified based on the constitutive PI3K-Akt-mTOR signaling of their leukemic cells; a functional, proteomic, and transcriptomic comparison. Expert Opin. Targets 2018, 22, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Stosić-Grujicić, S.; Basara, N.; Milenković, P.; Dinarello, C.A. Modulation of acute myeloblastic leukemia (AML) cell proliferation and blast colony formation by antisense oligomer for IL-1 beta converting enzyme (ICE) and IL-1 receptor antagonist (IL-1ra). J. Chemother. 1995, 7, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Sohn, S.K.; Kim, D.H.; Baek, J.H.; Lee, N.Y.; Suh, J.S.; Chae, S.C.; Lee, K.S.; Lee, K.B. Clinical implications of angiogenic factors in patients with acute or chronic leukemia: Hepatocyte growth factor levels have prognostic impact, especially in patients with acute myeloid leukemia. Leuk Lymphoma 2005, 46, 885–891. [Google Scholar]
- Guo, J.R.; Li, W.; Wu, Y.; Wu, L.Q.; Li, X.; Guo, Y.F.; Zheng, X.H.; Lian, X.L.; Huang, H.F.; Chen, Y.Z. Hepatocyte growth factor promotes proliferation, invasion, and metastasis of myeloid leukemia cells through PI3K-AKT and MAPK/ERK signaling pathway. Am. J. Transl. Res. 2016, 8, 3630–3644. [Google Scholar] [PubMed]
- Kentsis, A.; Reed, C.; Rice, K.L.; Sanda, T.; Rodig, S.J.; Tholouli, E.; Christie, A.; Valk, P.J.; Delwel, R.; Ngo, V.; et al. Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia. Nat. Med. 2012, 18, 1118–1122. [Google Scholar] [CrossRef]
- Reikvam, H.; Nepstad, I.; Bruserud, Ø.; Hatfield, K.J. Pharmacological targeting of the PI3K/mTOR pathway alters the release of angioregulatory mediators both from primary human acute myeloid leukemia cells and their neighboring stromal cells. Oncotarget 2013, 4, 830–843. [Google Scholar] [CrossRef]
- Aref, S.; El-Sherbiny, M.; Mabed, M.; Menessy, A.; El-Refaei, M. Urokinase plasminogen activator receptor and soluble matrix metalloproteinase-9 in acute myeloid leukemia patients: A possible relation to disease invasion. Hematology 2003, 8, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Xian, J.; Shao, H.; Chen, X.; Zhang, S.; Quan, J.; Zou, Q.; Jin, H.; Zhang, L. Nucleophosmin mutants promote adhesion, migration and invasion of human leukemia THP-1 cells through MMPs op-regulation via Ras/ERK MAPK signaling. Int. J. Biol. Sci. 2016, 12, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Stefanidakis, M.; Karjalainen, K.; Jaalouk, D.E.; Gahmberg, C.G.; O’Brien, S.; Pasqualini, R.; Arap, W.; Koivunen, E. Role of leukemia cell invadosome in extramedullary infiltration. Blood 2009, 114, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.; Edwards, D.K.t.; Eide, C.A.; Newell, L.; Traer, E.; Medeiros, B.C.; Pollyea, D.A.; Deininger, M.W.; Collins, R.H.; Tyner, J.W.; et al. Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep. 2017, 18, 3204–3218. [Google Scholar] [CrossRef]
- Bruserud, Ø. Effects of endogenous interleukin 1 on blast cells derived from acute myelogenous leukemia patients. Leuk Res. 1996, 20, 65–73. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Ryningen, A.; Wergeland, L.; Glenjen, N.I.; Gjertsen, B.T. Osteoblasts increase proliferation and release of pro-angiogenic interleukin 8 by native human acute myelogenous leukemia blasts. Haematologica 2004, 89, 391–402. [Google Scholar] [PubMed]
- Hatfield, K.; Ryningen, A.; Corbascio, M.; Bruserud, Ø. Microvascular endothelial cells increase proliferation and inhibit apoptosis of native human acute myelogenous leukemia blasts. Int. J. Cancer 2006, 119, 2313–2321. [Google Scholar] [CrossRef] [PubMed]
- Ryningen, A.; Wergeland, L.; Glenjen, N.; Gjertsen, B.T.; Bruserud, Ø. In vitro crosstalk between fibroblasts and native human acute myelogenous leukemia (AML) blasts via local cytokine networks results in increased proliferation and decreased apoptosis of AML cells as well as increased levels of proangiogenic Interleukin 8. Leuk Res. 2005, 29, 185–196. [Google Scholar] [CrossRef]
- Santos, S.C.; Dias, S. Internal and external autocrine VEGF/KDR loops regulate survival of subsets of acute leukemia through distinct signaling pathways. Blood 2004, 103, 3883–3889. [Google Scholar] [CrossRef] [PubMed]
- Langley, G.R.; Smith, L.J.; McCulloch, E.A. Adherent cells in cultures of blast progenitors in acute myeloblastic leukemia. Leuk Res. 1986, 10, 953–959. [Google Scholar] [CrossRef]
- Levine, J.H.; Simonds, E.F.; Bendall, S.C.; Davis, K.L.; Amir el, A.D.; Tadmor, M.D.; Litvin, O.; Fienberg, H.G.; Jager, A.; Zunder, E.R.; et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015, 162, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, P.; Yan, A.; Guo, Z.; Ban, Y.; Li, J.; Chen, S.; Yang, H.; He, Y.; Li, J.; et al. ASXL1 interacts with the cohesin complex to maintain chromatid separation and gene expression for normal hematopoiesis. Sci. Adv. 2017, 3, e1601602. [Google Scholar] [CrossRef]
- Bullinger, L.; Döhner, K.; Döhner, H. Genomics of acute myeloid leukemia diagnosis and pathways. J. Clin. Oncol. 2017, 35, 934–946. [Google Scholar] [CrossRef]
- Thota, S.; Viny, A.D.; Makishima, H.; Spitzer, B.; Radivoyevitch, T.; Przychodzen, B.; Sekeres, M.A.; Levine, R.L.; Maciejewski, J.P. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood 2014, 124, 1790–1798. [Google Scholar] [CrossRef]
- Rau, R.E.; Rodriguez, B.A.; Luo, M.; Jeong, M.; Rosen, A.; Rogers, J.H.; Campbell, C.T.; Daigle, S.R.; Deng, L.; Song, Y.; et al. DOT1L as a therapeutic target for the treatment of DNMT3A-mutant acute myeloid leukemia. Blood 2016, 128, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Reikvam, H.; Tamburini, J.; Skrede, S.; Holdhus, R.; Poulain, L.; Ersvær, E.; Hatfield, K.J.; Bruserud, Ø. Antileukaemic effect of PI3K-mTOR inhibitors in acute myeloid leukaemia-gene expression profiles reveal CDC25B expression as determinate of pharmacological effect. Br. J. Haematol. 2014, 164, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Dysvik, B.; Jonassen, I. J-Express: Exploring gene expression data using Java. Bioinformatics 2001, 17, 369–370. [Google Scholar] [CrossRef] [PubMed]
- Aasebø, E.; Mjaavatten, O.; Vaudel, M.; Farag, Y.; Selheim, F.; Berven, F.; Bruserud, Ø.; Hernandez-Valladares, M. Freezing effects on the acute myeloid leukemia cell proteome and phosphoproteome revealed using optimal quantitative workflows. J. Proteom. 2016, 145, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Valladares, M.; Aasebø, E.; Mjaavatten, O.; Vaudel, M.; Bruserud, Ø.; Berven, F.; Selheim, F. Reliable FASP-based procedures for optimal quantitative proteomic and phosphoproteomic analysis on samples from acute myeloid leukemia patients. Biol. Proced. Online 2016, 18, 13. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Aasebø, E.; Selheim, F.; Berven, F.S.; Bruserud, Ø. Selecting sample preparation workflows for mass spectrometry-based proteomic and phosphoproteomic analysis of patient samples with acute myeloid leukemia. Proteomes 2016, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Aasebø, E.; Vaudel, M.; Mjaavatten, O.; Gausdal, G.; Van der Burgh, A.; Gjertsen, B.T.; Døskeland, S.O.; Bruserud, Ø.; Berven, F.S.; Selheim, F. Performance of super-SILAC based quantitative proteomics for comparison of different acute myeloid leukemia (AML) cell lines. Proteomics 2014, 14, 1971–1976. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Arntzen, M.Ø.; Koehler, C.J.; Barsnes, H.; Berven, F.S.; Treumann, A.; Thiede, B. IsobariQ: Software for isobaric quantitative proteomics using IPTL, iTRAQ, and TMT. J. Proteome Res. 2011, 10, 913–920. [Google Scholar] [CrossRef]



| Patient Characteristics, Cell Morphology | Disease Etiology and Cell Morphology | Cell Genetics | |||
|---|---|---|---|---|---|
| Age | De novo AML | 45 | Cytogenetics | ||
| Median (yrs) | 64 | Favorable | 7 | ||
| Range (yrs) | 18–92 | Secondary AML | Intermediate | 5 | |
| MDS | 10 | Normal | 35 | ||
| Gender | CMML | 2 | Adverse | 15 | |
| Females | 33 | CML | 1 | n.d. | 6 |
| Males | 35 | CLL | 1 | ||
| MF | 3 | Flt3 mutations | |||
| FAB classification | PV | 1 | ITD 2 | 16 | |
| M0 | 4 | Chemotherapy | 2 | Wild-type 2 | 39 |
| M1 | 17 | n.d. | 13 | ||
| M2 | 13 | AML relapse1 | 7 | ||
| M4 | 15 | NPM1 mutations | |||
| M5 | 14 | CD34 receptor | Mutated | 20 | |
| n.d. | 5 | Negative (≤20%) | 21 | Wild-type | 33 |
| Positive (>20%) | 41 | n.d. | 15 | ||
| n.d. | 6 | ||||
| Flt3 plus NPM1 mutation | 9 | ||||
| Number of Colonies | |||
|---|---|---|---|
| Not Observed | 1–200 | >200 | |
| Colony number | 0 | 19 (1.5–180) | 1367 (285–35,050) |
| Viable cell number (106) | 0.26 (0–3.06) | 0.18 (0.01–2.25) | 2.46 (0.30–53.5) |
| Variable | Adj. HR | 95% CI | p-Value |
|---|---|---|---|
| Age (≥60 years) 1 | 5.67 | 1.49–21.56 | 0.011 |
| Colony number (≥200) | 5.82 | 1.72–19.66 | 0.005 |
| Flt3-ITD | 1.83 | 0.48–6.92 | 0.377 |
| NPM1-insertion | 2.33 | 0.65–8.39 | 0.194 |
| Etiology (de novo) | 0.44 | 0.05–3.89 | 0.458 |
| Favorable cytogenetics | 0.78 | 0.11–5.61 | 0.802 |
| Unfavorable/intermediate cytogenetics | 0.58 | 0.11–3.00 | 0.513 |
| Classification Based on Protein Function | Gene Expression in AML Population with Colony Formation | |
|---|---|---|
| Upregulated | Downregulated | |
| Cell cycle, mitosis | KIAA1383/MTR120; LMLN; PARD6A; GRB10 | |
| Transcription | (RPS4Y1) | |
| Apoptosis | C3orf38? | |
| Chaperone | HSPA12A | |
| Signaling, transport | APOL1; GRB10; PPADDC3/PLPP7 | P2XR4; TMLHE |
| Metabolism | NUDT19; TMLHE | |
| Unknown | LOC642082; LOC648605 | |
| Biological Process | Proteomic Analysis | Phosphoproteomic Analysis | ||
|---|---|---|---|---|
| Increased ≥200 Colonies | Decreased ≥200 Colonies | Increased ≥200 Colonies | Decreased ≥200 Colonies | |
| mRNA splicing | RBMXL1; TXNL4A; ZNF830 | PCBP1 2 | PAPOLA; PRPF31; PRPF40A | |
| Transcription | MED1; POLR3C; ZNF830 | SMG1 2 | DOT1L 2; TCEAL3; ZNF521 2 | CBFB 2; ISL2; LEO1 2; MYSM1 |
| Transcription repressor | SPEN; ZNF521 2 | AEBP2 2; GATAD2B | ||
| Translation | ABCF1; EIF2B2; FARSB; MRPL3 | DNAJC2; EIF5B | ||
| Kinase | CAMK2D; DGKZ; SMG1 2 | NEK1; PRKD2 | ||
| Chaperone | DNAJC2; DNAJC5 | |||
| Metabolism | CPD 1; HEXB; MPDU1 | USP4 | CAST 2; CPD 1; DOT1L 2 | CTPS1; MYO9B; PPP6R3; RASGRP2 |
| Transporter, ion channel | CLIC4; CPD 1; CRIP1; RALGAPA2 | ABCF1; CLCN3; RBP4 | CPD | ATP13A1; OSBP; SCAP |
| Signaling | RALGAPA2; VIM 1,2 | CAP1; MRC2 | VIM 1,2; ZFYVE26 | LAT 2; LSP12; mTOR 2; RNF31; TMPO |
| Cytoskeleton | VIM 1,2 | AAMP; CAP1; MYOF | SP100; VIM 1,2; ZFYVE26 | DBNL; MYO9B; SH3BP1; TMPO |
| Apoptosis | MED1; RMDN3; TMEM173 | GRAMD4; MAP1S 2 | ||
| Cell cycle/mitosis | KIF4A | NEK1 | ||
| Proliferation regulation | FAM53C; SCRIB | |||
| Others | C19orf19/TRIR; MOB4 | BOD1L1; CAD; IRF2BP2; TTC7A | ||
| Unknown | GPALPP1 | MAP7D1 1 | MAP7D1 1; TOR4A | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brenner, A.K.; Aasebø, E.; Hernandez-Valladares, M.; Selheim, F.; Berven, F.; Grønningsæter, I.-S.; Bartaula-Brevik, S.; Bruserud, Ø. The Capacity of Long-Term In Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome. Cancers 2019, 11, 73. https://doi.org/10.3390/cancers11010073
Brenner AK, Aasebø E, Hernandez-Valladares M, Selheim F, Berven F, Grønningsæter I-S, Bartaula-Brevik S, Bruserud Ø. The Capacity of Long-Term In Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome. Cancers. 2019; 11(1):73. https://doi.org/10.3390/cancers11010073
Chicago/Turabian StyleBrenner, Annette K., Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik, and Øystein Bruserud. 2019. "The Capacity of Long-Term In Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome" Cancers 11, no. 1: 73. https://doi.org/10.3390/cancers11010073
APA StyleBrenner, A. K., Aasebø, E., Hernandez-Valladares, M., Selheim, F., Berven, F., Grønningsæter, I.-S., Bartaula-Brevik, S., & Bruserud, Ø. (2019). The Capacity of Long-Term In Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome. Cancers, 11(1), 73. https://doi.org/10.3390/cancers11010073






