Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance
Abstract
1. Introduction
2. Results
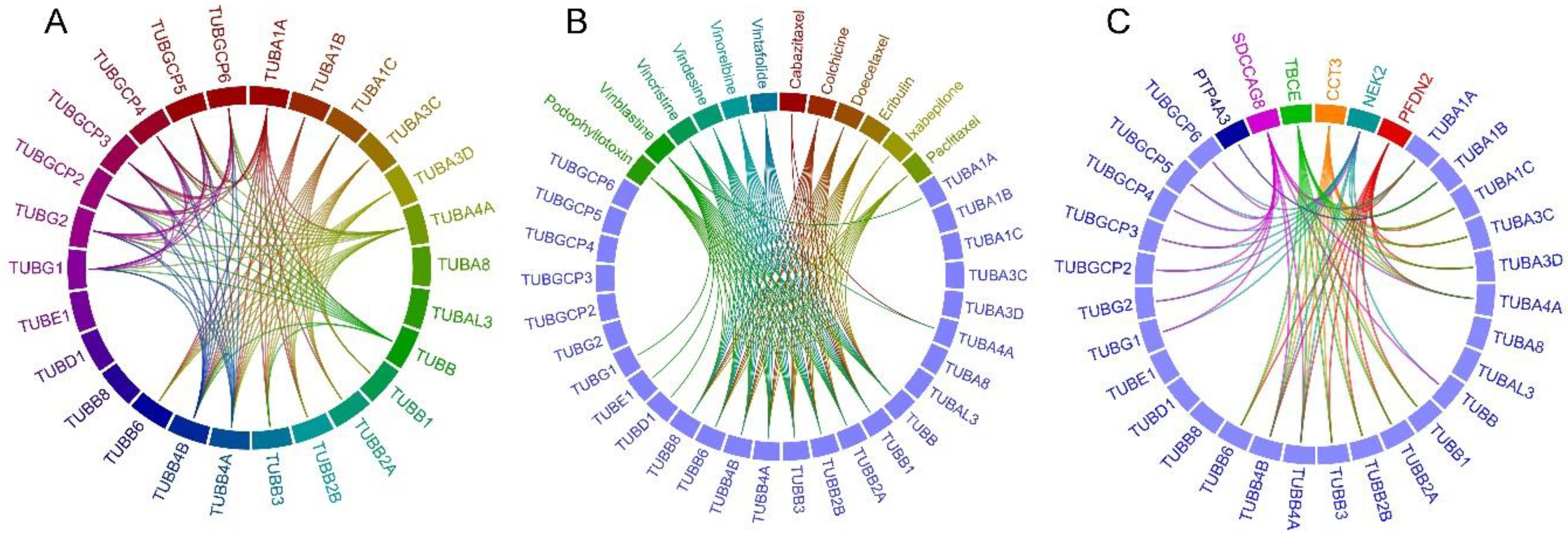
2.1. Tubulin-Tubulin Interactions in BC
2.2. Drug-Tubulin Interactions in BC
2.3. Frequently Altered Neighbor Genes
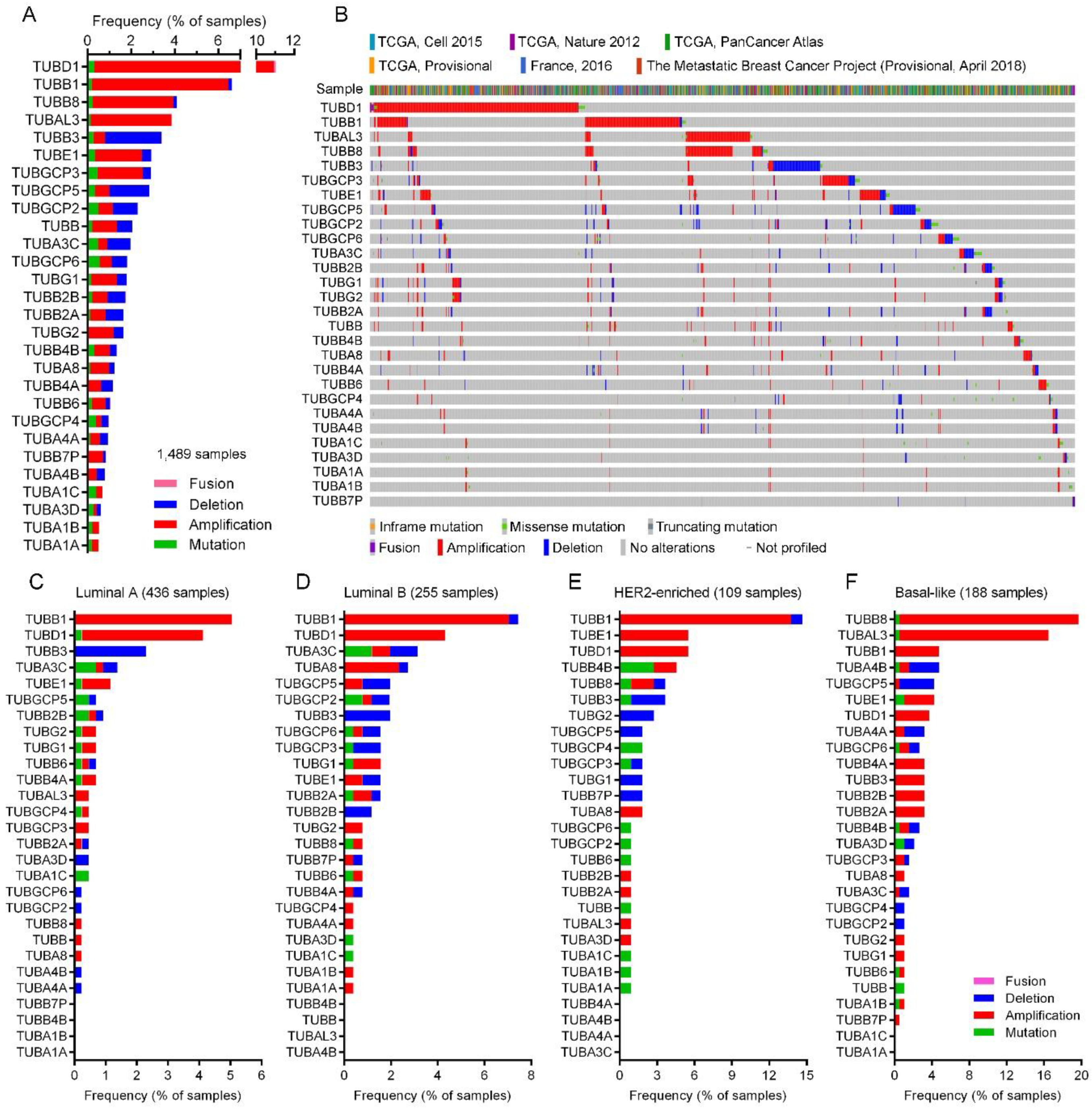
2.4. Tubulin Gene Alterations in BC
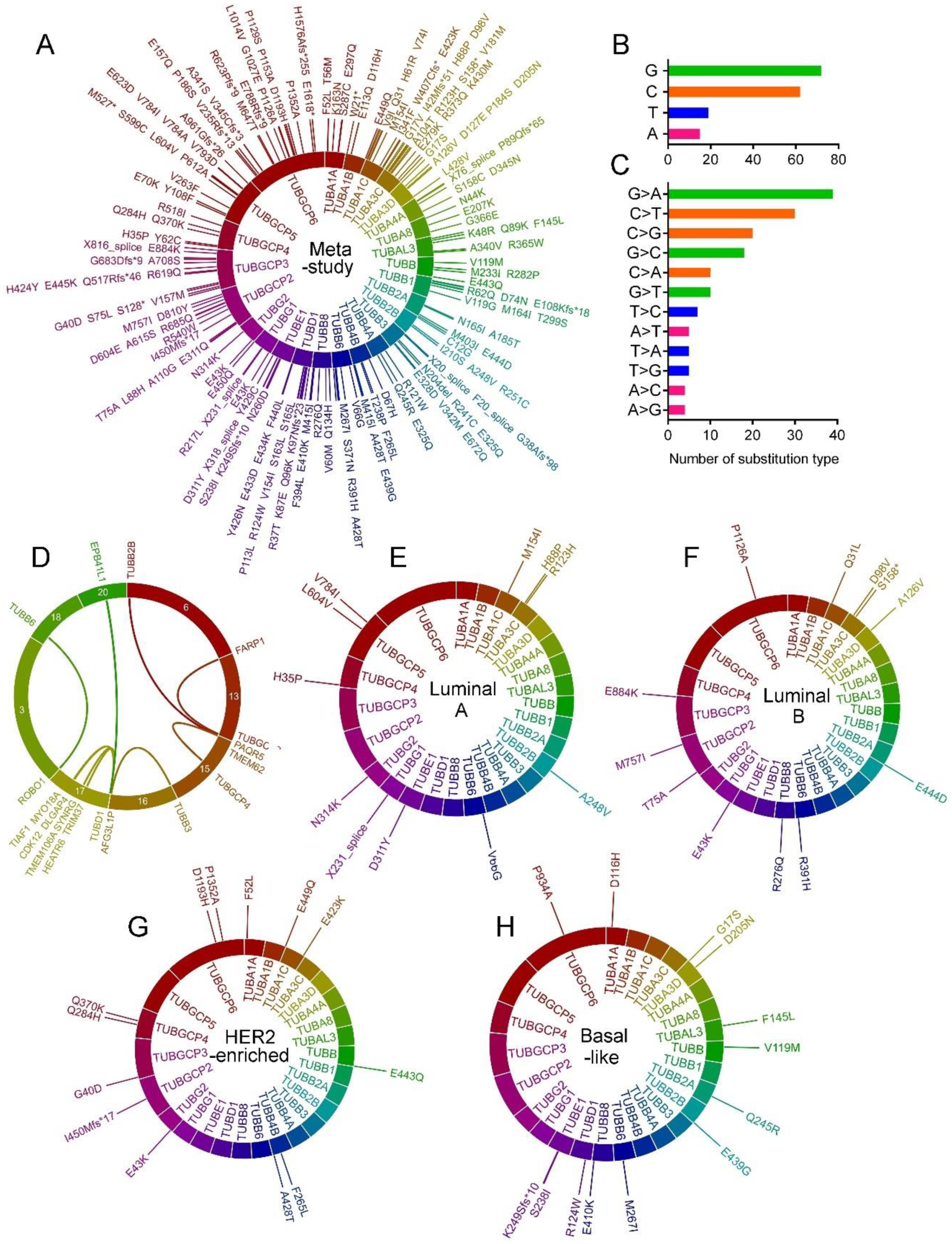
2.5. Tubulin Gene Mutations in BC
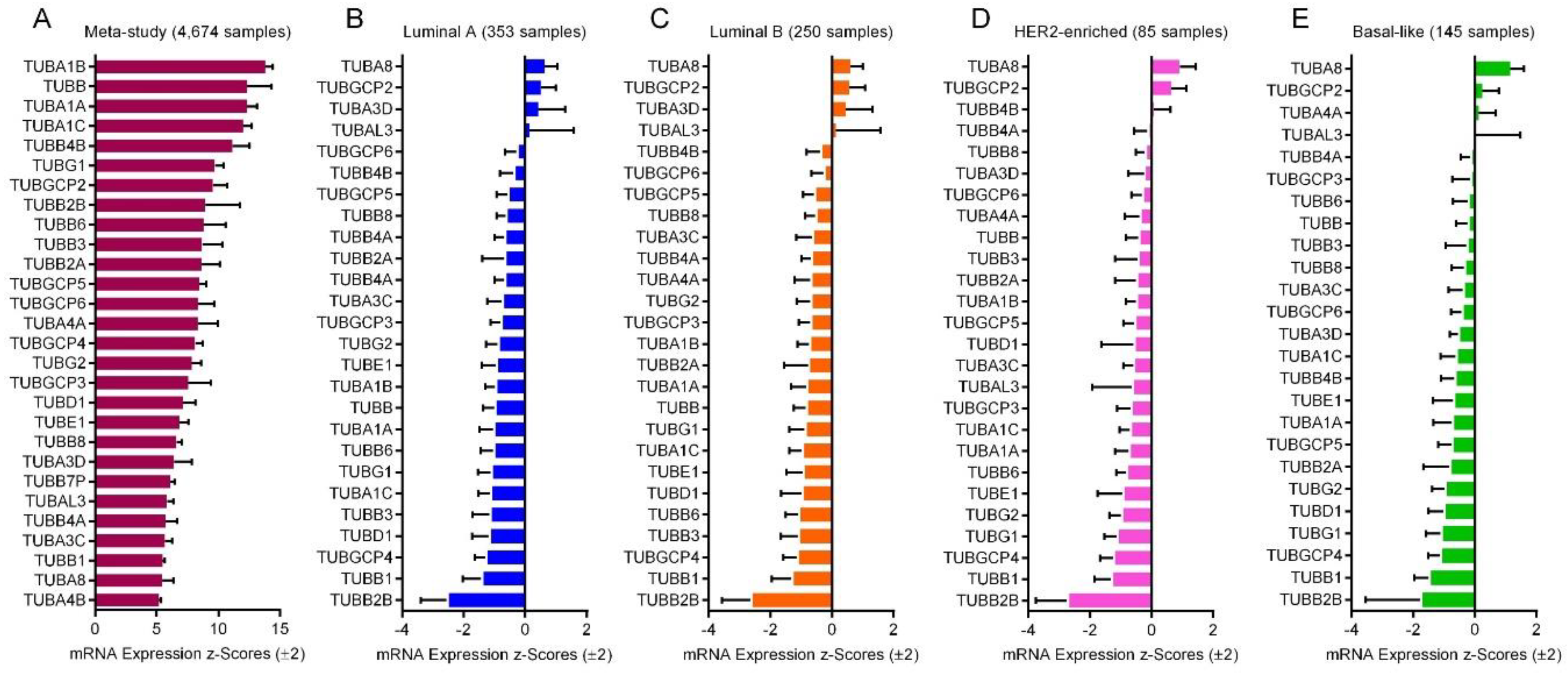
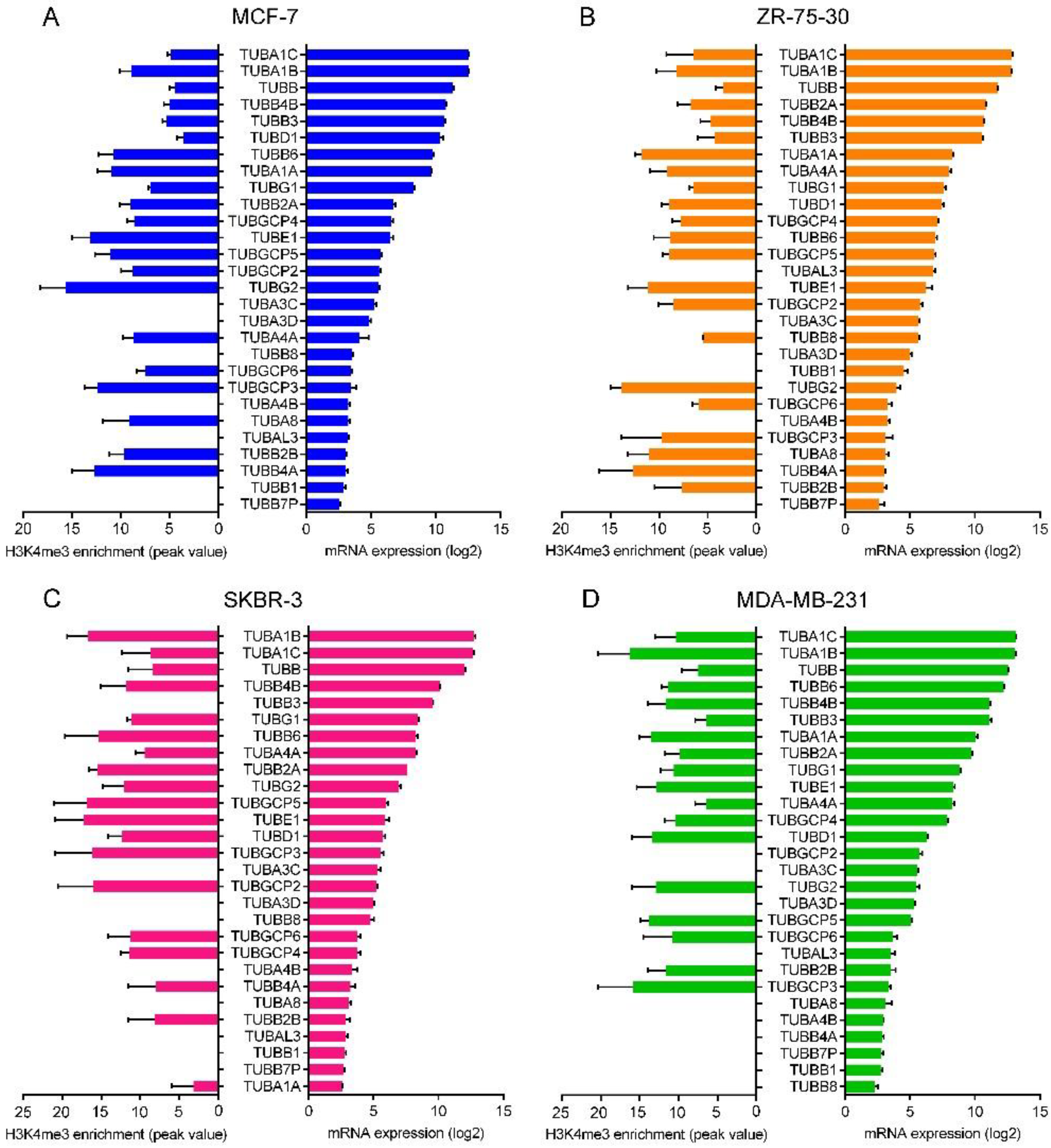
2.6. mRNA Expression of the Tubulin Genes
2.7. Correlation between the Promoter Activity and the Expression of Tubulin Genes
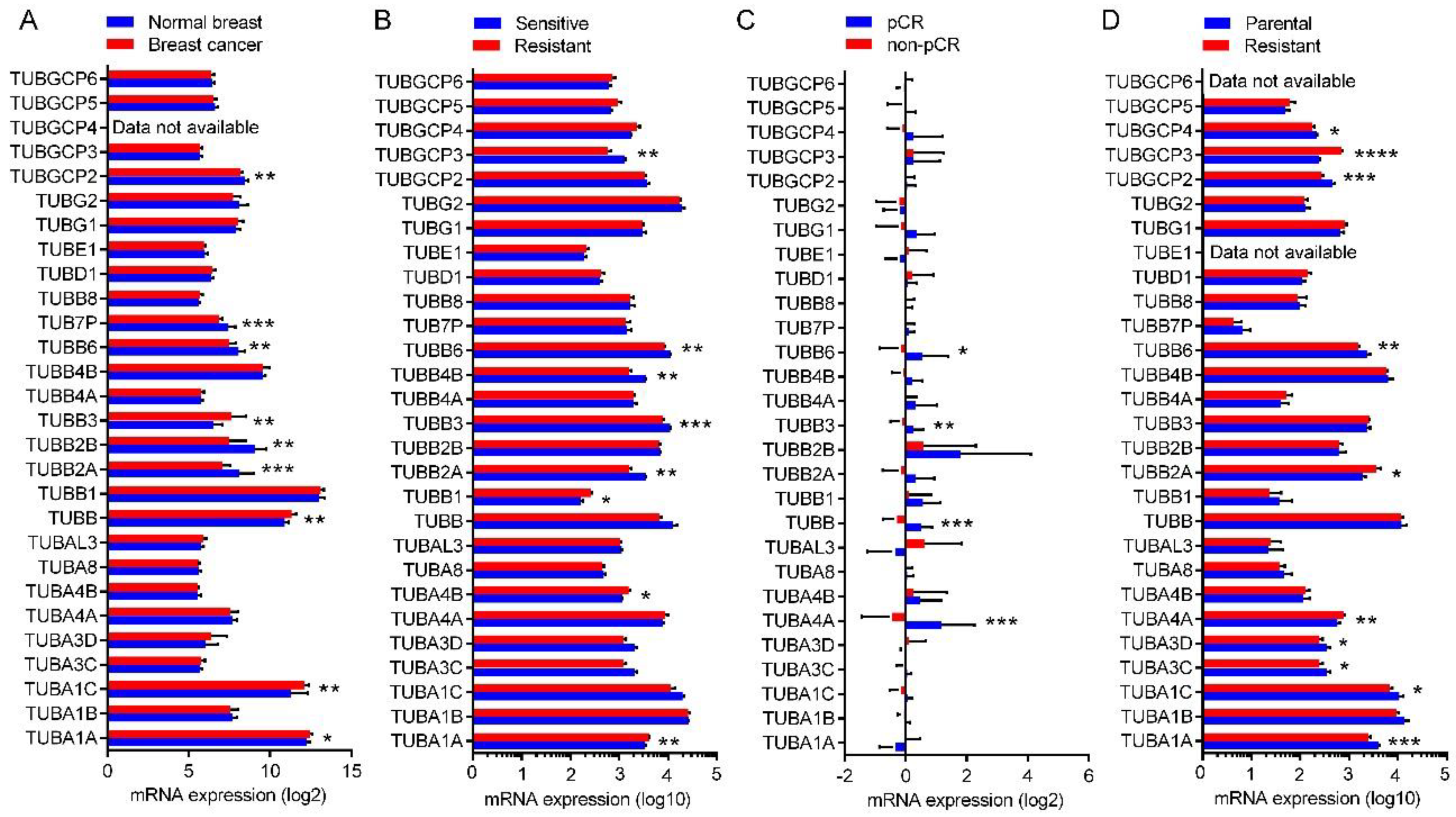
2.8. Expression of the Tubulin Genes in Normal Breast and BC Tumors
2.9. Expression of the Tubulin Genes in Taxane-Sensitive and Resistant BC
3. Discussion
4. Materials and Methods
4.1. BC Tumor Genomic Data
4.2. Interaction Network Analysis
4.3. ChIP-Seq Data
4.4. Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- DeSantis, C.E.; Ma, J.; Sauer, A.G.; Newman, L.A.; Jemal, A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J. Clin. 2017, 67, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; Van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Kamangar, F.; Dores, G.M.; Anderson, W.F. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J. Clin. Oncol. 2006, 24, 2137–2150. [Google Scholar] [CrossRef] [PubMed]
- Yardley, D.A. Drug resistance and the role of combination chemotherapy in improving patient outcomes. Int. J. Breast Cancer 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Smalley, M.; Piggott, L.; Clarkson, R. Breast cancer stem cells: Obstacles to therapy. Cancer Lett. 2013, 338, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Jassem, J.; Pienkowski, T.; Pluzanska, A.; Jelic, S.; Gorbunova, V.; Berzins, J.; Nagykalnai, T.; Biganzoli, L.; Aloe, A.; Astier, L.; Munier, S. Doxorubicin and paclitaxel versus fluorouracil, doxorubicin and cyclophosphamide as first-line therapy for women with advanced breast cancer: long-term analysis of the previously published trial. Onkologie 2009, 32, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Nabholtz, J.-M.; Falkson, C.; Campos, D.; Szanto, J.; Martin, M.; Chan, S.; Pienkowski, T.; Zaluski, J.; Pinter, T.; Krzakowski, M.; et al. Docetaxel and doxorubicin compared with doxorubicin and cyclophosphamide as first-line chemotherapy for metastatic breast cancer: results of a randomized, multicenter, phase III trial. J. Clin. Oncol. 2003, 21, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Zelnak, A. Overcoming taxane and anthracycline resistance. Breast J. 2010, 16, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Rivera, E. Implications of anthracycline-resistant and taxane-resistant metastatic breast cancer and new therapeutic options. Breast J. 2010, 16, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Downing, K.H.; Nogales, E. Crystallographic structure of tubulin: Implications for dynamics and drug binding. Cell Struct. Funct. 1999, 24, 269–275. [Google Scholar] [CrossRef] [PubMed]
- McGrogan, B.T.; Gilmartin, B.; Carney, D.N.; McCann, A. Taxanes, microtubules and chemoresistant breast cancer. Biochim. Biophys. Acta 2008, 1785, 96–132. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr. Med. Chem. Anti-Cancer Agents 2002, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dumontet, C.; Jordan, M.A. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat. Rev. Drug Discov. 2010, 9, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.L.; Gray, H.; Payne, C.M.; Gillies, R.J. Docetaxel induces cell death through mitotic catastrophe in human breast cancer cells. Mol. Cancer Ther. 2005, 4, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Tangutur, A.D.; Kumar, D.; Krishna, K.V.; Kantevari, S. Microtubule targeting agents as cancer chemotherapeutics: An overview of molecular hybrids as stabilizing and destabilizing agents. Curr. Top. Med. Chem. 2017, 17, 2523–2537. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, R.B.G.; Gigant, B.; Curmi, P.A.; Jourdain, I.; Lachkar, S.; Sobel, A.; Knossow, M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004, 428, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.-S.; Koh, C.-G.; Li, H.-Y. Mitosis-targeted anti-cancer therapies: Where they stand. Cell Death Dis. 2012, 3, e411. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.C.; Uniyal, S.; Zhou, H.; Morris, V.L.; Chan, B.M.C. Threshold levels of ERK activation for chemotactic migration differ for NGF and EGF in rat pheochromocytoma PC12 cells. Mol. Cell. Biochem. 2005, 271, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Berrieman, H.K.; Lind, M.J.; Cawkwell, L. Do β-tubulin mutations have a role in resistance to chemotherapy? Lancet. Oncol. 2004, 5, 158–164. [Google Scholar] [CrossRef]
- Burkhart, C.A.; Kavallaris, M.; Band Horwitz, S. The role of β-tubulin isotypes in resistance to antimitotic drugs. Biochim. Biophys. Acta 2001, 1471, O1–O9. [Google Scholar] [CrossRef]
- Lal, S.; Mahajan, A.; Chen, W.N.; Chowbay, B. Pharmacogenetics of target genes across doxorubicin disposition pathway: A review. Curr. Drug Metab. 2010, 11, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.; Briasoulis, E.; Linardou, H.; Bafaloukos, D.; Papadimitriou, C. Taxane resistance in breast cancer: Mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev. 2012, 38, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, A.; Braguer, D.; Kamath, K.; Martello, L.; Briand, C.; Horwitz, S.; Wilson, L.; Jordan, M.A. Resistance to Taxol in lung cancer cells associated with increased microtubule dynamics. Proc. Natl. Acad. Sci. USA 2001, 98, 11737–11742. [Google Scholar] [CrossRef] [PubMed]
- Kienitz, A.; Vogel, C.; Morales, I.; Muller, R.; Bastians, H. Partial downregulation of MAD1 causes spindle checkpoint inactivation and aneuploidy, but does not confer resistance towards taxol. Oncogene 2005, 24, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; Carmena, M.; Sambade, C.; Earnshaw, W.C.; Wheatley, S.P. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 2003, 116, 2987–2998. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.F.; Donaldson, K.L.; Fairchild, C.; Lee, F.Y.; Foster, S.A.; Demers, G.W.; Galloway, D.A. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat. Med. 1996, 2, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Shitashige, M.; Toi, M.; Yano, T.; Shibata, M.; Matsuo, Y.; Shibasaki, F. Dissociation of Bax from a Bcl-2/Bax heterodimer triggered by phosphorylation of serine 70 of Bcl-2. J. Biochem. 2001, 130, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, K.N. Microtubule-targeted anticancer agents and apoptosis. Oncogene 2003, 22, 9075–9086. [Google Scholar] [CrossRef] [PubMed]
- Janke, C. The tubulin code: Molecular components, readout mechanisms, and functions. J. Cell Biol. 2014, 206, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, S.; Natarajan, K.; Curiel, J.; Janke, C.; Liu, J. The emerging role of the tubulin code: From the tubulin molecule to neuronal function and disease. Cytoskeleton 2016, 73, 521–550. [Google Scholar] [CrossRef] [PubMed]
- Gadadhar, S.; Bodakuntla, S.; Natarajan, K.; Janke, C. The tubulin code at a glance. J. Cell Sci. 2017, 130, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Luduena, R.F. Are tubulin isotypes functionally significant. Mol. Biol. Cell 1993, 4, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Mariani, M.; Karki, R.; Spennato, M.; Pandya, D.; He, S.; Andreoli, M.; Fiedler, P.; Ferlini, C. Class III β-tubulin in normal and cancer tissues. Gene 2015, 563, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.A.; Lee, M.G.; Cowan, N.J. Five mouse tubulin isotypes and their regulated expression during development. J. Cell Biol. 1985, 101, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.C.; Cleveland, D.W. Differential utilization of β-tubulin isotypes in differentiating neurites. J. Cell Biol. 1989, 109, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Renthal, R.; Schneider, B.G.; Miller, M.M.; Luduena, R.F. Beta IV is the major β-tubulin isotype in bovine cilia. Cell Motil. Cytoskelet. 1993, 25, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Villasante, A.; Lewis, S.A.; Cowan, N.J. The mammalian β-tubulin repertoire: Hematopoietic expression of a novel, heterologous β-tubulin isotype. J. Cell Biol. 1986, 103, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Lecine, P.; Italiano, J.E.J.; Kim, S.W.; Villeval, J.L.; Shivdasani, R.A. Hematopoietic-specific β1 tubulin participates in a pathway of platelet biogenesis dependent on the transcription factor NF-E2. Blood 2000, 96, 1366–1373. [Google Scholar] [PubMed]
- Kunishima, S.; Kobayashi, R.; Itoh, T.J.; Hamaguchi, M.; Saito, H. Mutation of the β1-tubulin gene associated with congenital macrothrombocytopenia affecting microtubule assembly. Blood 2009, 113, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Vo, T.; Hajar, A.; Li, S.; Chen, X.; Parissenti, A.M.; Brindley, D.N.; Wang, Z. Multiple mechanisms underlying acquired resistance to taxanes in selected docetaxel-resistant MCF-7 breast cancer cells. BMC Cancer 2014, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Kavallaris, M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer 2010, 10, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Derry, W.B.; Wilson, L.; Khan, I.A.; Luduena, R.F.; Jordan, M.A. Taxol differentially modulates the dynamics of microtubules assembled from unfractionated and purified β-tubulin isotypes. Biochemistry 1997, 36, 3554–3562. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A. Increased levels of tyrosinated α-, βIII-, and βIV-tubulin isotypes in paclitaxel-resistant MCF-7 breast cancer cells. Biochem. Biophys. Res. Commun. 2002, 293, 598–601. [Google Scholar] [CrossRef]
- Haber, M.; Burkhart, C.A.; Regl, D.L.; Madafiglio, J.; Norris, M.D.; Horwitz, S.B. Altered expression of Mβ2, the class II β-tubulin isotype, in a murine J774.2 cell line with a high level of taxol resistance. J. Biol. Chem. 1995, 270, 31269–31275. [Google Scholar] [CrossRef] [PubMed]
- Iseri, O.D.; Kars, M.D.; Gunduz, U. Drug resistant MCF-7 cells have altered expression levels of β-tubulin isotypes and mutations in TUBB gene. Int. J. Hematol. Oncol. 2010, 20, 75–83. [Google Scholar]
- Kavallaris, M.; Kuo, D.Y.; Burkhart, C.A.; Regl, D.L.; Norris, M.D.; Haber, M.; Horwitz, S.B. Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific β-tubulin isotypes. J. Clin. Investig. 1997, 100, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Mozzetti, S.; Ferlini, C.; Concolino, P.; Filippetti, F.; Raspaglio, G.; Prislei, S.; Gallo, D.; Martinelli, E.; Ranelletti, F.O.; Ferrandina, G.; et al. Class III β-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin. Cancer Res. 2005, 11, 298–305. [Google Scholar] [PubMed]
- Panda, D.; Miller, H.P.; Banerjee, A.; Ludueña, R.F.; Wilson, L. Microtubule dynamics in vitro are regulated by the tubulin isotype composition. Proc. Natl. Acad. Sci. USA 1994, 91, 11358–11362. [Google Scholar] [CrossRef] [PubMed]
- Kamath, K.; Wilson, L.; Cabral, F.; Jordan, M.A. βIII-tubulin induces paclitaxel resistance in association with reduced effects on microtubule dynamic instability. J. Biol. Chem. 2005, 280, 12902–12907. [Google Scholar] [CrossRef] [PubMed]
- Seve, P.; Dumontet, C. Is class III β-tubulin a predictive factor in patients receiving tubulin-binding agents? Lancet. Oncol. 2008, 9, 168–175. [Google Scholar] [CrossRef]
- Hari, M.; Yang, H.; Zeng, C.; Canizales, M.; Cabral, F. Expression of class III β-tubulin reduces microtubule assembly and confers resistance to paclitaxel. Cell Motil. Cytoskelet. 2003, 56, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Seve, P.; Reiman, T.; Lai, R.; Hanson, J.; Santos, C.; Johnson, L.; Dabbagh, L.; Sawyer, M.; Dumontet, C.; Mackey, J.R. Class III β-tubulin is a marker of paclitaxel resistance in carcinomas of unknown primary site. Cancer Chemother. Pharmacol. 2007, 60, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Galmarini, C.M.; Treilleux, I.; Cardoso, F.; Bernard-Marty, C.; Durbecq, V.; Gancberg, D.; Bissery, M.-C.; Paesmans, M.; Larsimont, D.; Piccart, M.J.; et al. Class III β-tubulin isotype predicts response in advanced breast cancer patients randomly treated either with single-agent doxorubicin or docetaxel. Clin. Cancer Res. 2008, 14, 4511–4516. [Google Scholar] [CrossRef] [PubMed]
- Aoki, D.; Oda, Y.; Hattori, S.; Taguchi, K.; Ohishi, Y.; Basaki, Y.; Oie, S.; Suzuki, N.; Kono, S.; Tsuneyoshi, M.; et al. Overexpression of class III β-tubulin predicts good response to taxane-based chemotherapy in ovarian clear cell adenocarcinoma. Clin. Cancer Res. 2009, 15, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Akasaka, K.; Maesawa, C.; Shibazaki, M.; Maeda, F.; Takahashi, K.; Akasaka, T.; Masuda, T. Loss of class III β-tubulin induced by histone deacetylation is associated with chemosensitivity to paclitaxel in malignant melanoma cells. J. Investig. Dermatol. 2009, 129, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Tame, M.A.; Manjon, A.G.; Belokhvostova, D.; Raaijmakers, J.A.; Medema, R.H. TUBB3 overexpression has a negligible effect on the sensitivity to taxol in cultured cell lines. Oncotarget 2017, 8, 71536–71547. [Google Scholar] [CrossRef] [PubMed]
- Dekker, C.; Stirling, P.C.; McCormack, E.A.; Filmore, H.; Paul, A.; Brost, R.L.; Costanzo, M.; Boone, C.; Leroux, M.R.; Willison, K.R. The interaction network of the chaperonin CCT. EMBO J. 2008, 27, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Rellos, P.; Ivins, F.J.; Baxter, J.E.; Pike, A.; Nott, T.J.; Parkinson, D.-M.; Das, S.; Howell, S.; Fedorov, O.; Shen, Q.Y.; et al. Structure and regulation of the human Nek2 centrosomal kinase. J. Biol. Chem. 2007, 282, 6833–6842. [Google Scholar] [CrossRef] [PubMed]
- Cappello, P.; Blaser, H.; Gorrini, C.; Lin, D.C.C.; Elia, A.J.; Wakeham, A.; Haider, S.; Boutros, P.C.; Mason, J.M.; Miller, N.A.; et al. Role of Nek2 on centrosome duplication and aneuploidy in breast cancer cells. Oncogene 2014, 33, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rai, M.; Wang, C.; Gonzalez, C.; Wang, H. Prefoldin and Pins synergistically regulate asymmetric division and suppress dedifferentiation. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Insolera, R.; Shao, W.; Airik, R.; Hildebrandt, F.; Shi, S.-H. SDCCAG8 regulates pericentriolar material recruitment and neuronal migration in the developing cortex. Neuron 2014, 83, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Huang, Y.; Rommelaere, H.; Vandekerckhove, J.; Ampe, C.; Cowan, N.J. Pathway leading to correctly folded β-tubulin. Cell 1996, 86, 287–296. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.; Van Jaarsveld, M.T.M.; Hollestelle, A.; Prager-van der Smissen, W.J.C.; Heine, A.A.J.; Boersma, A.W.M.; Liu, J.; Helmijr, J.; Ozturk, B.; Smid, M.; et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013, 15, R33. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; De, S.; Bebek, G.; Orloff, M.S.; Wesolowski, R.; Downs-Kelly, E.; Budd, G.T.; Stark, G.R.; Eng, C. Specific kinesin expression profiles associated with taxane resistance in basal-like breast cancer. Breast Cancer Res. Treat. 2012, 131, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Chakravarthy, A.B.; Donach, M.; Spicer, D.; Lymberis, S.; Singh, B.; Bauer, J.A.; Hochman, T.; Goldberg, J.D.; Muggia, F.; et al. Preoperative concurrent paclitaxel-radiation in locally advanced breast cancer: Pathologic response correlates with five-year overall survival. Breast Cancer Res. Treat. 2010, 124, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.A.; Chakravarthy, A.B.; Rosenbluth, J.M.; Mi, D.; Seeley, E.H.; De Matos Granja-Ingram, N.; Olivares, M.G.; Kelley, M.C.; Mayer, I.A.; Meszoely, I.M.; et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin. Cancer Res. 2010, 16, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Schork, N.J.; Marschke, K.B.; Ng, S.-C.; Hermann, T.W.; Zhang, J.; Sanders, J.M.; Tooker, P.; Malo, N.; Zapala, M.A.; et al. Identification of polymorphisms associated with hypertriglyceridemia and prolonged survival induced by bexarotene in treating non-small cell lung cancer. Anticancer Res. 2011, 31, 2303–2311. [Google Scholar] [PubMed]
- Jozwik, K.M.; Chernukhin, I.; Serandour, A.A.; Nagarajan, S.; Carroll, J.S. FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep. 2016, 17, 2715–2723. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Xu, W.; Guo, R.; Rong, B.; Gu, L.; Wang, Z.; He, C.; Zheng, L.; Hu, X.; Hu, Z.; et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 2016, 165, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Chaligne, R.; Popova, T.; Mendoza-Parra, M.-A.; Saleem, M.-A.M.; Gentien, D.; Ban, K.; Piolot, T.; Leroy, O.; Mariani, O.; Gronemeyer, H.; et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 2015, 25, 488–503. [Google Scholar] [CrossRef] [PubMed]
- Rhie, S.K.; Hazelett, D.J.; Coetzee, S.G.; Yan, C.; Noushmehr, H.; Coetzee, G.A. Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genom. 2014, 15, 331. [Google Scholar] [CrossRef] [PubMed]
- Beghin, A.; Galmarini, C.M.; Dumontet, C. Tubulin folding pathways: Implication in the regulation of microtubule dynamics. Curr. Cancer Drug Targets 2007, 7, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, G.; Cowan, N.J.; Cabral, F. Mutations affecting β-tubulin folding and degradation. J. Biol. Chem. 2006, 281, 13628–13635. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Zhao, J.; Feng, J. Parkin binds to α/β tubulin and increases their ubiquitination and degradation. J. Neurosci. 2003, 23, 3316–3324. [Google Scholar] [CrossRef] [PubMed]
- Nami, B.; Donmez, H.; Kocak, N. Tunicamycin-induced endoplasmic reticulum stress reduces in vitro subpopulation and invasion of CD44+ / CD24- phenotype breast cancer stem cells. Exp. Toxicol. Pathol. 2016, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fabritius, M.; Ip, C. Chemotherapeutic sensitization by endoplasmic reticulum stress: Increasing the efficacy of taxane against prostate cancer. Cancer Biol. Ther. 2009, 8, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Rodvold, J.J.; Chiu, K.T.; Hiramatsu, N.; Nussbacher, J.K.; Galimberti, V.; Mahadevan, N.R.; Willert, K.; Lin, J.H.; Zanetti, M. Intercellular transmission of the unfolded protein response promotes survival and drug resistance in cancer cells. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Avril, T.; Vauléon, E.; Chevet, E. Endoplasmic reticulum stress signaling and chemotherapy resistance in solid cancers. Oncogenesis 2017, 6, e373. [Google Scholar] [CrossRef] [PubMed]
- Seve, P.; Mackey, J.; Isaac, S.; Tredan, O.; Souquet, P.-J.; Perol, M.; Lai, R.; Voloch, A.; Dumontet, C. Class III β-tubulin expression in tumor cells predicts response and outcome in patients with non-small cell lung cancer receiving paclitaxel. Mol. Cancer Ther. 2005, 4, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Nami, B.; Wang, Z. HER2 in breast cancer stemness: A negative feedback loop towards trastuzumab resistance. Cancers 2017, 9. [Google Scholar] [CrossRef]
- Prieto-Vila, M.; Takahashi, R.; Usuba, W.; Kohama, I.; Ochiya, T. Drug resistance driven by cancer stem cells and their niche. Int. J. Mol. Sci. 2017, 18, 2574. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, L.N.; Chow, E.K.-H. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Strasser, A. Is tumor growth sustained by rare cancer stem cells or dominant clones? Cancer Res. 2008, 68, 4018–4021. [Google Scholar] [CrossRef] [PubMed]
- Nami, B.; Ghasemi-Dizgah, A.; Vaseghi, A. Overexpression of molecular chaperons GRP78 and GRP94 in CD44hi/CD24lo breast cancer stem cells. Bioimpacts 2016, 6, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Fillmore, C.M.; Kuperwasser, C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Qiu, Q.; Khanna, A.; Todd, N.W.; Deepak, J.; Xing, L.; Wang, H.; Liu, Z.; Su, Y.; Stass, S.A.; et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol. Cancer Res. 2009, 7, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, S.; Wei, X. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine 2012, 7, 597–615. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat. Rev. 2018, 69, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Miragaya, J.; Palafox, M.; Paré, L.; Yoldi, G.; Ferrer, I.; Vila, S.; Galván, P.; Pellegrini, P.; Pérez-Montoyo, H.; Igea, A.; et al. Resistance to taxanes in triple-negative breast cancer associates with the dynamics of a CD49f+ tumor-initiating population. Stem Cell Reports 2017, 8, 1392–1407. [Google Scholar] [CrossRef] [PubMed]
- De Beca, F.F.; Caetano, P.; Gerhard, R.; Alvarenga, C.A.; Gomes, M.; Paredes, J.; Schmitt, F. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J. Clin. Pathol. 2013, 66, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitao, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, p11. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Lefebvre, C.; Bachelot, T.; Filleron, T.; Pedrero, M.; Campone, M.; Soria, J.-C.; Massard, C.; Levy, C.; Arnedos, M.; Lacroix-Triki, M.; et al. Mutational profile of metastatic breast cancers: A retrospective analysis. PLoS Med. 2016, 13, e1002201. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Zheng, R.; Zang, C.; Zhu, M.; Wu, J.; Shi, X.; Taing, L.; et al. Cistrome data browser: A data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017, 45, D658–D662. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
| GEO Dataset | GEO Samples | Data Type | Platform | Ref. |
|---|---|---|---|---|
| GSE41313 | GSM1014277, GSM1014278, GSM1014279, GSM1014371, GSM1014372, GSM1014373 GSM1014322, GSM1014323, GSM1014289, GSM1014290, GSM1014291 | Log2 RMA signal of array expression profiling for MCF-7, ZR-75-30, SKBR-3 and MD-MB-231 cell lines. | Affymetrix HT HG-U133+ PM Array Plate | [67] |
| GSE22796 | All samples | Log2 quantile normalized signal of beadchip expression profiling for 8 normal breast and 16 BC tumor tissues. | Illumina HumanRef-8 v3.0 expression beadchip | [68] |
| GSE99225 | All samples | Log10 DEVA 1.2 software compute signal intensities of array expression profiling for 2 taxane-sensitive and 2 taxane-resistant BC tumors. | NimbleGen Homo sapiens HG18 expression array | N/A |
| GSE22513 | All samples | Log2 RMA signal of array expression profiling for 4 BC tumors from patients with pathologic complete response (pCR) and 13 BC tumors from patients with non-pCR. | Affymetrix Human Genome U133 Plus 2.0 Array | [69,70] |
| GSE12791 | GSM320837, GSM320838, GSM320841, GSM320842, GSM320845, GSM320846, GSM320849, GSM320850 | Log10 MAS 5.0 signal intensity of array expression profiling for paclitaxel-sensitive parental and paclitaxel-resistant MDA-MB-231 cells. | Affymetrix Human Genome U133A Array | [71] |
| GSE81714 | GSM2171845, GSM2171846, GSM2171847, GSM2171848 | ChIP-seq signal profiling for H3K4me3 in MCF-7 cell line. | Illumina Genome Analyzer II | [72] |
| GSE71327 | GSM1832644, GSM2029585 | ChIP-seq signal profiling for H3K4me3 in ZR-75-30 cell line. | Illumina HiSeq 2500 | [73] |
| GSE62966 | GSM1537290, GSM1537285 | ChIP-seq signal profiling for H3K4me3 in SKBR-3 cell line. | Illumina HiSeq 2000/2500 | [74] |
| GSE49651 | GSM1204472, GSM1204473 | ChIP-seq signal profiling for H3K4me3 in MDA-MB-231 cell line. | Illumina HiSeq 1000 | [75] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nami, B.; Wang, Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 2018, 10, 274. https://doi.org/10.3390/cancers10080274
Nami B, Wang Z. Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers. 2018; 10(8):274. https://doi.org/10.3390/cancers10080274
Chicago/Turabian StyleNami, Babak, and Zhixiang Wang. 2018. "Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance" Cancers 10, no. 8: 274. https://doi.org/10.3390/cancers10080274
APA StyleNami, B., & Wang, Z. (2018). Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers, 10(8), 274. https://doi.org/10.3390/cancers10080274






