Clinical Impact of Epithelial-to-Mesenchymal Transition Regulating MicroRNAs in Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Results
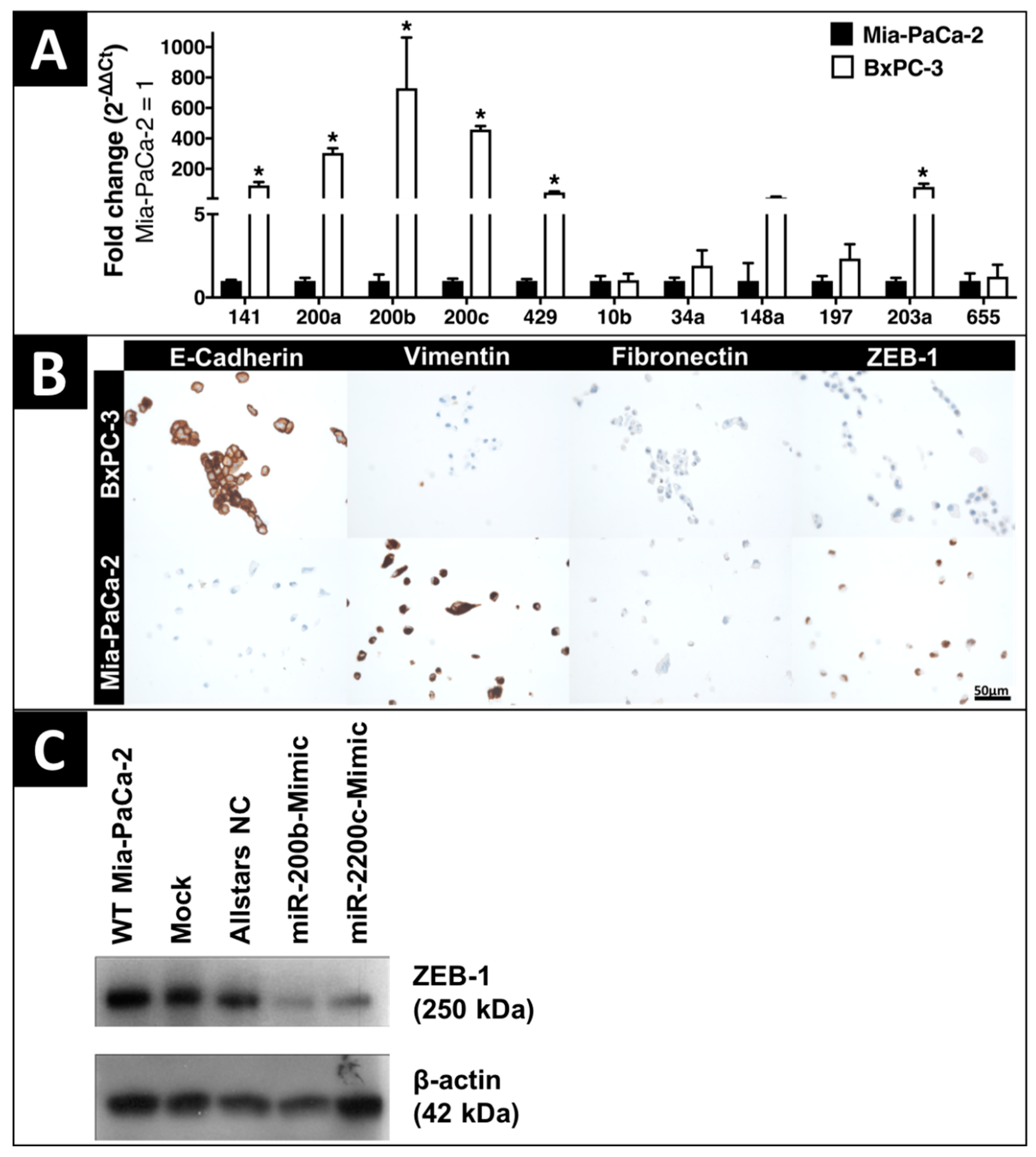
2.1. In-Vitro Expression of EMT-Regulating MicroRNAs and Proteins
2.2. Clinicopathologic Characteristics
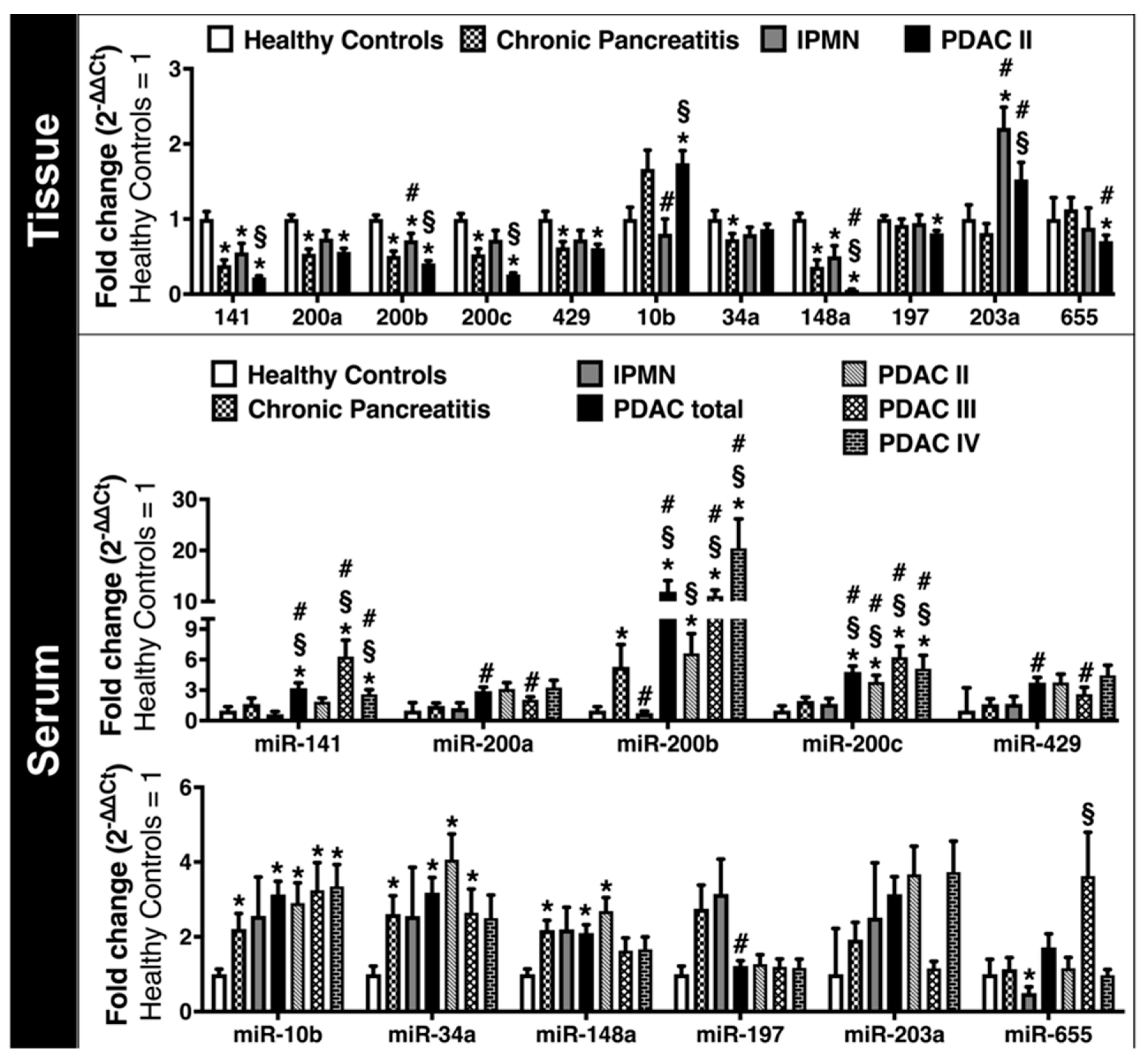
2.3. Expression of EMT-Regulating MicroRNAs in Clinical Solid and Liquid Biopsies
2.4. Expression of EMT-Marker Proteins in Human Pancreatic Tissue and Blood Serum Samples
2.5. Diagnostic Impact of EMT-Regulating MicroRNAs
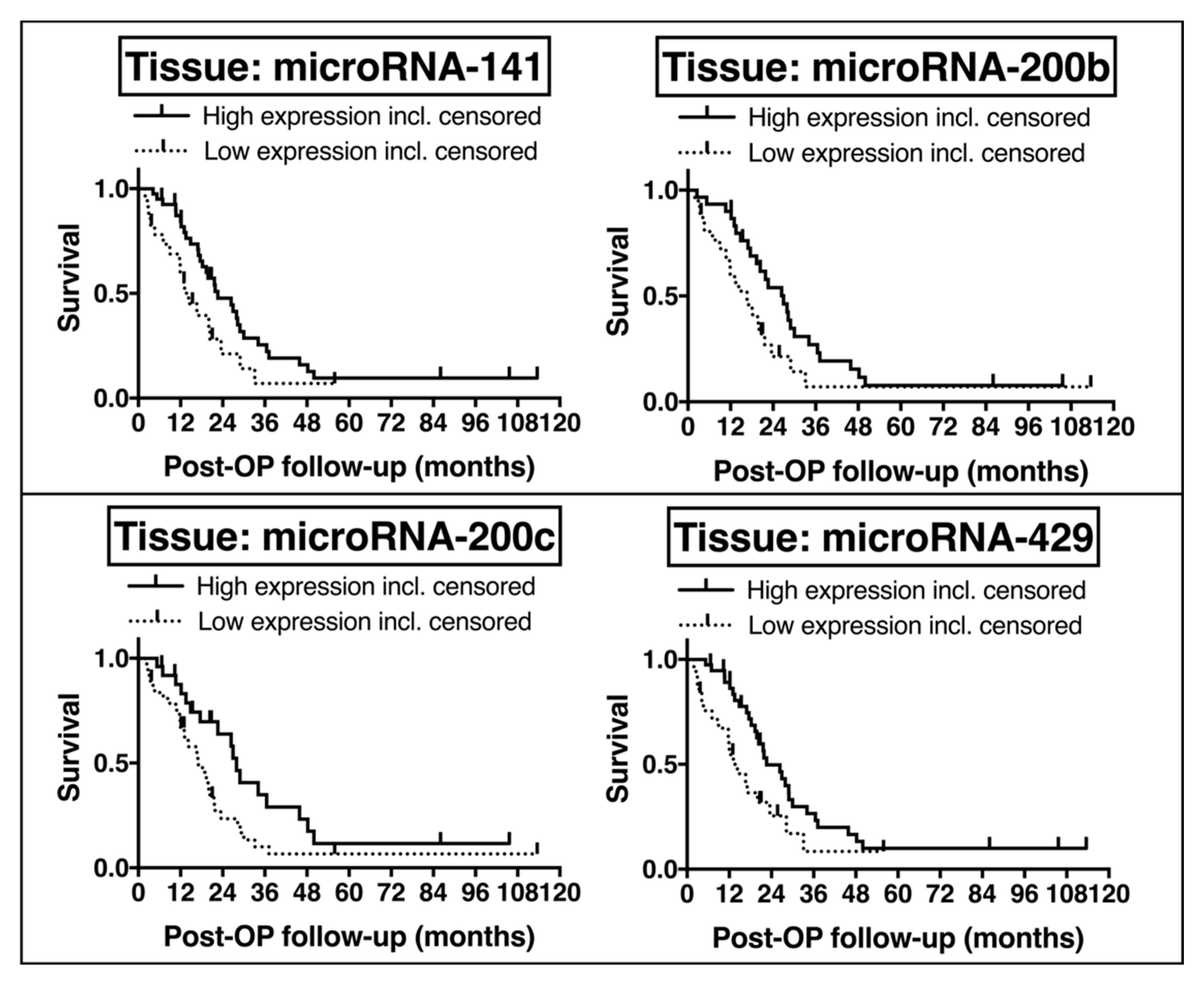
2.6. Potential Prognostic Impact of EMT-Regulating MicroRNAs
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Selection of MicroRNAs and Their Target Proteins
4.3. RNA Isolation and Quantification of EMT-Regulating MicroRNAs
4.4. Tissue Array Immunohistochemistry
4.5. Cell Lines and MicroRNA Transfection
4.6. Western Blotting
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Statistical Analysis
4.9. Limitations of This Study
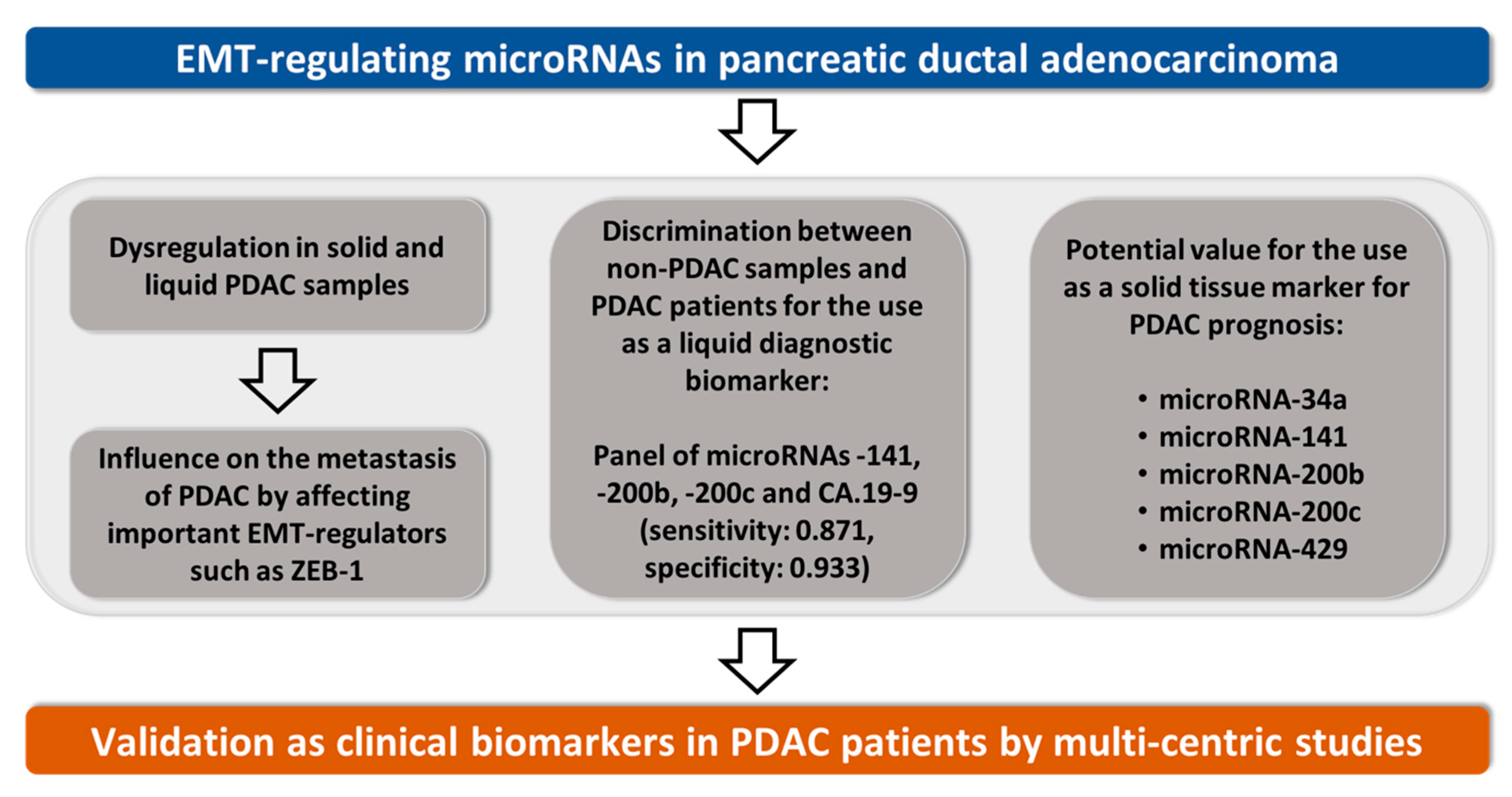
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.L.; Malvezzi, M.; Carioli, G.; Negri, E.; La Vecchia, C.; Boffetta, P.; Bosetti, C. Global Trends in Pancreatic Cancer Mortality From 1980 Through 2013 and Predictions for 2017. Clin. Gastroenterol. Hepatol. 2016, 14, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-H.; Liu, C.; Cheng, H.; Lu, Y.; Qin, Y.; Xu, Y.-F.; Xu, J.; Long, J.; Liu, L.; Ni, Q.-X.; et al. Epithelial-mesenchymal transition in pancreatic cancer: Is it a clinically significant factor? Biochim. Biophys. Acta 2015, 1855, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tian, X.-J.; Xing, J. Signal Transduction Pathways of EMT Induced by TGF-β, SHH, and WNT and Their Crosstalks. J. Clin. Med. 2016, 5, e41. [Google Scholar] [CrossRef] [PubMed]
- Träger, M.M.; Dhayat, S.A. Epigenetics of epithelial-to-mesenchymal transition in pancreatic carcinoma. Int. J. Cancer 2017, 141, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Wang, J.; Wen, Z.; Lai, M.; Zhang, H. Molecular mechanisms of microRNAs in regulating epithelial-mesenchymal transitions in human cancers. Cancer Lett. 2016, 371, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Bajdak, K.; Meidhof, S.; Burk, U.; Niedermann, G.; Firat, E.; Wellner, U.; Dimmler, A.; Faller, G.; Schubert, J.; et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011, 30, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Omura, N.; Hong, S.-M.; Vincent, A.; Walter, K.; Griffith, M.; Borges, M.; Goggins, M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010, 70, 5226–5237. [Google Scholar] [CrossRef] [PubMed]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Mongroo, P.S.; Rustgi, A.K. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010, 10, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Tang, Y.; Cheng, Y.-S. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the Notch signaling pathway. Sci. Rep. 2017, 7, e38232. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.-H.; Gibbons, D.L.; Chakravarti, D.; Creighton, C.J.; Rizvi, Z.H.; Adams, H.P.; Pertsemlidis, A.; Gregory, P.A.; Wright, J.A.; Goodall, G.J.; et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J. Clin. Investig. 2012, 122, 3170–3183. [Google Scholar] [CrossRef] [PubMed]
- Alemar, B.; Izetti, P.; Gregório, C.; Macedo, G.S.; Castro, M.A.A.; Osvaldt, A.B.; Matte, U.; Ashton-Prolla, P. miRNA-21 and miRNA-34a Are Potential Minimally Invasive Biomarkers for the Diagnosis of Pancreatic Ductal Adenocarcinoma. Pancreas 2016, 45, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, Y.; Su, J.; Liang, H.; Zhang, C.-Y.; Chen, X.; Yao, W. MicroRNA-148a Suppresses the Proliferation and Migration of Pancreatic Cancer Cells by Down-regulating ErbB3. Pancreas 2016, 45, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Banerjee, P.; Liu, X.; Yu, J.; Gibbons, D.L.; Wu, P.; Scott, K.L.; Diao, L.; Zheng, X.; Wang, J.; et al. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J. Clin. Investig. 2018, 128, 1267–1282. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Banerjee, P.; Guo, H.-F.; Ireland, S.; Pankova, D.; Ahn, Y.; Nikolaidis, I.M.; Liu, X.; Zhao, Y.; Xue, Y.; et al. Epithelial-to-mesenchymal transition drives a pro-metastatic Golgi compaction process through scaffolding protein PAQR11. J. Clin. Investig. 2017, 127, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Liu, Z.; Xiao, J.; Tu, Y.; Wan, Z.; Xiong, H.; Li, Y.; Xiao, W. MicroRNA-148a suppresses epithelial-mesenchymal transition and invasion of pancreatic cancer cells by targeting Wnt10b and inhibiting the Wnt/β-catenin signaling pathway. Oncol. Rep. 2017, 38, 301–308. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.A.; Fitzgerald, T.L.; Yang, L.V.; Lertpiriyapong, K.; Steelman, L.S.; Abrams, S.L.; Montalto, G.; Cervello, M.; Neri, L.M.; Cocco, L.; et al. Roles of GSK-3 and microRNAs on epithelial mesenchymal transition and cancer stem cells. Oncotarget 2016, 8, 14221–14250. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Jiang, X.; Chen, Z.; Song, X.; Wu, L.; Zong, D.; Song, D.; Yin, L.; Wang, D.; Chen, C.; et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, e138. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, L.; Tang, X.; Bai, W. miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncol. Lett. 2017, 14, 7687–7690. [Google Scholar] [CrossRef] [PubMed]
- Harazono, Y.; Muramatsu, T.; Endo, H.; Uzawa, N.; Kawano, T.; Harada, K.; Inazawa, J.; Kozaki, K. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS ONE 2013, 8, e62757. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Gore, J.; Deitz, S.; Korc, M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene 2014, 33, 4664–4674. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Satoh, K.; Miura, S.; Hirota, M.; Kanno, A.; Masamune, A.; Kikuta, K.; Kume, K.; Unno, J.; Egawa, S.; et al. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J. Cell. Physiol. 2013, 228, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Chitkara, D.; Mittal, A.; Mahato, R.I. miRNAs in pancreatic cancer: Therapeutic potential, delivery challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 34–52. [Google Scholar] [CrossRef] [PubMed]
- Ballehaninna, U.K.; Chamberlain, R.S. Biomarkers for pancreatic cancer: Promising new markers and options beyond CA 19-9. Tumor Biol. 2013, 34, 3279–3292. [Google Scholar] [CrossRef] [PubMed]
- Gayral, M.; Jo, S.; Hanoun, N.; Vignolle-Vidoni, A.; Lulka, H.; Delpu, Y.; Meulle, A.; Dufresne, M.; Humeau, M.; Chalret du Rieu, M.; et al. MicroRNAs as emerging biomarkers and therapeutic targets for pancreatic cancer. World J. Gastroenterol. 2014, 20, 11199–11209. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Sethi, S.; Bluth, M.H. Clinical Implication of MicroRNAs in Molecular Pathology: An Update for 2018. Clin. Lab. Med. 2018, 38, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Dhayat, S.A.; Abdeen, B.; Köhler, G.; Senninger, N.; Haier, J.; Mardin, W.A. MicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage II. Clin. Epigenet. 2015, 7, e132. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Dubaybo, H.; Brand, R.E.; Sarkar, F.H. Differential Expression of MicroRNAs in Tissues and Plasma Co-exists as a Biomarker for Pancreatic Cancer. J. Cancer Sci. Ther. 2015, 7, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Calatayud, D.; Dehlendorff, C.; Boisen, M.K.; Hasselby, J.P.; Schultz, N.A.; Werner, J.; Immervoll, H.; Molven, A.; Hansen, C.P.; Johansen, J.S. Tissue MicroRNA profiles as diagnostic and prognostic biomarkers in patients with resectable pancreatic ductal adenocarcinoma and periampullary cancers. Biomark. Res. 2017, 5, e8. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Kundranda, M. Novel Diagnostic and Predictive Biomarkers in Pancreatic Adenocarcinoma. Int. J. Mol. Sci. 2017, 18, e667. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.M.; Yeo, C.J.; Brody, J.R. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J. Surg. Oncol. 2013, 107, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ballehaninna, U.K.; Chamberlain, R.S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J. Gastrointest. Oncol. 2012, 3, 105–119. [Google Scholar] [PubMed]
- Lee, J.S.; Ahn, Y.-H.; Won, H.S.; Sun, D.S.; Kim, Y.H.; Ko, Y.H. Prognostic Role of the MicroRNA-200 Family in Various Carcinomas: A Systematic Review and Meta-Analysis. BioMed Res. Int. 2017, 2017, 1928021. [Google Scholar] [CrossRef] [PubMed]
- Senfter, D.; Madlener, S.; Krupitza, G.; Mader, R.M. The microRNA-200 family: Still much to discover. Biomol. Concepts 2016, 7, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Shen, J.; Jiang, C.-P.; Liu, B.-R. How to explain the contradiction of microRNA 200c expression and survival in solid tumors? A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 3687–3690. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, N.N.; Aigner, M.; Thorlund, K.; Mollberg, N.; Motschall, E.; Jensen, K.; Diener, M.K.; Büchler, M.W.; Koch, M.; Weitz, J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology 2010, 138, 1714–1726. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-B.; Yang, X.-Q.; Yang, S.; Wang, B.-C.; Feng, M.-H.; Tu, J.-C. A higher number of circulating tumor cells (CTC) in peripheral blood indicates poor prognosis in prostate cancer patients—A meta-analysis. Asian Pac. J. Cancer Prev. 2011, 12, 2629–2635. [Google Scholar] [PubMed]
- Zhang, L.; Riethdorf, S.; Wu, G.; Wang, T.; Yang, K.; Peng, G.; Liu, J.; Pantel, K. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin. Cancer Res. 2012, 18, 5701–5710. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.N.; Hamar, P.; Guo, C.; Basar, E.; Perdigão-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Hur, K.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 2014, 259, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, Z.; Fillmore, R.; Xi, Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014, 344, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A Double-Negative Feedback Loop between ZEB1-SIP1 and the microRNA-200 Family Regulates Epithelial-Mesenchymal Transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle Georget. Tex 2015, 14, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Guo, X.; Yan, J.; Deng, K. The role of miR-148a in gastric cancer. J. Cancer Res. Clin. Oncol. 2014, 140, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Huang, S.; He, R.; Rong, M.; Dang, Y.; Chen, G. Decreased expression and clinical significance of miR-148a in hepatocellular carcinoma tissues. Eur. J. Med. Res. 2014, 19, e68. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Fang, Y.; Deng, X.; Chen, H.; Jin, J.; Lu, X.; Peng, C.; Li, H.; Shen, B. The Interplay Between miR-148a and DNMT1 Might be Exploited for Pancreatic Cancer Therapy. Cancer Investig. 2015, 33, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, M.; Zang, W.; Chen, X.; Wang, Y.; Li, P.; Du, Y.; Zhao, G.; Li, L. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. 2014, 35, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, X.; Chai, J.; Chen, P.; Ren, P.; Gong, M. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014, 35, 12467–12472. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, B.; Lu, H.; Chen, Y.; Lu, C.; Zhu, R.; Liu, S.; Yi, Q.; Li, J.; Song, C. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015, 36, 3035–3042. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yu, J.; Ma, Y.; Wang, F.; Liu, H. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol. Lett. 2016, 12, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Zhang, X.; Liang, H.; Luo, D.; Rong, M.; Dang, Y.; Chen, G. Prognostic significance of MiR-34a in solid tumors: A systemic review and meta-analysis with 4030 patients. Int. J. Clin. Exp. Med. 2015, 8, 17377–17391. [Google Scholar] [PubMed]
- Wang, J.; Dan, G.; Zhao, J.; Ding, Y.; Ye, F.; Sun, H.; Jiang, F.; Cheng, J.; Yuan, F.; Zou, Z. The predictive effect of overexpressed miR-34a on good survival of cancer patients: A systematic review and meta-analysis. OncoTargets Ther. 2015, 8, 2709–2719. [Google Scholar]
- Jamieson, N.B.; Morran, D.C.; Morton, J.P.; Ali, A.; Dickson, E.J.; Carter, C.R.; Sansom, O.J.; Evans, T.R.J.; McKay, C.J.; Oien, K.A. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2012, 18, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Hao, X.; Zhang, M.; Tang, W.; Yang, M.; Li, L.; Xiang, D.; DeSano, J.T.; Bommer, G.T.; Fan, D.; et al. MicroRNA miR-34 Inhibits Human Pancreatic Cancer Tumor-Initiating Cells. PLoS ONE 2009, 4, e6816. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G. miR-34—A microRNA replacement therapy is headed to the clinic. Front. Genet. 2012, 3, e120. [Google Scholar] [CrossRef] [PubMed]
- Karasek, P.; Gablo, N.; Hlavsa, J.; Kiss, I.; Vychytilova-Faltejskova, P.; Hermanova, M.; Kala, Z.; Slaby, O.; Prochazka, V. Pre-operative Plasma miR-21-5p Is a Sensitive Biomarker and Independent Prognostic Factor in Patients with Pancreatic Ductal Adenocarcinoma Undergoing Surgical Resection. Cancer Genom. Proteom. 2018, 15, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, P.; Wang, Y.; Wang, X. Blood-Derived microRNAs for Pancreatic Cancer Diagnosis: A Narrative Review and Meta-Analysis. Front. Physiol. 2018, 9, e685. [Google Scholar] [CrossRef] [PubMed]
- Duell, E.J.; Lujan-Barroso, L.; Sala, N.; Deitz McElyea, S.; Overvad, K.; Tjonneland, A.; Olsen, A.; Weiderpass, E.; Busund, L.-T.; Moi, L.; et al. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int. J. Cancer 2017, 141, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Yang, S.; Lin, Y.; Zhang, J. An eight-miRNA signature expression-based risk scoring system for prediction of survival in pancreatic adenocarcinoma. Cancer Biomark. Sect. Dis. Mark. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Xu, C.; Yuan, W.; Wang, C.; Zhao, P.; Chen, L.; Ma, J. Evaluation of Plasma MicroRNAs as Diagnostic and Prognostic Biomarkers in Pancreatic Adenocarcinoma: miR-196a and miR-210 Could Be Negative and Positive Prognostic Markers, Respectively. BioMed Res. Int. 2017, 2017, e6495867. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Tang, W.; Xie, Y.; Wang, S.; Chen, Y.; Qi, J.; Qiao, Y.; Ma, J. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget 2016, 7, 80033–80045. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, H.; Kim, H.W.; Lee, J.-C.; Paik, K.-H.; Kang, J.; Kim, J.; Yoon, Y.-S.; Han, H.-S.; Sohn, I.; et al. High Expression of MicroRNA-196a Indicates Poor Prognosis in Resected Pancreatic Neuroendocrine Tumor. Medicine (Baltimore) 2015, 94, e2224. [Google Scholar] [CrossRef] [PubMed]
- Dhayat, S.A.; Mardin, W.A.; Seggewiß, J.; Ströse, A.J.; Matuszcak, C.; Hummel, R.; Senninger, N.; Mees, S.T.; Haier, J. MicroRNA Profiling Implies New Markers of Gemcitabine Chemoresistance in Mutant p53 Pancreatic Ductal Adenocarcinoma. PLoS ONE 2015, 10, e0143755. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Raimondo, M.; Guha, S.; Chen, J.; Diao, L.; Dong, X.; Wallace, M.B.; Killary, A.M.; Frazier, M.L.; Woodward, T.A.; et al. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J. Cancer 2014, 5, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y.; Li, J.; Zhang, Z.; Huang, C.; Lian, G.; Yang, K.; Chen, S.; Lin, Y.; Wang, L.; et al. Co-delivery of microRNA-21 antisense oligonucleotides and gemcitabine using nanomedicine for pancreatic cancer therapy. Cancer Sci. 2017, 108, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ohuchida, K.; Mizumoto, K.; Sato, N.; Kayashima, T.; Fujita, H.; Nakata, K.; Tanaka, M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol. Cancer 2010, 9, e169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-F.; Xu, L.-Y.; Li, E.-M. A family of pleiotropically acting microRNAs in cancer progression, miR-200: Potential cancer therapeutic targets. Curr. Pharm. Des. 2014, 20, 1896–1903. [Google Scholar] [CrossRef] [PubMed]
- Gilles, M.-E.; Hao, L.; Huang, L.; Rupaimoole, R.; Lopez-Casas, P.P.; Pulver, E.; Jeong, J.C.; Muthuswamy, S.K.; Hidalgo, M.; Bhatia, S.N.; et al. Personalized RNA Medicine for Pancreatic Cancer. Clin. Cancer Res. 2018, 24, 1734–1747. [Google Scholar] [CrossRef] [PubMed]
- Pai, P.; Rachagani, S.; Are, C.; Batra, S.K. Prospects of miRNA-Based Therapy for Pancreatic Cancer. Curr. Drug Targets 2013, 14, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, D.; Campbell, N.R.; Karikari, C.; Chivukula, R.; Kent, O.A.; Mendell, J.T.; Maitra, A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 2011, 10, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic Delivery of Tumor Suppressor microRNA Mimics Using a Neutral Lipid Emulsion Inhibits Lung Tumors in Mice. Mol. Ther. 2011, 19, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, X.; Luo, J.; Liu, M.; Xi, S.; Guo, J.; Kong, Y.; Wu, M.; Gao, J.; Xie, Z.; et al. Targeted Expression of miR-34a Using the T-VISA System Suppresses Breast Cancer Cell Growth and Invasion. Mol. Ther. 2012, 20, 2326–2334. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Chitkara, D.; Kumar, V.; Behrman, S.W.; Mahato, R.I. miRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Lett. 2013, 334, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Tan, C. Combination of microRNA therapeutics with small-molecule anticancer drugs: Mechanism of action and co-delivery nanocarriers. Adv. Drug Deliv. Rev. 2015, 81, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Mondal, G.; Almawash, S.; Chaudhary, A.K.; Mahato, R.I. EGFR-Targeted Cationic Polymeric Mixed Micelles for Codelivery of Gemcitabine and miR-205 for Treating Advanced Pancreatic Cancer. Mol. Pharm. 2017, 14, 3121–3133. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, S.; Sun, Y.L. Epithelial-Mesenchymal Transition in Pancreatic Cancer: A Review. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Dhayat, S.A.; Hüsing, A.; Senninger, N.; Schmidt, H.H.; Haier, J.; Wolters, H.; Kabar, I. Circulating microRNA-200 Family as Diagnostic Marker in Hepatocellular Carcinoma. PLoS ONE 2015, 10, e0140066. [Google Scholar] [CrossRef] [PubMed]
- Dhayat, S.A.; Mardin, W.A.; Köhler, G.; Bahde, R.; Vowinkel, T.; Wolters, H.; Senninger, N.; Haier, J.; Mees, S.T. The microRNA-200 family-A potential diagnostic marker in hepatocellular carcinoma? MicroRNA-200 Family Expression in HCC. J. Surg. Oncol. 2014, 110, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Detre, S.; Saclani Jotti, G.; Dowsett, M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995, 48, 876–878. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Reporting recommendations for tumour marker prognostic studies (REMARK). Br. J. Cancer 2005, 93, 387–391. [Google Scholar] [CrossRef] [PubMed]


| Category | Number of PDAC Patients | Median Overall Survival (Months) | 95% CI | p-Value |
|---|---|---|---|---|
| Total | 96 | 17.1 | 12.6–21.6 | |
| Age (years) | 0.097 | |||
| ≤60 | 35 | 21.8 | 9.5–34.2 | |
| >60 | 61 | 14.9 | 9.9–19.9 | |
| Gender | 0.594 | |||
| Female | 57 | 16.5 | 11.7–21.3 | |
| Male | 39 | 18.3 | 12.621.6 | |
| Body Mass Index | 0.932 | |||
| ≤25 | 57 | 16.5 | 12.0–21.0 | |
| >25 | 37 | 18.3 | 11.4–25.1 | |
| Smoker | 0.245 | |||
| Yes | 22 | 11.8 | 6.1–17.4 | |
| No | 74 | 18.3 | 14.7–21.8 | |
| Alcohol abusus | 0.100 | |||
| Yes | 5 | |||
| No | 91 | 17 | 13.0–20.9 | |
| Pre-surgical Diabetes mellitus | 0.865 | |||
| Yes | 31 | 14.9 | 7.1–22.4 | |
| No | 65 | 17.1 | 14.0–20.3 | |
| Pre-surgical pancreatitis | 0.094 | |||
| Yes | 15 | 13.1 | 7.9–14.8 | |
| No | 81 | 18.3 | 14.7–21.8 | |
| Pre-surgical CA.19-9 (U/mL) | 0.866 | |||
| ≤30 | 14 | 16.6 | 7.1–26.3 | |
| >30 | 49 | 14.3 | 9.3–19.3 | |
| UICC Stage | <0.001 | |||
| IIa | 16 | 26.9 | 20.9–21.9 | |
| IIb | 53 | 17.1 | 12.8–21.5 | |
| III | 11 | 12.5 | 11.1–13.9 | |
| IV | 16 | 5.1 | 1.8–8.4 | |
| Grading | 0.006 | |||
| G1 and G2 | 46 | 23.6 | 15.5–31.6 | |
| G3 | 34 | 13.1 | 8.4–17.9 | |
| Metastasis | <0.001 | |||
| M0 | 80 | 20.1 | 16.0–24.2 | |
| M1 | 16 | 5.1 | 1.8–8.5 | |
| Nodal invasion | 0.002 | |||
| Nx | 9 | 6.0 | 3.9–8.2 | |
| N0 | 19 | 26.4 | 21.1–31.7 | |
| N1 | 68 | 16.7 | 12.6–20.7 | |
| Lymphatic invasion | 0.730 | |||
| L0 | 39 | 20.4 | 14.7–26.0 | |
| L1 | 31 | 20.1 | 12.0–28.2 | |
| Perineural invasion | 0.061 | |||
| Pn0 | 17 | 29 | 15.6–42.3 | |
| Pn1 | 48 | 19.8 | 13.3–26.4 | |
| Vene invasion | 0.800 | |||
| V0 | 59 | 20.1 | 15.4–24.7 | |
| V1 | 11 | 21.6 | 9.3–33.9 | |
| Resection margin | 0.521 | |||
| R0 | 51 | 21.6 | 18.4–24.9 | |
| R1 | 19 | 17 | 11.5–22.4 | |
| Tumor size (cm) | 0.382 | |||
| ≤3 | 45 | 20.4 | 15.9–24.8 | |
| >3 | 21 | 28 | 13.7-42.2 | |
| Type of surgery | <0.001 | |||
| Pancreatic head resection | 53 | 21.8 | 15.3–28.3 | |
| Pancreatic left resection | 9 | 19.8 | 11.9–27.8 | |
| Total Pancreatectomy | 9 | 20.1 | 9.7–30.4 | |
| Excisional biopsy | 25 | 7.9 | 2.7–13.2 | |
| Type of chemotherapy | <0.001 | |||
| Adjuvant | 62 | 21.6 | 18.1–25.2 | |
| Palliative | 25 | 11.9 | 10.5–13.4 | |
| No chemotherapy | 9 | 1.9 | 1.7–2.3 |
| Non-PDAC vs. PDAC | ||||
|---|---|---|---|---|
| microRNA | Tissue | Serum | ||
| AUC | p-Value | AUC | p-Value | |
| miR-10b | 0.656 | 0.018 | 0.663 | 0.010 |
| miR-34a | 0.503 | 0.963 | 0.640 | 0.026 |
| miR-141 | 0.726 | <0.001 | 0.682 | 0.004 |
| miR-148a | 0.885 | <0.001 | 0.534 | 0.554 |
| miR-197 | 0.674 | 0.010 | 0.627 | 0.049 |
| miR-200a | 0.542 | 0.434 | 0.658 | 0.013 |
| miR-200b | 0.723 | <0.001 | 0.792 | <0.001 |
| miR-200c | 0.838 | <0.001 | 0.780 | <0.001 |
| miR-203a | 0.550 | 0.455 | 0.627 | 0.047 |
| miR-429 | 0.537 | 0.490 | 0.650 | 0.019 |
| miR-655 | 0.646 | 0.026 | 0.548 | 0.452 |
| AUC | p-Value | Sensitivity | Specificity | |
| CA.19-9 | 0.834 | <0.001 | 0.781 | 0.870 |
| EMT-regulating microRNA | Target/Function | Ref. |
|---|---|---|
| EMT-Suppressive MicroRNAs | ||
| miR-34a | Blocks Snail1 and Notch1 | [17] |
| miR-148a | Inhibits the Wnt/β-Catenin pathway | [23] |
| miR-200-family (-141, -200a, -200b, -200c, -429) | Block ZEB-1 and ZEB-2 | [14] |
| miR-203a | Inhibits the Wnt/β-Catenin pathway | [24] |
| miR-655 | Blocks ZEB-1 and TGF-β-R2 | [27] |
| EMT-Promoting MicroRNAs | ||
| miR-10b | Promotes TGF-β-signaling | [28] |
| miR-197 | Blocks p120 catenin (a cooperator of E-cadherin) | [29] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhayat, S.A.; Traeger, M.M.; Rehkaemper, J.; Stroese, A.J.; Steinestel, K.; Wardelmann, E.; Kabar, I.; Senninger, N. Clinical Impact of Epithelial-to-Mesenchymal Transition Regulating MicroRNAs in Pancreatic Ductal Adenocarcinoma. Cancers 2018, 10, 328. https://doi.org/10.3390/cancers10090328
Dhayat SA, Traeger MM, Rehkaemper J, Stroese AJ, Steinestel K, Wardelmann E, Kabar I, Senninger N. Clinical Impact of Epithelial-to-Mesenchymal Transition Regulating MicroRNAs in Pancreatic Ductal Adenocarcinoma. Cancers. 2018; 10(9):328. https://doi.org/10.3390/cancers10090328
Chicago/Turabian StyleDhayat, Sameer Abdallah, Max Michael Traeger, Jan Rehkaemper, Anda Jana Stroese, Konrad Steinestel, Eva Wardelmann, Iyad Kabar, and Norbert Senninger. 2018. "Clinical Impact of Epithelial-to-Mesenchymal Transition Regulating MicroRNAs in Pancreatic Ductal Adenocarcinoma" Cancers 10, no. 9: 328. https://doi.org/10.3390/cancers10090328
APA StyleDhayat, S. A., Traeger, M. M., Rehkaemper, J., Stroese, A. J., Steinestel, K., Wardelmann, E., Kabar, I., & Senninger, N. (2018). Clinical Impact of Epithelial-to-Mesenchymal Transition Regulating MicroRNAs in Pancreatic Ductal Adenocarcinoma. Cancers, 10(9), 328. https://doi.org/10.3390/cancers10090328






