Non-Coding RNAs in Glioma
Abstract
1. Introduction
2. Expression of Non-Coding RNAs in Cell Proliferation, Migration, Invasion and Apoptosis
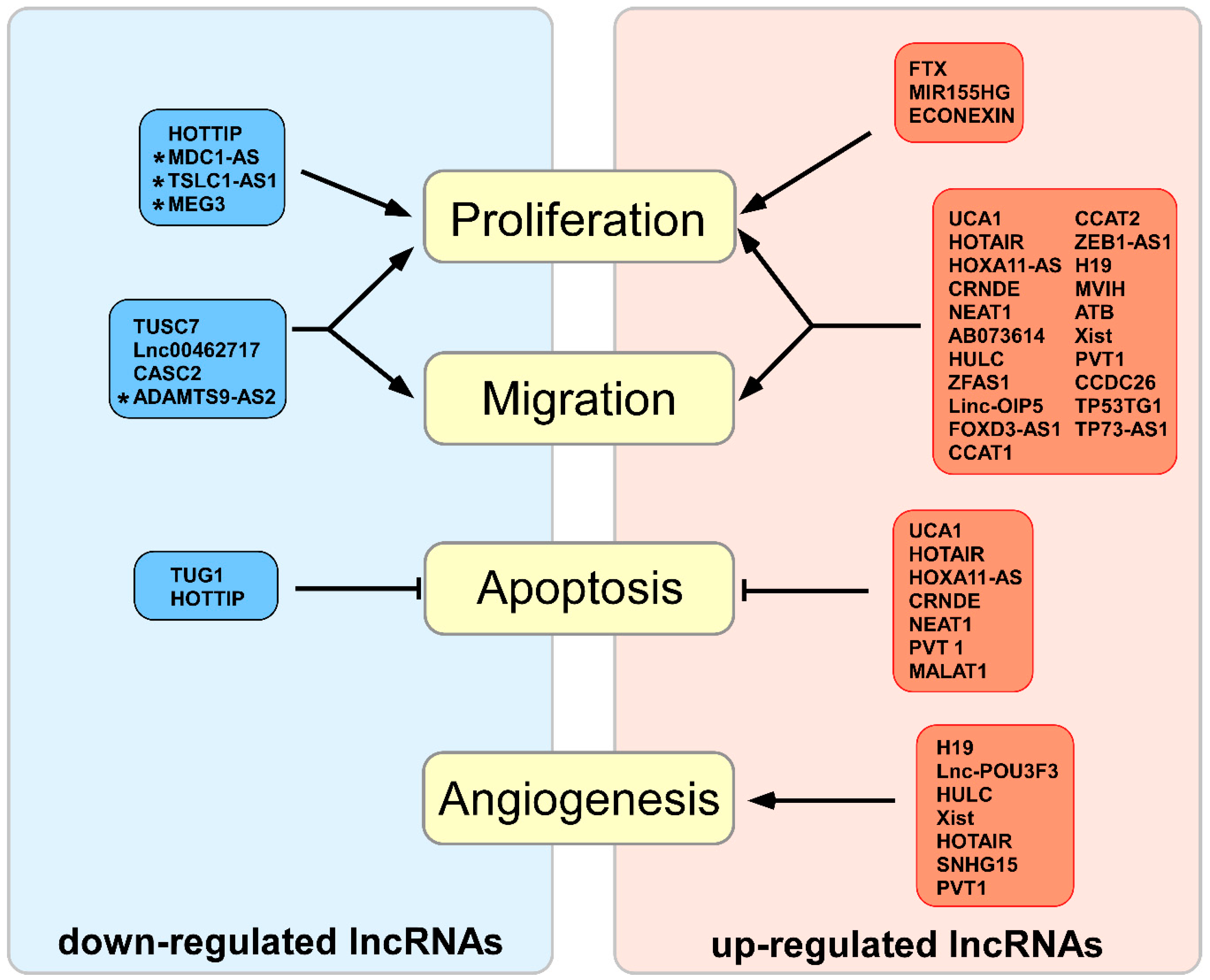
2.1. Up-Regulated lncRNAs in Biological Glioma Processes
2.1.1. AB073614 lncRNA
2.1.2. ATB, H19, ZEB1-AS lncRNAs
2.1.3. CCAT1, CCAT2, CCND2-AS1 lncRNAs
2.1.4. CRNDE lncRNA
2.1.5. CCDC26, FER1L4, miR210HG, MVIH, SPRY4-IT1, TP53TG1, lncRNAs
2.1.6. ECONEXIN, TP73-AS1, Xist lncRNAs
2.1.7. FOXD3-AS1, Linc-OIP5, ZFAS1lncRNAs
2.1.8. FTX lncRNA
2.1.9. HOTAIR, HOXA11-AS, UCA1 lncRNAs
2.1.10. HULC lncRNA
2.1.11. MIR155HG lncRNA
2.1.12. NEAT1 lncRNA
2.1.13. PVT I lncRNA
2.2. Down-Regulated lncRNAs in Biological Glioma Processes
2.2.1. ADAMTS9-AS2, MDC1-AS, MEG3, TSLC1-AS1 lncRNAs
2.2.2. CASC2 lncRNA
2.2.3. HOTTIP lncRNA
2.2.4. Lnc00462717 lncRNA
2.2.5. MALAT1 lncRNA
2.2.6. TUG1 lncRNA
2.2.7. TUSC7 lncRNA
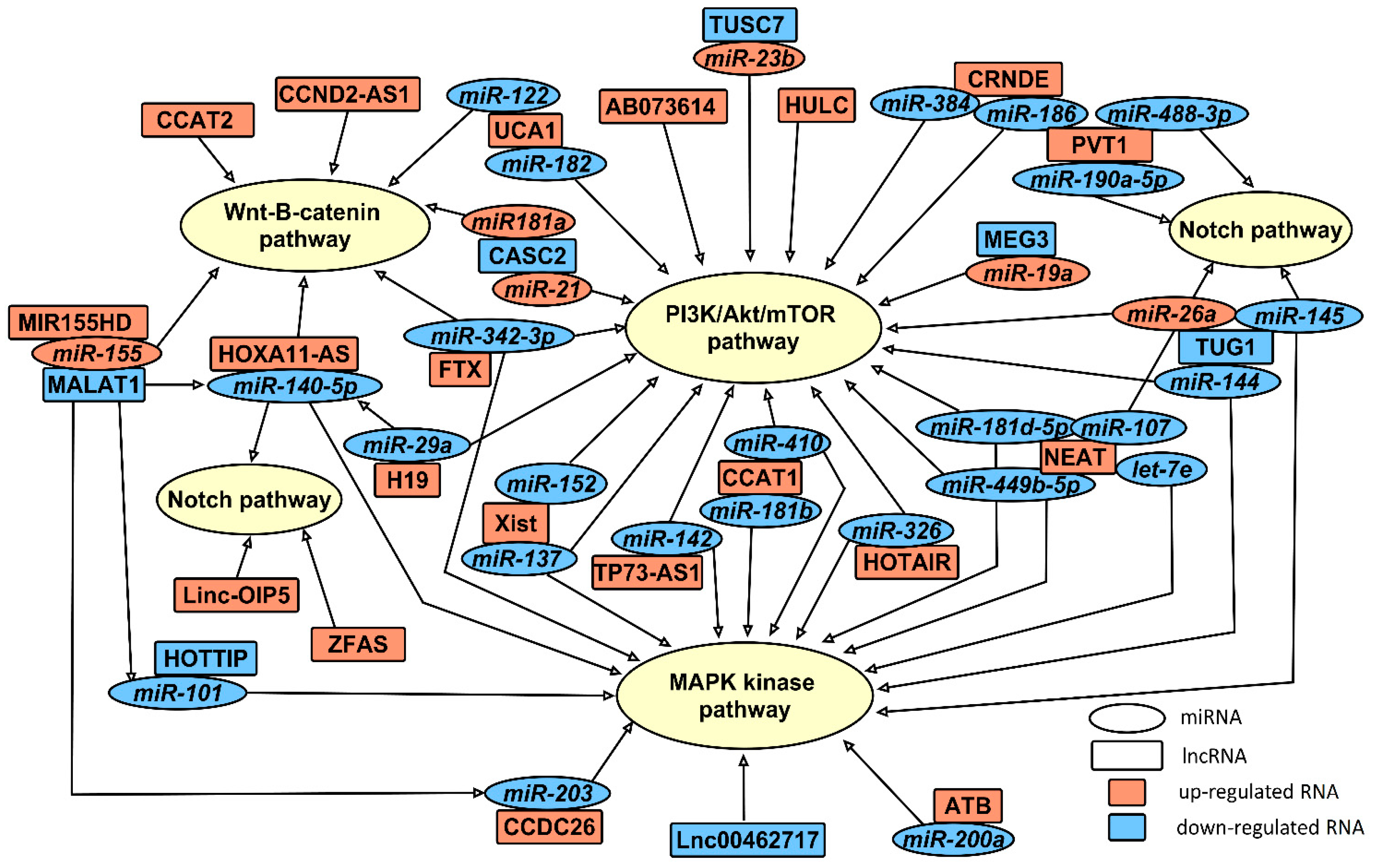
2.3. miRNAs Involved in Gliomagenesis in Association with lncRNAs
2.3.1. let-7e
2.3.2. miR-19a
2.3.3. miR-21
2.3.4. miR-23b
2.3.5. miR-26a and miR-145
2.3.6. miR-29
2.3.7. miR-101
2.3.8. miRNA-107
2.3.9. miR-122
2.3.10. miR-137
2.3.11. miR-140
2.3.12. miR-142
2.3.13. miR-144
2.3.14. miR-152
2.3.15. miR-155
2.3.16. miR-181
2.3.17. miR-182
2.3.18. miR-186, miR-190a-5p, miR-488-3p
2.3.19. miR-200a
2.3.20. miR-203
2.3.21. miR-326
2.3.22. miR-342-3p
2.3.23. miR-384
2.3.24. miR-429
2.3.25. miR-449b-5p
2.4. The Potential of Non-Coding RNAs for the Prediction of Resistance to Chemotherapy
2.4.1. H19 lncRNA
2.4.2. Xist lncRNA, miR-29c
2.4.3. miR-181a, miR-193a-5p
2.4.4. MALAT1 lncRNA, miR-203
2.5. Non-Coding RNAs in Angiogenesis
2.5.1. H19 lncRNA, miR-29a, miR-675-5p
2.5.2. HULC, POU3F3 lncRNAs
2.5.3. Xist lncRNA, miR-137, miR-429
2.5.4. HOTAIR lncRNA
2.5.5. PVT1, SNHG15 lncRNAs, miR-153, miR-186
2.6. Non-Coding RNAs and Regulation of Blood-Tumor-Barrier
3. Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alfonso, J.C.L.; Talkenberger, K.; Seifert, M.; Klink, B.; Hawkins-Daarud, A.; Swanson, K.R.; Hatzikirou, H.; Deutsch, A. The biology and mathematical modelling of glioma invasion: A review. J. R. Soc. Interface 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, T.; Gao, L.; Zheng, J.; Shao, A.; Zhang, J. Efficacy and safety of long-term therapy for high-grade glioma with temozolomide: A meta-analysis. Oncotarget 2017, 8, 51758–51765. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Qin, Z.; Chen, Z.; Xie, L.; Wang, R.; Zhao, H. Tumor Microenvironment in Treatment of Glioma. Open Med. 2017, 12, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Crocetti, E.; Trama, A.; Stiller, C.; Caldarella, A.; Soffietti, R.; Jaal, J.; Weber, D.C.; Ricardi, U.; Slowinski, J.; Brandes, A. Epidemiology of glial and non-glial brain tumours in Europe. Eur. J. Cancer 2012, 48, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Buckner, J.C.; Shaw, E.G.; Pugh, S.L.; Chakravarti, A.; Gilbert, M.R.; Barger, G.R.; Coons, S.; Ricci, P.; Bullard, D.; Brown, P.D.; et al. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N. Engl. J. Med. 2016, 374, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- van den Bent, M.J.; Baumert, B.; Erridge, S.C.; Vogelbaum, M.A.; Nowak, A.K.; Sanson, M.; Brandes, A.A.; Clement, P.M.; Baurain, J.F.; Mason, W.P.; et al. Interim results from the CATNON trial (EORTC study 26053-22054) of treatment with concurrent and adjuvant temozolomide for 1p/19q non-co-deleted anaplastic glioma: A phase 3, randomised, open-label intergroup study. Lancet 2017, 390, 1645–1653. [Google Scholar] [CrossRef]
- Jaeckle, K.; Vogelbaum, M.; Ballman, K.; Anderson, S.K.; Giannini, C.; Aldape, K.; Cerhan, J.; Wefel, J.S.; Nordstrom, D.; Jenkins, R.; et al. CODEL (Alliance-N0577; EORTC-26081/22086; NRG-1071; NCIC-CEC-2): Phase III Randomized Study of RT vs. RT+TMZ vs. TMZ for Newly Diagnosed 1p/19q-Codeleted Anaplastic Oligodendroglial Tumors. Analysis of Patients Treated on the Original Protocol Design (PL02.005). Neurology 2016, 86, PL02-005. [Google Scholar]
- De Sanctis, V.; Mazzarella, G.; Osti, M.F.; Valeriani, M.; Alfo, M.; Salvati, M.; Banelli, E.; Tombolini, V.; Enrici, R.M. Radiotherapy and sequential temozolomide compared with radiotherapy with concomitant and sequential temozolomide in the treatment of newly diagnosed glioblastoma multiforme. Anti-Cancer Drugs 2006, 17, 969–975. [Google Scholar] [CrossRef]
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017, 318, 2306–2316. [Google Scholar] [CrossRef]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Stahlhut, C.; Slack, F.J. MicroRNAs and the cancer phenotype: Profiling, signatures and clinical implications. Genome Med. 2013, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Bolha, L.; Ravnik-Glavac, M.; Glavac, D. Long Noncoding RNAs as Biomarkers in Cancer. Dis. Mark. 2017, 2017, 7243968. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Fejes-Toth, K.; Sotirova, V.; Sachidanandam, R.; Assaf, G.; Hannon, G.J.; Kapranov, P.; Foissac, S.; Willingham, A.T.; Duttagupta, R.; Dumais, E.; et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 2009, 457, 1028–1032. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Yu, Y.; Nangia-Makker, P.; Farhana, L.; Majumdar, A.P.N. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol. Cancer 2017, 16, 155. [Google Scholar] [CrossRef]
- Bayoumi, A.S.; Sayed, A.; Broskova, Z.; Teoh, J.P.; Wilson, J.; Su, H.; Tang, Y.L.; Kim, I.M. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int. J. Mol. Sci. 2016, 17, 356. [Google Scholar] [CrossRef] [PubMed]
- Homami, A.; Ghazi, F. MicroRNAs as biomarkers associated with bladder cancer. Med. J. Islam. Repub. Iran 2016, 30, 475. [Google Scholar] [PubMed]
- Misawa, A.; Takayama, K.I.; Inoue, S. Long non-coding RNAs and prostate cancer. Cancer Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liu, W.; Cui, J.; Li, J.; Li, C. Analysis of long non-coding RNA expression profiles in ovarian cancer. Oncol. Lett. 2017, 14, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zou, X.; He, J.; Mao, Y. Identification of long non-coding RNAs biomarkers associated with progression of endometrial carcinoma and patient outcomes. Oncotarget 2017, 8, 52604–52613. [Google Scholar] [CrossRef] [PubMed]
- Switlik, W.Z.; Szemraj, J. Circulating miRNAs as non-invasive biomarkers for non-small cell lung cancer diagnosis, prognosis and prediction of treatment response. Postepy Higieny i Medycyny Doswiadczalnej (Online) 2017, 71, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Waseem, M.; Ahmad, M.K.; Srivatava, V.K.; Rastogi, N.; Serajuddin, M.; Kumar, S.; Mishra, D.P.; Sankhwar, S.N.; Mahdi, A.A. Evaluation of miR-711 as Novel Biomarker in Prostate Cancer Progression. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shen, J.; Hodges, T.R.; Song, R.; Fuller, G.N.; Heimberger, A.B. Serum microRNA profiling in patients with glioblastoma: A survival analysis. Mol. Cancer 2017, 16, 59. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, X.; Hu, Y.; Shi, Z.; Zhou, Q.; Zheng, J.; Wang, Y. Long non-coding RNA expression profile in cervical cancer tissues. Oncol. Lett. 2017, 14, 1379–1386. [Google Scholar] [CrossRef]
- Inamura, K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs: Clinical Relevance in Lung Cancer. Cells 2017, 6, 12. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, J. LncRNAs: Macromolecules with big roles in neurobiology and neurological diseases. Metab. Brain Dis. 2017, 32, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.G.; Sanders, A.J.; Katoh, M.; Ungefroren, H.; Gieseler, F.; Prince, M.; Thompson, S.K.; Zollo, M.; Spano, D.; Dhawan, P.; et al. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin. Cancer Biol. 2015, 35, S244–S275. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.A.; Gutman, D.A.; Chisolm, C.; Appin, C.; Kong, J.; Rong, Y.; Kurc, T.; Van Meir, E.G.; Saltz, J.H.; Moreno, C.S.; et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am. J. Pathol. 2012, 180, 2108–2119. [Google Scholar] [CrossRef] [PubMed]
- Iwadate, Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol. Lett. 2016, 11, 1615–1620. [Google Scholar] [CrossRef] [PubMed]
- Pareja, F.; Macleod, D.; Shu, C.; Crary, J.F.; Canoll, P.D.; Ross, A.H.; Siegelin, M.D. PI3K and Bcl-2 inhibition primes glioblastoma cells to apoptosis through downregulation of Mcl-1 and Phospho-BAD. Mol. Cancer Res. MCR 2014, 12, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.A.; Yellen, P.; Xu, L.; Saqcena, M. Regulation of G1 Cell Cycle Progression: Distinguishing the Restriction Point from a Nutrient-Sensing Cell Growth Checkpoint(s). Genes Cancer 2010, 1, 1124–1131. [Google Scholar] [CrossRef]
- Hao, N.B.; He, Y.F.; Li, X.Q.; Wang, K.; Wang, R.L. The role of miRNA and lncRNA in gastric cancer. Oncotarget 2017, 8, 81572–81582. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.M.; Song, Y.L. Knockdown of long noncoding RNA AB073614 inhibits glioma cell proliferation and migration via affecting epithelial-mesenchymal transition. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3997–4002. [Google Scholar]
- Wang, Y.; Kuang, H.; Xue, J.; Liao, L.; Yin, F.; Zhou, X. LncRNA AB073614 regulates proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway. Biomed. Pharmacother. 2017, 93, 1230–1237. [Google Scholar] [CrossRef]
- Hu, L.; Lv, Q.L.; Chen, S.H.; Sun, B.; Qu, Q.; Cheng, L.; Guo, Y.; Zhou, H.H.; Fan, L. Up-Regulation of Long Non-Coding RNA AB073614 Predicts a Poor Prognosis in Patients with Glioma. Int. J. Environ. Res. Public Health 2016, 13, 433. [Google Scholar] [CrossRef]
- Yao, J.; Zhou, B.; Zhang, J.; Geng, P.; Liu, K.; Zhu, Y.; Zhu, W. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 7935–7944. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.C.; Xiong, Z.; Zhu, G.N.; Wang, C.; Zong, G.; Wang, H.L.; Bian, E.B.; Zhao, B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J. Exp. Clin. Cancer Res. CR 2016, 35, 90. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, Y.H.; Yao, Y.L.; Li, Z.; Li, Z.Q.; Ma, J.; Xue, Y.X. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell. Signal. 2015, 27, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Shen, L.; Zhao, H.; Liu, Q.; Fu, J.; Guo, Y.; Peng, R.; Cheng, L. LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J. Cell. Biochem. 2017, 118, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y.; Zhu, G.; Tian, B.; Zeng, W.; Yang, Y.; Li, Z. Long noncoding RNA CASC2 predicts the prognosis of glioma patients and functions as a suppressor for gliomas by suppressing Wnt/beta-catenin signaling pathway. Neuropsychiatr. Dis. Treat. 2017, 13, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Shen, F.; Du, J.; Fang, X.; Li, X.; Su, J.; Wang, X.; Huang, X.; Liu, Z. Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed. Pharmacother. 2018, 97, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Guo, X.Q.; Zhang, Q.S.; Zhang, J.L.; Duan, Y.L.; Li, G.F.; Zheng, D.L. Long non-coding RNA CCAT1 promotes glioma cell proliferation via inhibiting microRNA-410. Biochem. Biophys. Res. Commun. 2016, 480, 715–720. [Google Scholar] [CrossRef]
- Cui, B.; Li, B.; Liu, Q.; Cui, Y. lncRNA CCAT1 Promotes Glioma Tumorigenesis by Sponging miR-181b. J. Cell. Biochem. 2017, 118, 4548–4557. [Google Scholar] [CrossRef]
- Guo, H.; Hu, G.; Yang, Q.; Zhang, P.; Kuang, W.; Zhu, X.; Wu, L. Knockdown of long non-coding RNA CCAT2 suppressed proliferation and migration of glioma cells. Oncotarget 2016, 7, 81806–81814. [Google Scholar] [CrossRef]
- Wang, S.; Hui, Y.; Li, X.; Jia, Q. Silencing of lncRNA-CCDC26 Restrains the Growth and Migration of Glioma Cells In Vitro and In Vivo Via Targeting miR-203. Oncol. Res. 2017. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, D.L.; Wan, L.; Yan, S.F.; Sun, Y.H. Highly expressed lncRNA CCND2-AS1 promotes glioma cell proliferation through Wnt/beta-catenin signaling. Biochem. Biophys. Res. Commun. 2017, 482, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.Y.; Lu, Y.Y.; Yang, J.K.; Deng, W.Y.; Zhou, Q.; Jiao, B.H. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3992–3996. [Google Scholar] [PubMed]
- Zheng, J.; Liu, X.; Wang, P.; Xue, Y.; Ma, J.; Qu, C.; Liu, Y. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 1199–1215. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Li, J.; Zhang, Y.; Yin, H.; Han, B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015, 367, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kiang, K.M.; Zhang, X.Q.; Zhang, G.P.; Li, N.; Cheng, S.Y.; Poon, M.W.; Pu, J.K.; Lui, W.M.; Leung, G.K. CRNDE Expression Positively Correlates with EGFR Activation and Modulates Glioma Cell Growth. Targeted Oncol. 2017, 12, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, S.; Katsushima, K.; Hatanaka, A.; Shinjo, K.; Ohka, F.; Wakabayashi, T.; Zong, H.; Natsume, A.; Kondo, Y. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene 2017, 36, 4629–4640. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Tang, H.; Nie, D.; Xia, L. Long non-coding RNA Fer-1-like family member 4 is overexpressed in human glioblastoma and regulates the tumorigenicity of glioma cells. Oncol. Lett. 2017, 14, 2379–2384. [Google Scholar] [CrossRef]
- Chen, Z.H.; Hu, H.K.; Zhang, C.R.; Lu, C.Y.; Bao, Y.; Cai, Z.; Zou, Y.X.; Hu, G.H.; Jiang, L. Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am. J. Transl. Res. 2016, 8, 4106–4119. [Google Scholar]
- Zhang, W.; Bi, Y.; Li, J.; Peng, F.; Li, H.; Li, C.; Wang, L.; Ren, F.; Xie, C.; Wang, P.; et al. Long noncoding RNA FTX is upregulated in gliomas and promotes proliferation and invasion of glioma cells by negatively regulating miR-342-3p. Lab. Investig. J. Tech. Methods Pathol. 2017, 97, 447–457. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, R.; Liu, Q.; Liu, D.; Du, P.; Yuan, J.; Peng, G.; Liao, Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch. Biochem. Biophys. 2016, 610, 1–7. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, Y.R.; Zeng, F.; Cao, H.Y.; Zhou, H.D.; Wang, Y.J. LncRNA H19 is overexpressed in glioma tissue, is negatively associated with patient survival, and promotes tumor growth through its derivative miR-675. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4891–4897. [Google Scholar] [PubMed]
- Li, C.; Lei, B.; Huang, S.; Zheng, M.; Liu, Z.; Li, Z.; Deng, Y. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am. J. Transl. Res. 2015, 7, 1747–1764. [Google Scholar] [PubMed]
- Jiang, P.; Wang, P.; Sun, X.; Yuan, Z.; Zhan, R.; Ma, X.; Li, W. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. OncoTargets Ther. 2016, 9, 3501–3509. [Google Scholar] [CrossRef][Green Version]
- Jiang, X.; Yan, Y.; Hu, M.; Chen, X.; Wang, Y.; Dai, Y.; Wu, D.; Wang, Y.; Zhuang, Z.; Xia, H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016, 124, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Cai, H.; Liu, X.; Chen, J.; Ma, J.; Wang, P.; Liu, Y.; Zheng, J.; Xue, Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016, 381, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, Z.; Tian, N.; Han, L.; Fu, Y.; Guo, Z.; Tian, Y. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol. Lett. 2016, 12, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Sa, L.; Li, Y.; Zhao, L.; Liu, Y.; Wang, P.; Liu, L.; Li, Z.; Ma, J.; Cai, H.; Xue, Y. The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB. Front. Mol. Neurosci. 2017, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Pastori, C.; Kapranov, P.; Penas, C.; Peschansky, V.; Volmar, C.H.; Sarkaria, J.N.; Bregy, A.; Komotar, R.; St Laurent, G.; Ayad, N.G.; et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. USA 2015, 112, 8326–8331. [Google Scholar] [CrossRef]
- Ke, J.; Yao, Y.L.; Zheng, J.; Wang, P.; Liu, Y.H.; Ma, J.; Li, Z.; Liu, X.B.; Li, Z.Q.; Wang, Z.H.; et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 2015, 6, 21934–21949. [Google Scholar] [CrossRef]
- Yang, B.; Wei, Z.Y.; Wang, B.Q.; Yang, H.C.; Wang, J.Y.; Bu, X.Y. Down-regulation of the long noncoding RNA-HOX transcript antisense intergenic RNA inhibits the occurrence and progression of glioma. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef]
- Ma, X.; Li, Z.; Li, T.; Zhu, L.; Li, Z.; Tian, N. Long non-coding RNA HOTAIR enhances angiogenesis by induction of VEGFA expression in glioma cells and transmission to endothelial cells via glioma cell derived-extracellular vesicles. Am. J. Transl. Res. 2017, 9, 5012–5021. [Google Scholar] [PubMed]
- Xu, L.M.; Chen, L.; Li, F.; Zhang, R.; Li, Z.Y.; Chen, F.F.; Jiang, X.D. Over-expression of the long non-coding RNA HOTTIP inhibits glioma cell growth by BRE. J. Exp. Clin. Cancer Res. CR 2016, 35, 162. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; He, T.; Li, Z.; Liu, H.; Ding, B. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed. Pharmacother. 2017, 95, 1504–1513. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yi, L.; Zhao, J.Z.; Jiang, Y.G. Long Noncoding RNA HOXA11-AS Functions as miRNA Sponge to Promote the Glioma Tumorigenesis Through Targeting miR-140-5p. DNA Cell Biol. 2017, 36, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, C.; Cai, J.; Yang, F.; Liang, T.; Yan, X.; Wang, H.; Wang, W.; Chen, J.; Jiang, T. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget 2017, 8, 53110–53123. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Tian, R.; Zhang, M.; Wu, J.; Ding, M.; He, J. High expression of long noncoding RNA HULC is a poor predictor of prognosis and regulates cell proliferation in glioma. OncoTargets Ther. 2017, 10, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, X.; Qi, L.; Cai, Y.; Yang, P.; Xuan, G.; Jiang, Y. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget 2016, 7, 14429–14440. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.W.; Wu, L.; Kuang, W.; Chen, Y.; Zhu, X.G.; Guo, H.; Lang, H.L. Knockdown of linc-OIP5 inhibits proliferation and migration of glioma cells through down-regulation of YAP-NOTCH signaling pathway. Gene 2017, 610, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Meng, M.; Zhao, X.; Kong, L. Long non-coding RNA ENST00462717 suppresses the proliferation, survival, and migration by inhibiting MDM2/MAPK pathway in glioma. Biochem. Biophys. Res. Commun. 2017, 485, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, P.; Yao, Y.; Liu, Y.; Li, Z.; Liu, X.; Li, Z.; Zhao, X.; Xi, Z.; Teng, H.; et al. Knockdown of long non-coding RNA MALAT1 increases the blood-tumor barrier permeability by up-regulating miR-140. Biochim. Biophys. Acta 2016, 1859, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Luo, W.; Wang, J.; Peng, T.; Sun, G.; Shi, J.; Li, Z.; Zhang, B. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem. Biophys. Res. Commun. 2017, 492, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Guo, S.; Jiang, S.; Xu, Y.; Li, J.; Li, L.; Xiang, J. Silencing of Long Non-Coding RNA MALAT1 Promotes Apoptosis of Glioma Cells. J. Korean Med. Sci. 2016, 31, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7, e2123. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wang, Y.; Li, J.; Lv, M.; Niu, H.; Tian, Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J. Cancer Res. 2016, 6, 2561–2574. [Google Scholar] [PubMed]
- Chen, W.; Xu, X.K.; Li, J.L.; Kong, K.K.; Li, H.; Chen, C.; He, J.; Wang, F.; Li, P.; Ge, X.S.; et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget 2017, 8, 22783–22799. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Zhu, J.; Xie, S.; Li, F.; Xu, Q. MDC1-AS, an antisense long noncoding RNA, regulates cell proliferation of glioma. Biomed. Pharmacother. 2016, 81, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Tong, G.F.; Qin, N.; Sun, L.W.; Xu, X.L. Long Noncoding RNA MEG3 Suppresses Glioma Cell Proliferation, Migration, and Invasion By Acting As Competing Endogenous RNA of MiR-19a. Oncol. Res. 2017. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Yu, T.; Nie, E.; Hu, Q.; Wu, W.; Zhi, T.; Jiang, K.; Wang, X.; Lu, X.; et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro-Oncology 2017, 19, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Dai, D.; Wang, J.; Zhang, D.; Zhang, Y.; Han, G.; Zhang, L.; Chen, C.; Li, X.; Li, Y.; et al. Long Noncoding RNA miR210HG as a Potential Biomarker for the Diagnosis of Glioma. PLoS ONE 2016, 11, e0160451. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.J.; Yue, M.; Zheng, Y.H.; Li, J.P.; Dong, X.Y. Long non-coding RNA MVIH acts as a prognostic marker in glioma and its role in cell migration and invasion. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4898–4904. [Google Scholar] [PubMed]
- Guo, J.; Cai, H.; Zheng, J.; Liu, X.; Liu, Y.; Ma, J.; Que, Z.; Gong, W.; Gao, Y.; Tao, W.; et al. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim. Biophys. Acta 2017, 1863, 2240–2254. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Jiang, B.; Ma, J.; Li, Q. Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2016, 124, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xiao, Z.; Du, X.; Huang, L.; Du, G. Silencing of the long non-coding RNA NEAT1 suppresses glioma stem-like properties through modulation of the miR-107/CDK6 pathway. Oncol. Rep. 2017, 37, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Zhen, L.; Yun-Hui, L.; Hong-Yu, D.; Jun, M.; Yi-Long, Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y.; Xue, Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget 2016, 7, 62208–62223. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wu, L.; Yang, Q.; Ye, M.; Zhu, X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene 2015, 554, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Wu, L.X.; Yang, Y.; Li, S.; Mei, Y.; Liu, Y.B.; Zhang, L.; Cheng, Y.; Zhou, H.H. lncRNAs PVT1 and HAR1A are prognosis biomarkers and indicate therapy outcome for diffuse glioma patients. Oncotarget 2017, 8, 78767–78780. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Chen, J.; Liu, X.; Gong, W.; Zheng, J.; Guo, X.; Liu, Y.; Liu, L.; Ma, J.; Wang, P.; et al. PVT1 regulates the malignant behaviors of human glioma cells by targeting miR-190a-5p and miR-488-3p. Biochim. Biophys. Acta 2018, 1864, 1783–1794. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, P.; Xue, Y.; Qu, C.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317694326. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Wang, H.; Yang, X. Long non-coding RNA PVT1 indicates a poor prognosis of glioma and promotes cell proliferation and invasion via target EZH2. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, D.L.; Pang, Q. Long noncoding RNA SPRY4-IT1 is a prognostic factor for poor overall survival and has an oncogenic role in glioma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3035–3039. [Google Scholar] [PubMed]
- Chen, X.; Gao, Y.; Li, D.; Cao, Y.; Hao, B. LncRNA-TP53TG1 Participated in the Stress Response Under Glucose Deprivation in Glioma. J. Cell. Biochem. 2017, 118, 4897–4904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jin, H.; Lou, F. The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yao, J.; Geng, P.; Fu, X.; Xue, J.; Zhang, Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014, 7, 3065–3072. [Google Scholar] [PubMed]
- Cai, H.; Xue, Y.; Wang, P.; Wang, Z.; Li, Z.; Hu, Y.; Li, Z.; Shang, X.; Liu, Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget 2015, 6, 19759–19779. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, M.; An, G.; Ma, Q. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp. Biol. Med. 2016, 241, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; An, G.; Zhang, M.; Ma, Q. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem. Biophys. Res. Commun. 2016, 477, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Guo, Y.; Hong, Y.; Xue, Y.X. Long Non-coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front. Cell. Neurosci. 2016, 10, 235. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, J.G.; Mi, W.Y.; Wu, H.; Zhang, S.R.; Meng, Q.; Zhang, S.T. Long Non-coding RNA UCA1 Targets miR-122 to Promote Proliferation, Migration, and Invasion of Glioma Cells. Oncol. Res. 2017. [Google Scholar] [CrossRef]
- Zhao, W.; Sun, C.; Cui, Z. A long noncoding RNA UCA1 promotes proliferation and predicts poor prognosis in glioma. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2017, 19, 735–741. [Google Scholar] [CrossRef]
- He, Z.; Wang, Y.; Huang, G.; Wang, Q.; Zhao, D.; Chen, L. The lncRNA UCA1 interacts with miR-182 to modulate glioma proliferation and migration by targeting iASPP. Arch. Biochem. Biophys. 2017, 623–624, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yuan, J.; Li, L.; Yang, Y.; Xu, X.; Wang, Y. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am. J. Transl. Res. 2017, 9, 1845–1855. [Google Scholar] [PubMed]
- Yu, H.; Xue, Y.; Wang, P.; Liu, X.; Ma, J.; Zheng, J.; Li, Z.; Li, Z.; Cai, H.; Liu, Y. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis 2017, 6, e303. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ma, J.; Xue, Y.; Wang, P.; Li, Z.; Liu, J.; Chen, L.; Xi, Z.; Teng, H.; Wang, Z.; et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015, 359, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Li, Z.; Ma, K.; Li, X.; Tian, N.; Duan, J.; Xiao, X.; Wang, Y. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J. Cancer 2017, 8, 4106–4116. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhao, H.; Peng, R.; Liu, Q.; Yuan, J.; Peng, G.; Liao, Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.L.; Hu, L.; Chen, S.H.; Sun, B.; Fu, M.L.; Qin, C.Z.; Qu, Q.; Wang, G.H.; He, C.J.; Zhou, H.H. A Long Noncoding RNA ZEB1-AS1 Promotes Tumorigenesis and Predicts Poor Prognosis in Glioma. Int. J. Mol. Sci. 2016, 17, 1431. [Google Scholar] [CrossRef]
- Gao, K.; Ji, Z.; She, K.; Yang, Q.; Shao, L. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed. Pharmacother. 2017, 87, 555–560. [Google Scholar] [CrossRef]
- Pervan, C.L. Smad-independent TGF-beta2 signaling pathways in human trabecular meshwork cells. Exp. Eye Res. 2017, 158, 137–145. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; He, J.; Zhang, C.; Chen, J.; Shi, D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol. Res. 2018. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Feng, Y.; Li, R.; Sun, X.; Du, W.; Piao, X.; Wang, H.; Yang, D.; Sun, Y.; et al. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int. J. Biochem. Cell Biol. 2012, 44, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Grieco, L.; Calzone, L.; Bernard-Pierrot, I.; Radvanyi, F.; Kahn-Perles, B.; Thieffry, D. Integrative modelling of the influence of MAPK network on cancer cell fate decision. PLoS Comput. Biol. 2013, 9, e1003286. [Google Scholar] [CrossRef]
- Sohun, M.; Shen, H. The implication and potential applications of high-mobility group box 1 protein in breast cancer. Ann. Transl. Med. 2016, 4, 217. [Google Scholar] [CrossRef] [PubMed]
- Zang, W.; Wang, T.; Wang, Y.; Chen, X.; Du, Y.; Sun, Q.; Li, M.; Dong, Z.; Zhao, G. Knockdown of long non-coding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget 2016, 7, 19960–19974. [Google Scholar] [CrossRef] [PubMed]
- Bid, H.K.; Roberts, R.D.; Manchanda, P.K.; Houghton, P.J. RAC1: An emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol. Cancer Ther. 2013, 12, 1925–1934. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, X.; Zhou, X.; Han, L.; Chen, L.; Shi, Z.; Zhang, A.; Ye, M.; Wang, Q.; Liu, C.; et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015, 6, 537–546. [Google Scholar] [CrossRef]
- Roth, P.; Wischhusen, J.; Happold, C.; Chandran, P.A.; Hofer, S.; Eisele, G.; Weller, M.; Keller, A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J. Neurochem. 2011, 118, 449–457. [Google Scholar] [CrossRef]
- Sun, J.; Ji, J.; Huo, G.; Song, Q.; Zhang, X. miR-182 induces cervical cancer cell apoptosis through inhibiting the expression of DNMT3a. Int. J. Clin. Exp. Pathol. 2015, 8, 4755–4763. [Google Scholar]
- Jin, Y.; Wang, J.; Han, J.; Luo, D.; Sun, Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/beta-cadherin signaling pathway. Exp. Cell Res. 2017. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, Y.; Zheng, Y. MiR-122/Wnt/beta-catenin regulatory circuitry sustains glioma progression. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 8565–8572. [Google Scholar] [CrossRef]
- Zhang, F.; Wan, M.; Xu, Y.; Li, Z.; Leng, K.; Kang, P.; Cui, Y.; Jiang, X. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/beta-catenin-signaling pathway. Biomed. Pharmacother. 2017, 94, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Lungu, G.; Covaleda, L.; Mendes, O.; Martini-Stoica, H.; Stoica, G. FGF-1-induced matrix metalloproteinase-9 expression in breast cancer cells is mediated by increased activities of NF-kappaB and activating protein-1. Mol. Carcinog. 2008, 47, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, Y.; Wu, C.; Zhou, L.; Han, X.; Wang, Q.; Xie, X.; Zhou, Y.; Du, Z. MicroRNA-140-5p inhibits cell proliferation and invasion by regulating VEGFA/MMP2 signaling in glioma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317697558. [Google Scholar] [CrossRef]
- Balasubramaniyan, V.; Bhat, K.P. Targeting MIR155HG in glioma: A novel approach. Neuro-Oncology 2017, 19, 1152–1153. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Yue, J.; Pfeffer, S.R.; Handorf, C.R.; Pfeffer, L.M. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J. Biol. Chem. 2011, 286, 39172–39178. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Shapiro, A.; Kosik, K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008, 68, 8164–8172. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Tang, M.K.; Yao, Y.; Tang, C.; Chui, Y.L.; Lee, K.K. BRE plays an essential role in preventing replicative and DNA damage-induced premature senescence. Sci. Rep. 2016, 6, 23506. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, W.; Liu, G.; Xie, S.; Li, Q.; Li, Y.; Lin, Z. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed. Pharmacother. 2017, 95, 711–720. [Google Scholar] [CrossRef]
- Guo, P.; Nie, Q.; Lan, J.; Ge, J.; Qiu, Y.; Mao, Q. C-Myc negatively controls the tumor suppressor PTEN by upregulating miR-26a in glioblastoma multiforme cells. Biochem. Biophys. Res. Commun. 2013, 441, 186–190. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, J.; Wang, Z.; Wu, G.; Liu, F. TFAM is directly regulated by miR-23b in glioma. Oncol. Rep. 2013, 30, 2105–2110. [Google Scholar] [CrossRef]
- Sun, J.; Jia, Z.; Li, B.; Zhang, A.; Wang, G.; Pu, P.; Chen, Z.; Wang, Z.; Yang, W. MiR-19 regulates the proliferation and invasion of glioma by RUNX3 via beta-catenin/Tcf-4 signaling. Oncotarget 2017, 8, 110785–110796. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tao, T.; Liu, N.; Luan, W.; Qian, J.; Li, R.; Hu, Q.; Wei, Y.; Zhang, J.; You, Y. PPARalpha, a predictor of patient survival in glioma, inhibits cell growth through the E2F1/miR-19a feedback loop. Oncotarget 2016, 7, 84623–84633. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, G.; Luo, W.; Sun, X.; Lin, J.; Wang, M.; Zhang, Y.; Luo, W.; Zhang, Y. MicroRNA21 promotes migration and invasion of glioma cells via activation of Sox2 and betacatenin signaling. Mol. Med. Rep. 2017, 15, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yu, Q.; Chen, B.; Lu, X.; Li, Q. The prognostic value of a seven-microRNA classifier as a novel biomarker for the prediction and detection of recurrence in glioma patients. Oncotarget 2016, 7, 53392–53413. [Google Scholar] [CrossRef] [PubMed]
- Hermansen, S.K.; Nielsen, B.S.; Aaberg-Jessen, C.; Kristensen, B.W. miR-21 Is Linked to Glioma Angiogenesis: A Co-Localization Study. J. Histochem. Cytochem. 2016, 64, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.B.; Liu, J.H.; Hu, J.; Xue, K. MiR-21 enhanced glioma cells resistance to carmustine via decreasing Spry2 expression. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5065–5071. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.B.; Rathod, S.S.; Karthik, S.; Kaur, N.; Muzumdar, D.; Shiras, A.S. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro-Oncology 2013, 15, 1302–1316. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, J.; Xu, T.; Zhou, D.D.; Zhang, L.; Wang, X. MicroRNA-145 induces apoptosis of glioma cells by targeting BNIP3 and Notch signaling. Oncotarget 2017, 8, 61510–61527. [Google Scholar] [CrossRef]
- Morgado, A.L.; Rodrigues, C.M.; Sola, S. MicroRNA-145 Regulates Neural Stem Cell Differentiation Through the Sox2-Lin28/let-7 Signaling Pathway. Stem Cells 2016, 34, 1386–1395. [Google Scholar] [CrossRef]
- Xu, G.; Li, J.Y. Differential expression of PDGFRB and EGFR in microvascular proliferation in glioblastoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 10577–10586. [Google Scholar] [CrossRef]
- Kim, H.; Huang, W.; Jiang, X.; Pennicooke, B.; Park, P.J.; Johnson, M.D. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc. Natl. Acad. Sci. USA 2010, 107, 2183–2188. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, B.; Sagardoy, A.; Nogueira, L.; Guruceaga, E.; Grande, L.; Huse, J.T.; Aznar, M.A.; Diez-Valle, R.; Tejada-Solis, S.; Alonso, M.M.; et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS ONE 2013, 8, e77098. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Wang, P.; Xue, Y.; Shang, C.; Liu, X.; Ma, J.; Li, Z.; Li, Z.; Bao, M.; Liu, Y. Overexpression of miR-29a reduces the oncogenic properties of glioblastoma stem cells by downregulating Quaking gene isoform 6. Oncotarget 2017, 8, 24949–24963. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Yang, Z.; Qiu, X.; Lv, R.; Liu, J.; Wu, M.; Liao, Y.; Liu, Q. miR-29c contribute to glioma cells temozolomide sensitivity by targeting O6-methylguanine-DNA methyltransferases indirectely. Oncotarget 2016, 7, 50229–50238. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, X.; Lei, Q.; Yu, Z.; Xu, G.; Tang, H.; Wang, W.; Wang, Z.; Li, G.; Wu, M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget 2015, 6, 7930–7943. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zheng, C.; Bai, E.; Yang, K. miR-101 inhibits glioma cell invasion via the downregulation of COX-2. Oncol. Lett. 2016, 12, 2538–2544. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, L.; Wang, Z.; Cheng, Y.; Zhang, P.; Wang, X.; Wen, W.; Yang, H.; Liu, H.; Jin, W.; et al. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget 2017, 8, 19244–19254. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Mingyi, M.; Qiu, X.; Qiu, Y. MicroRNA-101 reverses temozolomide resistance by inhibition of GSK3beta in glioblastoma. Oncotarget 2016, 7, 79584–79595. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, X.R.; Zhang, R.; Li, P.; Liu, Y.; Yan, K.; Jiang, X.D. MicroRNA-107 inhibits glioma cell migration and invasion by modulating Notch2 expression. J. Neuro-Oncol. 2013, 112, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, X.R.; Chen, F.F.; Liu, Y.; Li, P.; Zhang, R.; Yan, K.; Yi, Y.J.; Xu, Z.M.; Jiang, X.D. MicroRNA-107 inhibits U87 glioma stem cells growth and invasion. Cell. Mol. Neurobiol. 2013, 33, 651–657. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, W.; Zhou, Q.; Zhao, T.; Song, Y.; Chai, L.; Li, Y. Low-expression of microRNA-107 inhibits cell apoptosis in glioma by upregulation of SALL4. Int. J. Biochem. Cell Biol. 2013, 45, 1962–1973. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Z.Y.; Xu, S.Y.; Zhang, X.J.; Zhang, Y.; Luo, K.; Li, W.P. Upregulation of miR-107 Inhibits Glioma Angiogenesis and VEGF Expression. Cell. Mol. Neurobiol. 2016, 36, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Wei, Y.; Wang, J.; Ao, Q.; Gong, K.; Zuo, H. Decreased expression of microRNA-107 predicts poorer prognosis in glioma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 4461–4466. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Cao, S.; Ma, J.; Liu, Y.; Liu, X.; Zheng, J.; Chen, J.; Liu, L.; Cai, H.; Li, Z.; et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer 2017, 16, 171. [Google Scholar] [CrossRef] [PubMed]
- Yerukala Sathipati, S.; Huang, H.L.; Ho, S.Y. Estimating survival time of patients with glioblastoma multiforme and characterization of the identified microRNA signatures. BMC Genom. 2016, 17, 1022. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, G.; Cao, Y.; Shi, L.; Sun, L.; Wang, Y.; Chen, C.; Wan, Z.; Fu, L.; You, Y. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother. Radiopharm. 2013, 28, 327–334. [Google Scholar] [CrossRef]
- Li, H.Y.; Li, Y.M.; Li, Y.; Shi, X.W.; Chen, H. Circulating microRNA-137 is a potential biomarker for human glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3599–3604. [Google Scholar]
- Sun, J.; Zheng, G.; Gu, Z.; Guo, Z. MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J. Neuro-Oncol. 2015, 122, 481–489. [Google Scholar] [CrossRef]
- Liu, X.; Wang, S.; Yuan, A.; Yuan, X.; Liu, B. MicroRNA-140 represses glioma growth and metastasis by directly targeting ADAM9. Oncol. Rep. 2016, 36, 2329–2338. [Google Scholar] [CrossRef]
- Li, G.; Yang, H.; Han, K.; Zhu, D.; Lun, P.; Zhao, Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142-3p target ITGB8 in glioma. Biochem. Biophys. Res. Commun. 2018, 498, 254–261. [Google Scholar] [CrossRef]
- Qin, W.; Rong, X.; Dong, J.; Yu, C.; Yang, J. miR-142 inhibits the migration and invasion of glioma by targeting Rac1. Oncol. Rep. 2017, 38, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Yu, H.; Hu, M.; Xia, T.; Yue, X. miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J. Neurochem. 2015, 135, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chopp, M.; Lu, Y.; Buller, B.; Jiang, F. MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett. 2013, 329, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, H.; Yang, B.; Luo, W.; Liu, Z.; Wang, J.; Zuo, Y. miR-152 regulated glioma cell proliferation and apoptosis via Runx2 mediated by DNMT1. Biomed. Pharmacother. 2017, 92, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Tian, X.; Zhang, J.; Huang, Y.; Lin, X.; Chen, L.; Zhang, S. Regulation of human glioma cell apoptosis and invasion by miR-152-3p through targeting DNMT1 and regulating NF2: MiR-152-3p regulate glioma cell apoptosis and invasion. J. Exp. Clin. Cancer Res. CR 2017, 36, 100. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Che, S.; Wang, J.; Jiao, Y.; Wang, C.; Meng, Q. miR-155 contributes to the progression of glioma by enhancing Wnt/beta-catenin pathway. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 5323–5331. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zou, R.; Zhou, R.; Gong, C.; Wang, Z.; Cai, T.; Tan, C.; Fang, J. miR-155 Regulates Glioma Cells Invasion and Chemosensitivity by p38 Isforms In Vitro. J. Cell. Biochem. 2015, 116, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Peng, Y.; Liu, M.; Jiang, Y. MicroRNA-181b Inhibits Cellular Proliferation and Invasion of Glioma Cells via Targeting Sal-Like Protein 4. Oncol. Res. 2017, 25, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lu, X.; Wang, Y.; Sun, L.; Qian, C.; Yan, W.; Liu, N.; You, Y.; Fu, Z. MiR-181b suppresses proliferation of and reduces chemoresistance to temozolomide in U87 glioma stem cells. J. Biomed. Res. 2010, 24, 436–443. [Google Scholar] [CrossRef]
- Shi, Z.M.; Wang, X.F.; Qian, X.; Tao, T.; Wang, L.; Chen, Q.D.; Wang, X.R.; Cao, L.; Wang, Y.Y.; Zhang, J.X.; et al. MiRNA-181b suppresses IGF-1R and functions as a tumor suppressor gene in gliomas. RNA 2013, 19, 552–560. [Google Scholar] [CrossRef]
- Sun, Y.C.; Wang, J.; Guo, C.C.; Sai, K.; Wang, J.; Chen, F.R.; Yang, Q.Y.; Chen, Y.S.; Wang, J.; To, T.S.; et al. MiR-181b sensitizes glioma cells to teniposide by targeting MDM2. BMC Cancer 2014, 14, 611. [Google Scholar] [CrossRef]
- Ho, K.H.; Chen, P.H.; Hsi, E.; Shih, C.M.; Chang, W.C.; Cheng, C.H.; Lin, C.W.; Chen, K.C. Identification of IGF-1-enhanced cytokine expressions targeted by miR-181d in glioblastomas via an integrative miRNA/mRNA regulatory network analysis. Sci. Rep. 2017, 7, 732. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Shi, Z.M.; Wang, X.R.; Cao, L.; Wang, Y.Y.; Zhang, J.X.; Yin, Y.; Luo, H.; Kang, C.S.; Liu, N.; et al. MiR-181d acts as a tumor suppressor in glioma by targeting K-ras and Bcl-2. J. Cancer Res. Clin. Oncol. 2012, 138, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, J.; Hoadley, K.; Kushwaha, D.; Ramakrishnan, V.; Li, S.; Kang, C.; You, Y.; Jiang, C.; Song, S.W.; et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro-Oncology 2012, 14, 712–719. [Google Scholar] [CrossRef]
- Ma, J.; Yao, Y.; Wang, P.; Liu, Y.; Zhao, L.; Li, Z.; Li, Z.; Xue, Y. MiR-181a regulates blood-tumor barrier permeability by targeting Kruppel-like factor 6. J. Cerebr. Blood Flow Metab. 2014, 34, 1826–1836. [Google Scholar] [CrossRef]
- Kouri, F.M.; Ritner, C.; Stegh, A.H. miRNA-182 and the regulation of the glioblastoma phenotype—Toward miRNA-based precision therapeutics. Cell Cycle 2015, 14, 3794–3800. [Google Scholar] [CrossRef]
- Feng, Y.A.; Liu, T.E.; Wu, Y. microRNA-182 inhibits the proliferation and migration of glioma cells through the induction of neuritin expression. Oncol. Lett. 2015, 10, 1197–1203. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, A.; Wu, Y.; Morris, S.A.; Lin, K.; Amin, S.; Verhaak, R.; Fuller, G.; Xie, K.; Heimberger, A.B.; et al. miR-182-5p Induced by STAT3 Activation Promotes Glioma Tumorigenesis. Cancer Res. 2016, 76, 4293–4304. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, L.; Song, Z.; Guo, C.; Zhu, J.; Li, Z.; Zhu, S. Potential Diagnostic and Prognostic Value of Plasma Circulating MicroRNA-182 in Human Glioma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 855–862. [Google Scholar] [CrossRef]
- Zheng, J.; Li, X.D.; Wang, P.; Liu, X.B.; Xue, Y.X.; Hu, Y.; Li, Z.; Li, Z.Q.; Wang, Z.H.; Liu, Y.H. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget 2015, 6, 25339–25355. [Google Scholar] [CrossRef]
- Berthois, Y.; Delfino, C.; Metellus, P.; Fina, F.; Nanni-Metellus, I.; Al Aswy, H.; Pirisi, V.; Ouafik, L.; Boudouresque, F. Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: Evidence that miR200a-3p is regulated by O(6)-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol. Ther. 2014, 15, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Bai, Y.; Qiu, S.; Zheng, L.; Huang, L.; Liu, T.; Wang, X.; Liu, Y.; Xu, N.; Yan, X.; et al. MiR-203 downregulation is responsible for chemoresistance in human glioblastoma by promoting epithelial-mesenchymal transition via SNAI2. Oncotarget 2015, 6, 8914–8928. [Google Scholar] [CrossRef]
- Pal, D.; Mukhopadhyay, D.; Ramaiah, M.J.; Sarma, P.; Bhadra, U.; Bhadra, M.P. Regulation of Cell Proliferation and Migration by miR-203 via GAS41/miR-10b Axis in Human Glioblastoma Cells. PLoS ONE 2016, 11, e0159092. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Wang, Y.; Sims, M.; Cai, C.; He, P.; Hacker, H.; Yue, J.; Cheng, J.; Boop, F.A.; Pfeffer, L.M. MicroRNA203a suppresses glioma tumorigenesis through an ATM-dependent interferon response pathway. Oncotarget 2017, 8, 112980–112991. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xu, T.; Yan, Y.; Qin, R.; Wang, H.; Zhang, X.; Huang, Y.; Wang, Y.; Lu, Y.; Fu, D.; et al. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1). PLoS ONE 2013, 8, e68469. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Liu, X.; Chen, L.; Dou, Z.; Lei, X.; Chang, L.; Cai, J.; Cui, Y.; Yang, D.; Sun, Y.; et al. Targeting the SMO oncogene by miR-326 inhibits glioma biological behaviors and stemness. Neuro-Oncology 2015, 17, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wei, Y.; Kang, Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 5615–5620. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, P.; Li, A.; Jiang, W.; Wang, H.; Wang, J.; Xie, K. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J. Exp. Clin. Cancer Res. CR 2012, 31, 97. [Google Scholar] [CrossRef]
- Chen, L.; Xue, Y.; Zheng, J.; Liu, X.; Liu, J.; Chen, J.; Li, Z.; Xi, Z.; Teng, H.; Wang, P.; et al. MiR-429 Regulated by Endothelial Monocyte Activating Polypeptide-II (EMAP-II) Influences Blood-Tumor Barrier Permeability by Inhibiting the Expressions of ZO-1, Occludin and Claudin-5. Front. Mol. Neurosci. 2018, 11, 35. [Google Scholar] [CrossRef]
- Dong, H.; Hao, X.; Cui, B.; Guo, M. MiR-429 suppresses glioblastoma multiforme by targeting SOX2. Cell Biochem. Funct. 2017, 35, 260–268. [Google Scholar] [CrossRef]
- Hu, C.E.; Du, P.Z.; Zhang, H.D.; Huang, G.J. Long Noncoding RNA CRNDE Promotes Proliferation of Gastric Cancer Cells by Targeting miR-145. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 42, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sun, Y.; She, X.; Tu, C.; Cheng, X.; Wang, L.; Yu, Z.; Li, P.; Liu, Q.; Yang, H.; et al. CASC2c as an unfavorable prognosis factor interacts with miR-101 to mediate astrocytoma tumorigenesis. Cell Death Dis. 2017, 8, e2639. [Google Scholar] [CrossRef] [PubMed]
- Pratap, P.; Raza, S.T.; Abbas, S.; Mahdi, F. MicroRNA-associated carcinogenesis in lung carcinoma. J. Cancer Res. Ther. 2018, 14, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, H.; Liu, Y.; Zheng, S. Promotional effect of microRNA-194 on breast cancer cells via targeting F-box/WD repeat-containing protein 7. Oncol. Lett. 2018, 15, 4439–4444. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Tian, Y.; Chen, Y.; Zhang, G. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/beta-Catenin pathway. OncoTargets Ther. 2018, 11, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Chen, Z.; Wang, S.; Zhang, R.; Qiu, W.; Zhao, L.; Peng, C.; Xu, R.; Chen, W.; Wang, H.W.; et al. c-Myc-miR-29c-REV3L signalling pathway drives the acquisition of temozolomide resistance in glioblastoma. Brain J. Neurol. 2015, 138, 3654–3672. [Google Scholar] [CrossRef] [PubMed]
- Salvador, M.A.; Wicinski, J.; Cabaud, O.; Toiron, Y.; Finetti, P.; Josselin, E.; Lelievre, H.; Kraus-Berthier, L.; Depil, S.; Bertucci, F.; et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res. 2013, 19, 6520–6531. [Google Scholar] [CrossRef] [PubMed]
- Schouten, P.C.; Vollebergh, M.A.; Opdam, M.; Jonkers, M.; Loden, M.; Wesseling, J.; Hauptmann, M.; Linn, S.C. High XIST and Low 53BP1 Expression Predict Poor Outcome after High-Dose Alkylating Chemotherapy in Patients with a BRCA1-like Breast Cancer. Mol. Cancer Ther. 2016, 15, 190–198. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Jain, R.K.; di Tomaso, E.; Duda, D.G.; Loeffler, J.S.; Sorensen, A.G.; Batchelor, T.T. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 2007, 8, 610–622. [Google Scholar] [CrossRef]
- Knizhnik, A.V.; Roos, W.P.; Nikolova, T.; Quiros, S.; Tomaszowski, K.H.; Christmann, M.; Kaina, B. Survival and death strategies in glioma cells: Autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS ONE 2013, 8, e55665. [Google Scholar] [CrossRef]
- Shimizu, A.; Nakayama, H.; Wang, P.; Konig, C.; Akino, T.; Sandlund, J.; Coma, S.; Italiano, J.E., Jr.; Mammoto, A.; Bielenberg, D.R.; et al. Netrin-1 promotes glioblastoma cell invasiveness and angiogenesis by multiple pathways including activation of RhoA, cathepsin B, and cAMP-response element-binding protein. J. Biol. Chem. 2013, 288, 2210–2222. [Google Scholar] [CrossRef]
- Hardee, M.E.; Zagzag, D. Mechanisms of glioma-associated neovascularization. Am. J. Pathol. 2012, 181, 1126–1141. [Google Scholar] [CrossRef]
- Fischbach, C.; Kong, H.J.; Hsiong, S.X.; Evangelista, M.B.; Yuen, W.; Mooney, D.J. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc. Natl. Acad. Sci. USA 2009, 106, 399–404. [Google Scholar] [CrossRef]
- Huang, W.J.; Chen, W.W.; Zhang, X. Glioblastoma multiforme: Effect of hypoxia and hypoxia inducible factors on therapeutic approaches. Oncol. Lett. 2016, 12, 2283–2288. [Google Scholar] [CrossRef]
- Chang, Y.; Wu, Q.; Tian, T.; Li, L.; Guo, X.; Feng, Z.; Zhou, J.; Zhang, L.; Zhou, S.; Feng, G.; et al. The influence of SRPK1 on glioma apoptosis, metastasis, and angiogenesis through the PI3K/Akt signaling pathway under normoxia. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 6083–6093. [Google Scholar] [CrossRef]
- Koyanagi, T.; Saga, Y.; Takahashi, Y.; Suzuki, Y.; Suzuki, M.; Sato, Y. Downregulation of vasohibin-2, a novel angiogenesis regulator, suppresses tumor growth by inhibiting angiogenesis in endometrial cancer cells. Oncol. Lett. 2013, 5, 1058–1062. [Google Scholar] [CrossRef]
- Koyanagi, T.; Suzuki, Y.; Komori, K.; Saga, Y.; Matsubara, S.; Fujiwara, H.; Sato, Y. Targeting human vasohibin-2 by a neutralizing monoclonal antibody for anti-cancer treatment. Cancer Sci. 2017, 108, 512–519. [Google Scholar] [CrossRef]
- Xue, X.; Gao, W.; Sun, B.; Xu, Y.; Han, B.; Wang, F.; Zhang, Y.; Sun, J.; Wei, J.; Lu, Z.; et al. Vasohibin 2 is transcriptionally activated and promotes angiogenesis in hepatocellular carcinoma. Oncogene 2013, 32, 1724–1734. [Google Scholar] [CrossRef]
- Lo Dico, A.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p Acts on HIF-1alpha to Sustain Hypoxic Responses: A New Therapeutic Strategy for Glioma. Theranostics 2016, 6, 1105–1118. [Google Scholar] [CrossRef]
- Lang, H.L.; Hu, G.W.; Chen, Y.; Liu, Y.; Tu, W.; Lu, Y.M.; Wu, L.; Xu, G.H. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 959–972. [Google Scholar]
- Ma, Y.; Xue, Y.; Liu, X.; Qu, C.; Cai, H.; Wang, P.; Li, Z.; Li, Z.; Liu, Y. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol. Rep. 2017, 38, 3265–3277. [Google Scholar] [CrossRef]
- Bauer, H.; Traweger, A. Tight Junctions of the Blood-Brain Barrier—A Molecular Gatekeeper. CNS Neurol. Disord. Drug Targets 2016, 15, 1016–1029. [Google Scholar] [CrossRef]
- Luissint, A.C.; Artus, C.; Glacial, F.; Ganeshamoorthy, K.; Couraud, P.O. Tight junctions at the blood brain barrier: Physiological architecture and disease-associated dysregulation. Fluids Barriers CNS 2012, 9, 23. [Google Scholar] [CrossRef]
- Shen, S.; Yu, H.; Liu, X.; Liu, Y.; Zheng, J.; Wang, P.; Gong, W.; Chen, J.; Zhao, L.; Xue, Y. PIWIL1/piRNA-DQ593109 Regulates the Permeability of the Blood-Tumor Barrier via the MEG3/miR-330-5p/RUNX3 Axis. Mol. Ther. Nucleic Acids 2018, 10, 412–425. [Google Scholar] [CrossRef]
- Malzkorn, B.; Reifenberger, G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr. Opin. Oncol. 2016, 28, 494–501. [Google Scholar] [CrossRef]
- Kros, J.M.; Mustafa, D.M.; Dekker, L.J.; Sillevis Smitt, P.A.; Luider, T.M.; Zheng, P.P. Circulating glioma biomarkers. Neuro-Oncology 2015, 17, 343–360. [Google Scholar] [CrossRef]
- Cheng, W.; Ren, X.; Zhang, C.; Han, S.; Wu, A. Expression and prognostic value of microRNAs in lower-grade glioma depends on IDH1/2 status. J. Neuro-Oncol. 2017, 132, 207–218. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Kiang, K.M.; Wang, Y.C.; Pu, J.K.; Ho, A.; Cheng, S.Y.; Lee, D.; Zhang, P.D.; Chen, J.J.; Lui, W.M.; et al. IDH1 mutation-associated long non-coding RNA expression profile changes in glioma. J. Neuro-Oncol. 2015, 125, 253–263. [Google Scholar] [CrossRef]
- Jha, P.; Agrawal, R.; Pathak, P.; Kumar, A.; Purkait, S.; Mallik, S.; Suri, V.; Chand Sharma, M.; Gupta, D.; Suri, A.; et al. Genome-wide small noncoding RNA profiling of pediatric high-grade gliomas reveals deregulation of several miRNAs, identifies downregulation of snoRNA cluster HBII-52 and delineates H3F3A and TP53 mutant-specific miRNAs and snoRNAs. Int. J. Cancer 2015, 137, 2343–2353. [Google Scholar] [CrossRef]
- Cao, T.; Rajasingh, S.; Samanta, S.; Dawn, B.; Bittel, D.C.; Rajasingh, J. Biology and clinical relevance of noncoding sno/scaRNAs. Trends Cardiovasc. Med. 2017. [Google Scholar] [CrossRef]
- Xu, B.; Ye, M.H.; Lv, S.G.; Wang, Q.X.; Wu, M.J.; Xiao, B.; Kang, C.S.; Zhu, X.G. SNORD47, a box C/D snoRNA, suppresses tumorigenesis in glioblastoma. Oncotarget 2017, 8, 43953–43966. [Google Scholar] [CrossRef]
- Chen, L.; Han, L.; Wei, J.; Zhang, K.; Shi, Z.; Duan, R.; Li, S.; Zhou, X.; Pu, P.; Zhang, J.; et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci. Rep. 2015, 5, 8588. [Google Scholar] [CrossRef]
- Zheng, R.; Yao, Q.; Ren, C.; Liu, Y.; Yang, H.; Xie, G.; Du, S.; Yang, K.; Yuan, Y. Upregulation of Long Noncoding RNA Small Nucleolar RNA Host Gene 18 Promotes Radioresistance of Glioma by Repressing Semaphorin 5A. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 877–887. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Q.; Zhou, P.; Deng, D.; Xue, L.; Shao, N.; Peng, Y.; Zhi, F. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed. Pharmacother. 2017, 91, 906–911. [Google Scholar] [CrossRef]
- Ronchetti, D.; Mosca, L.; Cutrona, G.; Tuana, G.; Gentile, M.; Fabris, S.; Agnelli, L.; Ciceri, G.; Matis, S.; Massucco, C.; et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med. Genom. 2013, 6, 27. [Google Scholar] [CrossRef]
- Ronchetti, D.; Todoerti, K.; Tuana, G.; Agnelli, L.; Mosca, L.; Lionetti, M.; Fabris, S.; Colapietro, P.; Miozzo, M.; Ferrarini, M.; et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012, 2, e96. [Google Scholar] [CrossRef]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017, 10, 2. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, X.; Xue, Y.; Gong, W.; Ma, J.; Xi, Z.; Que, Z.; Liu, Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J. Hematol. Oncol. 2017, 10, 52. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110. [Google Scholar] [CrossRef]
- Yang, P.; Qiu, Z.; Jiang, Y.; Dong, L.; Yang, W.; Gu, C.; Li, G.; Zhu, Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget 2016, 7, 63449–63455. [Google Scholar] [CrossRef]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, K.; Li, C.; Yao, Y.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS ONE 2012, 7, e30999. [Google Scholar] [CrossRef]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef]
- Li, C.; Zhou, X.; Chen, J.; Lu, Y.; Sun, Q.; Tao, D.; Hu, W.; Zheng, X.; Bian, S.; Liu, Y.; et al. PIWIL1 destabilizes microtubule by suppressing phosphorylation at Ser16 and RLIM-mediated degradation of Stathmin1. Oncotarget 2015, 6, 27794–27804. [Google Scholar] [CrossRef]
- Mani, S.R.; Megosh, H.; Lin, H. PIWI proteins are essential for early Drosophila embryogenesis. Dev. Biol. 2014, 385, 340–349. [Google Scholar] [CrossRef]
- Tan, H.; Liao, H.; Zhao, L.; Lu, Y.; Jiang, S.; Tao, D.; Liu, Y.; Ma, Y. HILI destabilizes microtubules by suppressing phosphorylation and Gigaxonin-mediated degradation of TBCB. Sci. Rep. 2017, 7, 46376. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Xuan, J.; Gupta, S.; Krylov, S.N.; Ma, X.; Yang, Q.; Yang, B.B. Reciprocal regulation of miRNAs and piRNAs in embryonic development. Cell Death Differ. 2016, 23, 1458–1470. [Google Scholar] [CrossRef]
- He, X.; Chen, X.; Zhang, X.; Duan, X.; Pan, T.; Hu, Q.; Zhang, Y.; Zhong, F.; Liu, J.; Zhang, H.; et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015, 43, 3712–3725. [Google Scholar] [CrossRef]
| lncRNA | Target miRNA | Genes and/or Pathways | Samples | Biological Processes | References |
|---|---|---|---|---|---|
| AB073614 ↑ | - | E-cadherin, Vimentin | 80 glioma | Proliferation, invasion, migration | [38] |
| - | PI3K/AKT | GG-13, HGG-15 | Proliferation, invasion, migration, apoptosis | [39] | |
| - | - | 65 glioma | Poor survival | [40] | |
| ADAMTS9-AS2 ↓ | - | ADAMTS9, DNMT1 | LGG-46, HGG-24 | Proliferation, migration, invasion, correlates with survival | [41] |
| ATB ↑ | miR-200a | TGF-β2 | 79 glioma | Proliferation, migration, invasion | [42] |
| CASC2 ↓ | miR-21 | - | LGG-12, HGG-12 | Proliferation, migration, invasion, apoptosis | [43] |
| miR-181a | PTEN pathway | LGG-30, HGG-27 | Proliferation, chemoresistance to TMZ | [44] | |
| - | β-catenin, cyclin D1 and c-Myc | LGG-26, HGG-21 | Proliferation | [45] | |
| miR-193a-5p | mTOR | LGG-15, HGG-17 | TMZ sescitivity | [46] | |
| CCAT1 ↑ | miR-181b | FGFR3, PDGFRA | LGG 45, HGG 41 | Proliferation | [47] |
| miR-410 | - | 28 glioma | Proliferation | [48] | |
| CCAT2 ↑ | - | Wnt/β-catenin signal pathway | LGG-58, HGG-76 | Proliferation, cell cycle progression, migration | [49] |
| CCDC26 ↑ | miR-203 | - | 40 glioma | Proliferation, migration, | [50] |
| CCND2-AS1 ↑ | - | Wnt/β-catenin signaling | 54 glioma | Proliferation | [51] |
| CRNDE ↑ | - | - | LGG-46, HGG-118 | Proliferation | [52] |
| miR-384 | PIWIL4 | LGG-15, HGG-15 | Proliferation, invasion, migration, apoptosis | [53] | |
| - | mTOR | 37 glioma | Proliferation, invasion, migration | [54] | |
| - | - | LGG-5, HGG-14 | Proliferation, apoptosis | [55] | |
| ECONEXIN ↑ | miR-411-5p | TOP2A | 40 glioma | Proliferation | [56] |
| FER1L4 ↑ | - | - | LGG-335, HGG-21 | Proliferation, invasion, apoptosis | [57] |
| FOXD3-AS1 ↑ | - | FOXD3 | LGG-13, HGG-31 | Proliferation, invasion, migration | [58] |
| FTX ↑ | miR-342-3p | AEG1 | LGG-76, HGG-81 from data base; LGG-11, HGG-11 | Proliferation, invasion | [59] |
| H19 ↑ | miR-140 | iASPP | 28 glioma | Proliferation | [60] |
| miR-675 | - | LGG-15, HGG-20 | Proliferation | [61] | |
| miR-675 | CDK6 | LGG-4, HGG-4 | Proliferation | [62] | |
| - | MDR, MRP, ABCG2 | TMZ-Sensitive-44, TMZ-Resistant-25 | TMZ-Resistance | [63] | |
| miR-29a | - | 30 glioma | Angiogenesis | [64] | |
| miR-29a | VASH2 | LGG-5, HGG-5 | Angiogenesis | [65] | |
| HOTAIR ↑ | miR-148b-3p | - | 26 astrocytoma (7 grade II, 19 grade III), 50 oligodendroglioma (38 grade II, 12 grade III) and 81 GBM | Cell growth, proliferation, invasion | [66] |
| miR-148b-3p | ZO-1, OCLN, CLDN5, USF1 | - | Angiogenesis | [67] | |
| - | BRD4 | 17 glioma | Proliferation, invasion, migration, apoptosis | [68] | |
| miR-326 | FGF1, PI3K/AKT and MEK1/2 signal pathways | LGG-6, HGG-6 | Proliferation, invasion, migration | [69] | |
| - | - | 67 glioma | Proliferation, invasion, migration | [70] | |
| - | VEGFA | - | Angiogenesis | [71] | |
| HOTTIP ↓ | - | BRE, cycA and CDK2, p53 | LGG-37, HGG-48 | Proliferation, apoptosis | [72] |
| HOXA11-AS ↑ | miR-214-3p | EZH2 | 45 glioma | Proliferation, migration, invasion | [73] |
| miR-140-5p | - | LGG-13, HGG-30 | Proliferation, invasion, migration, apoptosis | [74] | |
| HOXA11-AS3 ↑ | - | - | LGG-24, HGG-23 | Proliferation | [75] |
| HULC ↑ | - | - | LGG-10, HGG-60 | Proliferation, patients overall survival | [76] |
| - | Survivin, c-Myc, Cyclin A/D1/E, p-Rb, Skp-1/2, CDK2/4 and EZH2, Bcl-2/Bax, caspase-3/8 | LGG-30, HGG-90 | Proliferation invasion, migration, angiogenesis, adhesion | [77] | |
| Linc-OIP5 ↑ | - | Yap, Notch | LGG-69, HGG-98 | Proliferation, migration | [78] |
| lnc00462717 ↓ | - | MDM2 | LGG-16, HGG-64 | Proliferation, migration, apoptosis | [79] |
| MALAT1 ↑ | miR-140 | Nuclear factor YA | LGG-8, HGG-8 | Proliferation, chemosensitivity, BTB permeability | [80] |
| miR-101 | STMN1, RAB5A, ATG4D | 32 glioma | Proliferation, migration, autophagy | [81] | |
| - | CCND1, MYC | LGG-37, HGG-37 | Proliferation, apoptosis | [82] | |
| MALAT1 ↓ | - | Ki-67, MMP2, MAPK | LGG-32, HGG-100 | Proliferation | [83] |
| miR-155 | FBXW7 | LGG-21, HGG-45 | Proliferation, correlates with survival | [84] | |
| miR-203 | TS | 180 glioma | Chemoresistance | [85] | |
| MDC1-AS ↓ | - | MDC1, CycB1/CDK2 | 15 glioma | Proliferation | [86] |
| MEG3 ↓ | miR-19a | PTEN | 40 glioma | Proliferation, apoptosis | [87] |
| MIR155HG ↑ | miR-155-5p, miR-155-3p | PCDH7, PCDH9, Wnt/B-catenin | 225 glioma | Proliferation | [88] |
| miR210HG ↑ | - | - | 28 glioma | Proliferation | [89] |
| MVIH ↑ | - | - | LGG-57, HGG-70 | Proliferation, invasion, migration | [90] |
| NEAT1 ↑ | miR-181d-5p | SOX5 | - | BTB | [91] |
| - | - | LGG-23, HGG-71 | Proliferation, invasion, migration | [92] | |
| miR-107 | CDK6 | LGG-5, HGG-24 | Proliferation, invasion, migration | [93] | |
| miR-449b-5p | c-met | LGG-10, HGG-5 | Proliferation, invasion, migration, apoptosis | [94] | |
| let-7e | PI3K/AKT/mTOR, MEK/ERK pathways | LGG-60, HGG-60 | Proliferation | [95] | |
| POU3F3 ↑ | - | - | LGG-37, HGG-45 | Proliferation, angiogenesis | [96] |
| PVT1 ↑ | - | - | 98 glioma | Patients survival, chemotherapy, radiotherapy response | [97] |
| miR-488-3p | MEF2C | LGG-9, HGG-10 | Proliferation, migration, invasion | [98] | |
| miR-186 | Atg7, Beclin1 | - | Proliferation, migration, angiogenesis | [99] | |
| miR-190a-5p | EZH2, JAGGED1 | LGG-40, HGG-40 | Cell cycle, apoptosis | [100] | |
| SPRY4-IT1 ↑ | - | - | LGG-73, HGG-90 | Proliferation, invasion, migration, apoptosis | [101] |
| TP53TG1 ↑ | - | - | LGG-12, HGG-12 | Proliferation, invasion, migration, apoptosis | [102] |
| TP73-AS1 ↑ | miR-142 | HMGB1 | LGG-26, HGG-21 | Proliferation, invasion | [103] |
| TSLC1-AS1 ↓ | - | TSLC1 | LGG-37, HGG-28 | Proliferation, migration, invasion | [104] |
| TUG1 ↑ | miR-144 | HSF2, ZO-1, OCLN, CLDN5 | LGG-5, HGG-5 | Angiogenesis | [105] |
| TUG1 ↓ | - | BCL-2, CASP3, CASP9 | LGG-57, HGG-63 | Proliferation, apoptosis | [106] |
| miR-26a | PTEN | LGG-9, HGG-11 | Proliferation, apoptosis | [107] | |
| TUSC7 ↓ | miR-23b | TUSC7 | LGG-19, HGG-20 | Proliferation, apoptosis, correlates with survival | [108] |
| UCA1 ↑ | miR-122 | - | 63 glioma | Proliferation, migration, invasion, apoptosis | [109] |
| - | cycD1 | LGG-22, HGG-42 | Proliferation | [110] | |
| miR-182 | iASPP | LGG-27, HGG-49 | Proliferation, migration, invasion, apoptosis | [111] | |
| Xist ↑ | miR-137 | Rac1 | LGG-9, HGG-21 | Proliferation | [112] |
| miR-137 | XCR7, ZO-2, FOXC1 | - | Angiogenesis | [113] | |
| miR-152 | - | Astrocytoma | Proliferation, migration, invasion, apoptosis | [114] | |
| miR-429 | - | 157 glioma | Proliferation, migration, invasion, angiogenesis | [115] | |
| miR-29c | MGMT, SP1 | LGG-33, HGG-36 | Chemosensitivity | [116] | |
| ZEB1-AS1 ↑ | - | CycD1 and CDK2 ZEB1, MMP2, MMP9, CDH2, Integrin-β1 | LGG-37, HGG-45 | Proliferation, invasion, migration, apoptosis | [117] |
| ZFAS1 ↑ | - | Notch signaling pathway | 46 glioma | Proliferation, migration, invasion, disease prognosis | [118] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rynkeviciene, R.; Simiene, J.; Strainiene, E.; Stankevicius, V.; Usinskiene, J.; Miseikyte Kaubriene, E.; Meskinyte, I.; Cicenas, J.; Suziedelis, K. Non-Coding RNAs in Glioma. Cancers 2019, 11, 17. https://doi.org/10.3390/cancers11010017
Rynkeviciene R, Simiene J, Strainiene E, Stankevicius V, Usinskiene J, Miseikyte Kaubriene E, Meskinyte I, Cicenas J, Suziedelis K. Non-Coding RNAs in Glioma. Cancers. 2019; 11(1):17. https://doi.org/10.3390/cancers11010017
Chicago/Turabian StyleRynkeviciene, Ryte, Julija Simiene, Egle Strainiene, Vaidotas Stankevicius, Jurgita Usinskiene, Edita Miseikyte Kaubriene, Ingrida Meskinyte, Jonas Cicenas, and Kestutis Suziedelis. 2019. "Non-Coding RNAs in Glioma" Cancers 11, no. 1: 17. https://doi.org/10.3390/cancers11010017
APA StyleRynkeviciene, R., Simiene, J., Strainiene, E., Stankevicius, V., Usinskiene, J., Miseikyte Kaubriene, E., Meskinyte, I., Cicenas, J., & Suziedelis, K. (2019). Non-Coding RNAs in Glioma. Cancers, 11(1), 17. https://doi.org/10.3390/cancers11010017






