POFUT1 as a Promising Novel Biomarker of Colorectal Cancer
Abstract
1. Introduction
2. Results
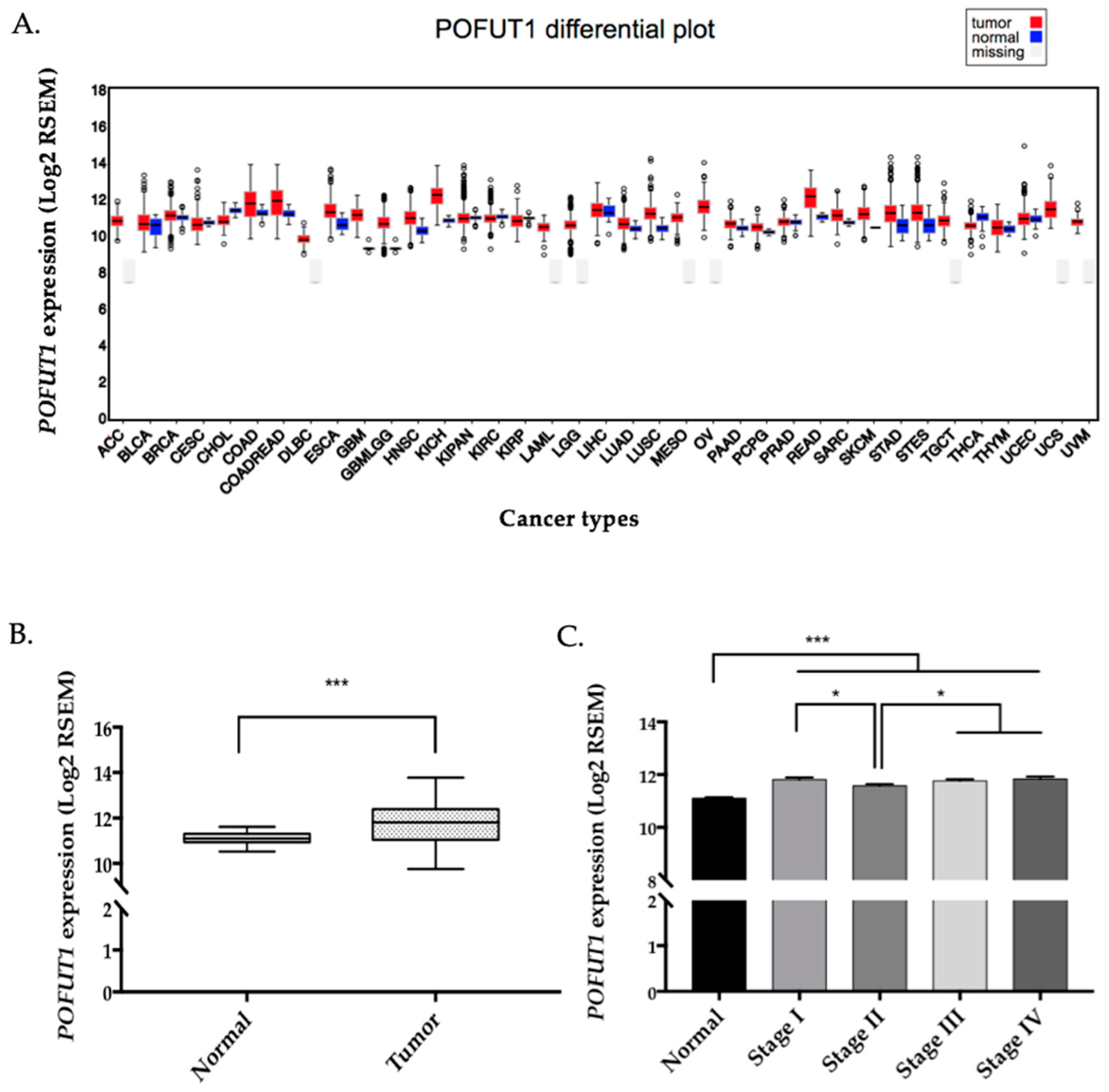
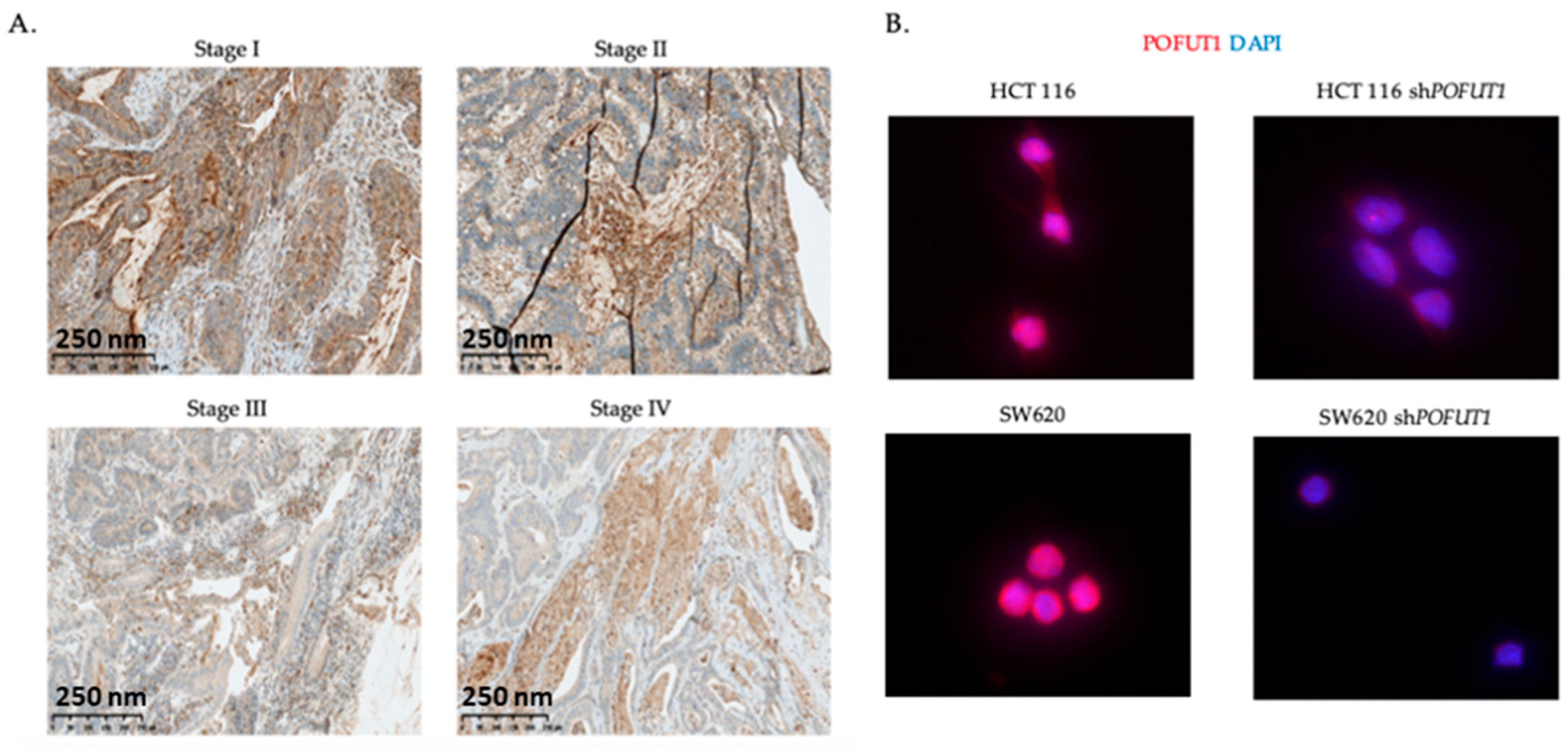
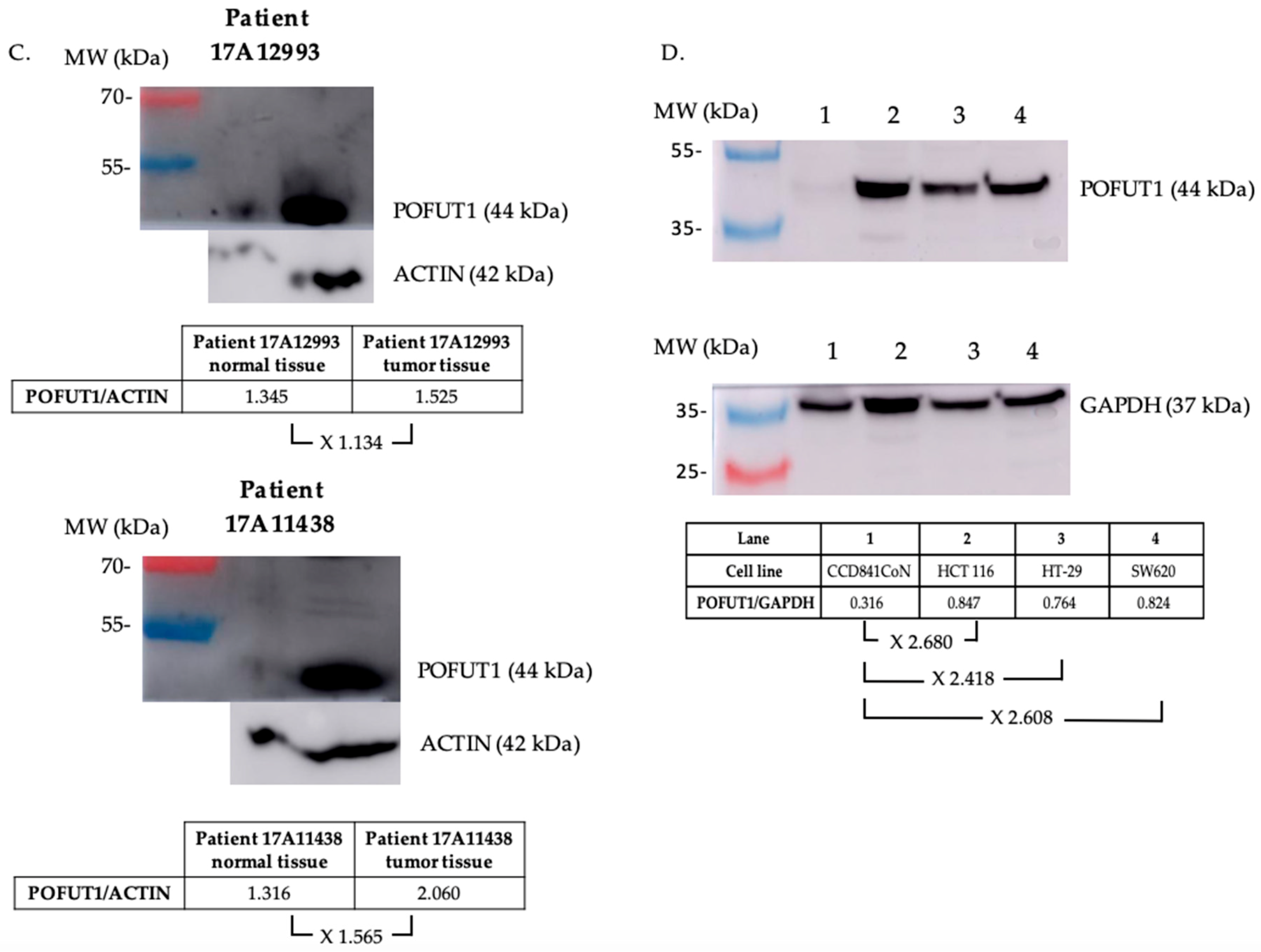
2.1. POFUT1 is Overexpressed in Human Colorectal Cancer Tissues
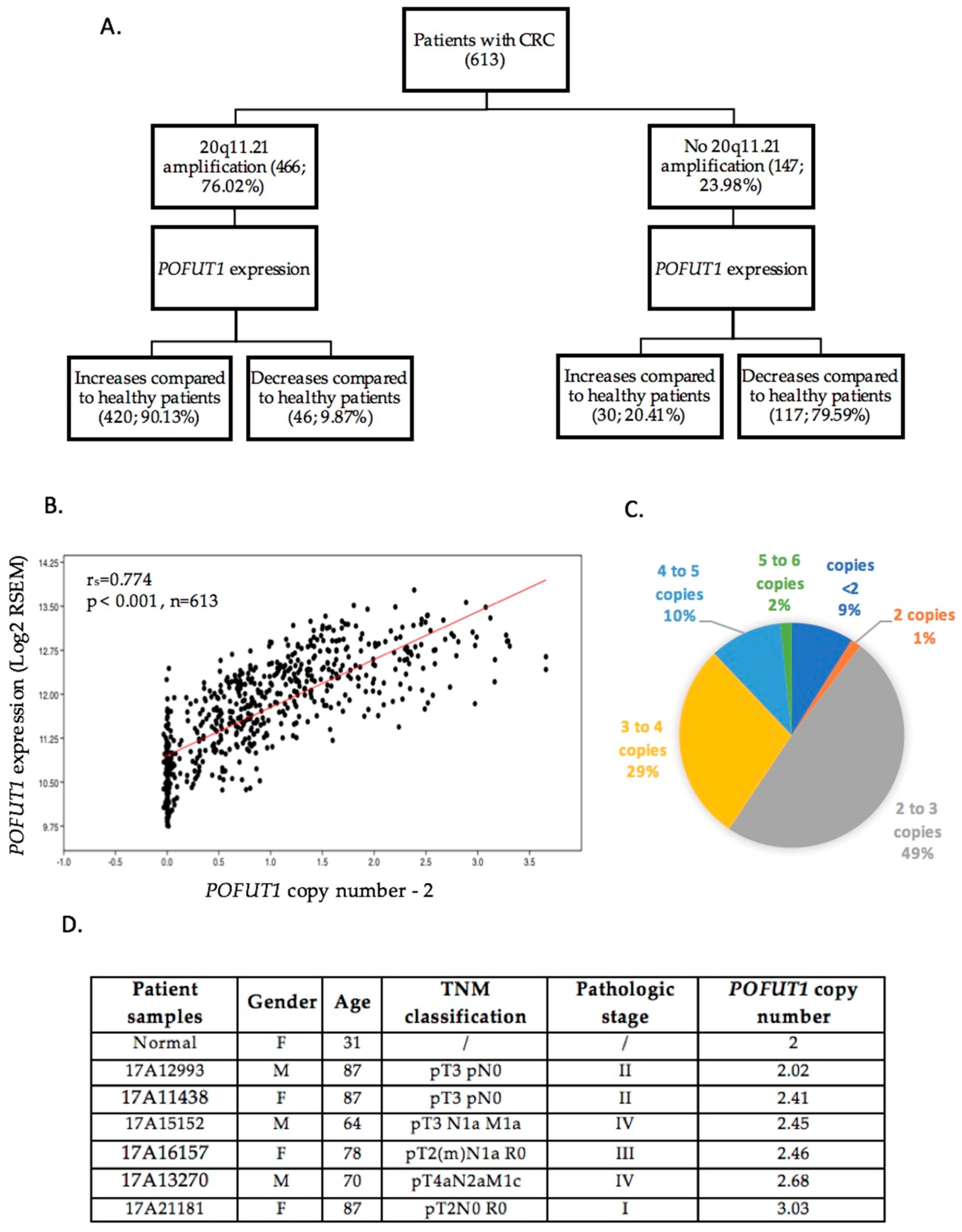
2.2. In CRC, 20q11.21 Chromosomic Region is Often Amplified, Which Induced POFUT1 Copy Number Alteration
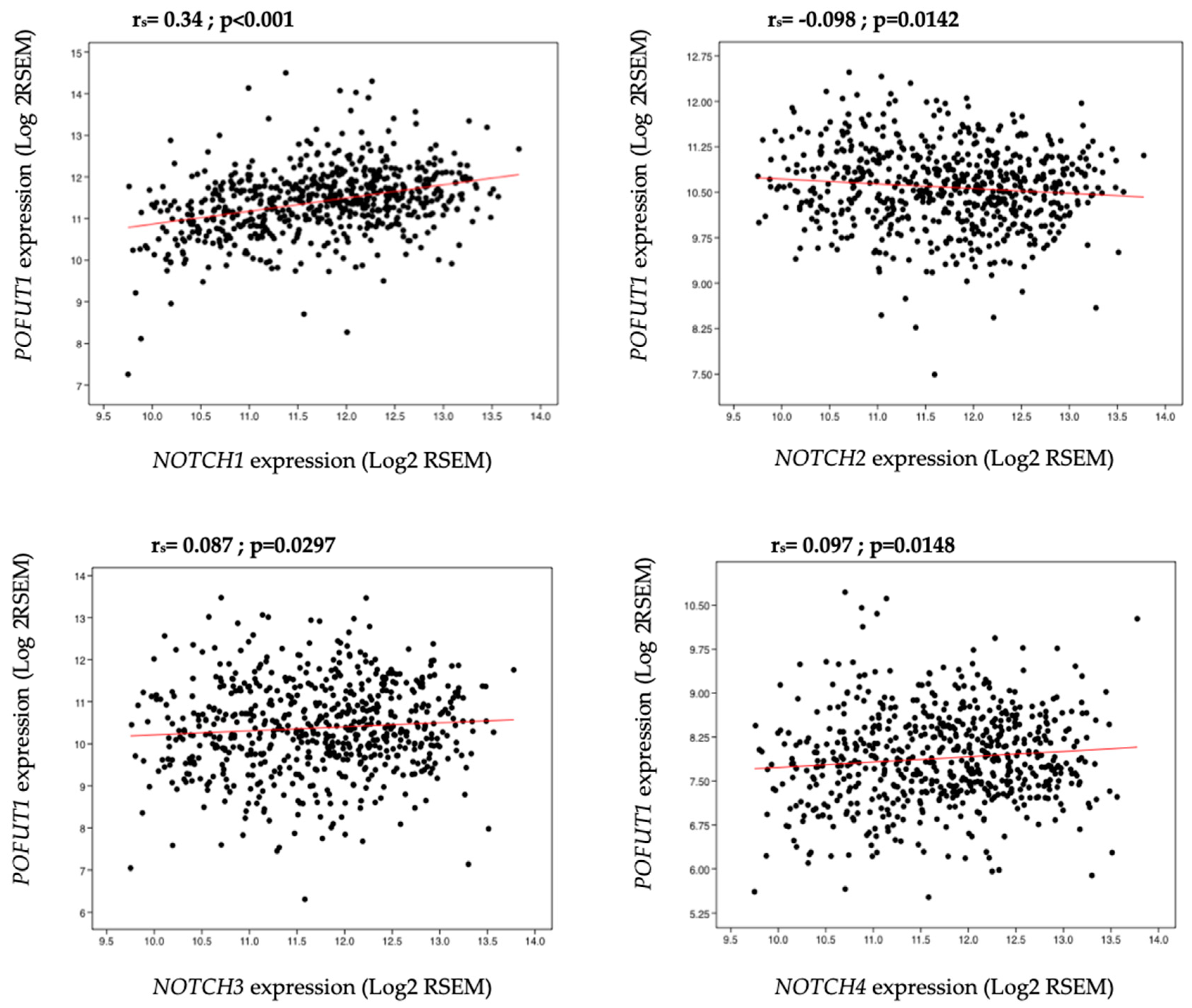
2.3. Correlation between POFUT1 and NOTCH Receptor Expressions
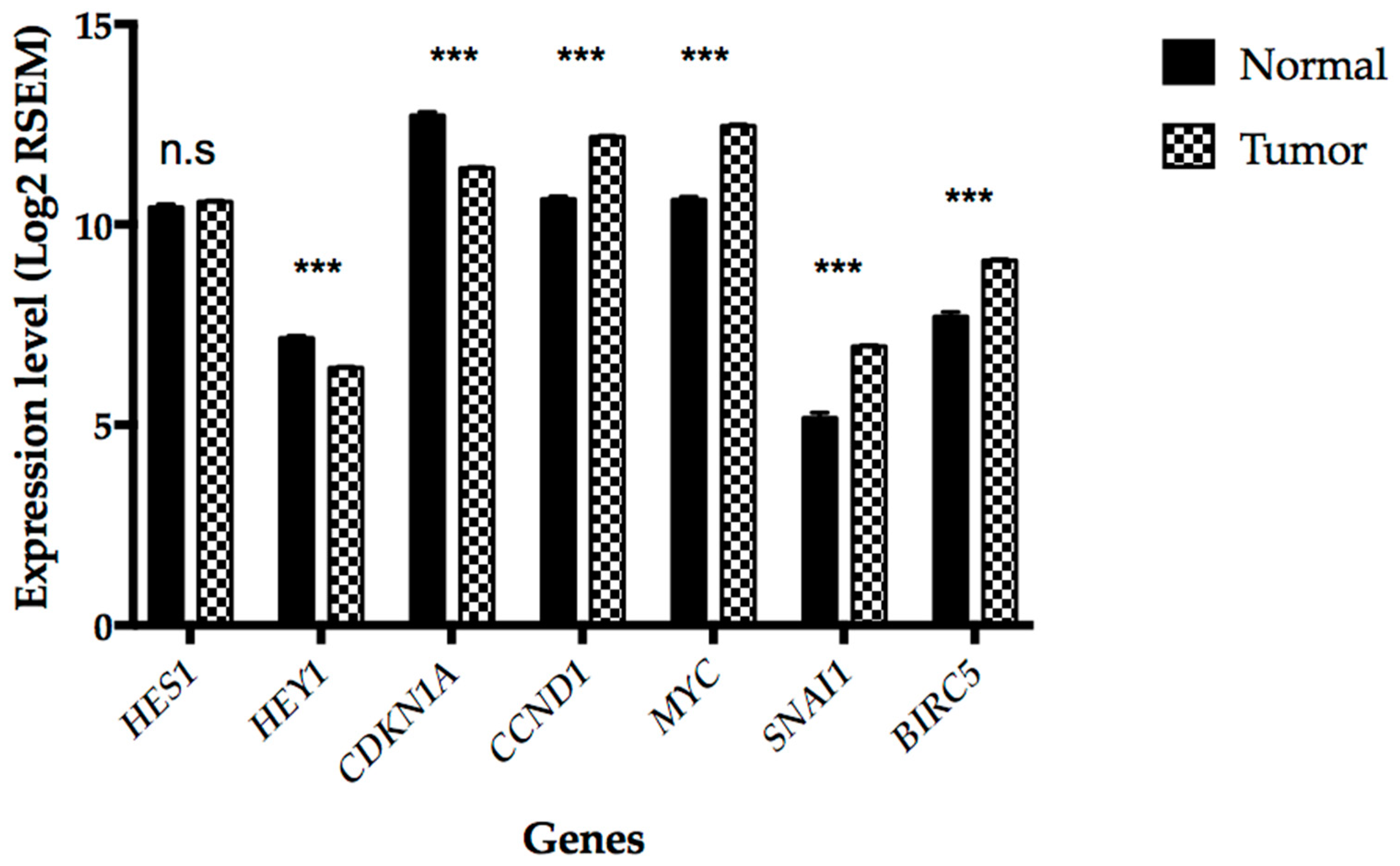
2.4. NOTCH Signaling Pathway is Deregulated in CRC
2.5. Relationship between POFUT1, NOTCH1 Expressions, and Clinical Features
3. Discussion
4. Materials and Methods
4.1. The Cancer Genome Atlas Data Analysis
4.2. Statistical Analysis
4.3. Genomic DNA Extraction and POFUT1 Copy Number Analysis
4.4. POFUT1 Labelling by Immunohistochemistry
4.5. POFUT1 Labeling by Immunofluorescence
4.6. Protein Extraction and Western Blot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524. [Google Scholar] [CrossRef] [PubMed]
- Hisamuddin, I.M.; Yang, V.W. Genetics of Colorectal Cancer. Med. Gen. Med. 2004, 6, 13. [Google Scholar]
- Dennis, J.W.; Granovsky, M.; Warren, C.E. Protein glycosylation in development and disease. BioEssays 1999, 21, 412–421. [Google Scholar] [CrossRef]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Shental-Bechor, D.; Levy, Y. Folding of glycoproteins: Toward understanding the biophysics of the glycosylation code. Curr. Opin. Struct. Biol. 2009, 19, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Freeze, H.H.; Stanley, P.; Bertozzi, C.R.; Hart, G.W.; Etzler, M.E. Essentials of Glycobiology, 2nd ed.; Chapter 6; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; ISBN 978-0-87969-770-9. [Google Scholar]
- Dennis, J.W.; Nabi, I.R.; Demetriou, M. Metabolism, Cell Surface Organization, and Disease. Cell 2009, 139, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Natsuka, S.; Lowe, J.B. Enzymes involved in mammalian oligosaccharide biosynthesis. Curr. Opin. Struct. Biol. 1994, 4, 683–691. [Google Scholar] [CrossRef]
- Kansas, G.S. Selectins and their ligands: Current concepts and controversies. Blood 1996, 88, 3259–3287. [Google Scholar] [PubMed]
- Aplin, J.D.; Jones, C.J. Fucose, placental evolution and the glycocode. Glycobiology 2012, 22, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Muinelo-Romay, L.; Vázquez-Martín, C.; Villar-Portela, S.; Cuevas, E.; Gil-Martín, E.; Fernández-Briera, A. Expression and enzyme activity of α(1,6)fucosyltransferase in human colorectal cancer: α(1,6)FT in Human CRC. Int. J. Cancer 2008, 123, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Gao, C.; Li, Y.; Sun, M.; Xu, J.; Li, H.; Jia, L.; Zhao, Y. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017, 8, e2968. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Nita-Lazar, A.; Haltiwanger, R.S. Two Distinct Pathways for O-fucosylation of Epidermal Growth Factor-like or Thrombospondin Type 1 Repeats. J. Biol. Chem. 2006, 281, 9385–9392. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Huang, Y.; Huang, X.; Wang, W.; Yan, Q.; Wei, L.; Xin, W.; Gerson, S.; Stanley, P.; Lowe, J.B.; et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood 2011, 117, 5652–5662. [Google Scholar] [CrossRef] [PubMed]
- Der Vartanian, A.; Audfray, A.; Al Jaam, B.; Janot, M.; Legardinier, S.; Maftah, A.; Germot, A. Protein O-fucosyltransferase 1 Expression Impacts Myogenic C2C12 Cell Commitment via the Notch Signaling Pathway. Mol. Cell. Biol. 2015, 35, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shao, L.; Shi, S.; Harris, R.J.; Spellman, M.W.; Stanley, P.; Haltiwanger, R.S. Modification of Epidermal Growth Factor-like Repeats with O-fucose: Molecular and expression of a novel GDP-fucose protein O-fucosyltransferase. J. Biol. Chem. 2001, 276, 40338–40345. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Haltiwanger, R.S. O-fucosylation of Notch Occurs in the Endoplasmic Reticulum. J. Biol. Chem. 2005, 280, 11289–11294. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Spellman, M.W. Purification and characterization of a GDP-fucose: Polypeptide fucosyltransferase from Chinese hamster ovary cells. J. Biol. Chem. 1998, 273, 8112–8118. [Google Scholar] [CrossRef] [PubMed]
- Rampal, R.; Luther, K.; Haltiwanger, R. Notch Signaling in Normal and Disease States: Possible Therapies Related to Glycosylation. Curr. Mol. Med. 2007, 7, 427–445. [Google Scholar] [CrossRef] [PubMed]
- Jafar-Nejad, H.; Leonardi, J.; Fernandez-Valdivia, R. Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology 2010, 20, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Muskavitch, M.A.T. Notch: The Past, the Present, and the Future. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 92, pp. 1–29. ISBN 978-0-12-380914-8. [Google Scholar]
- Shi, S.; Stanley, P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 2003, 100, 5234–5239. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Yu, H.; Hao, H.; Takeuchi, M.; Ito, A.; Li, H.; Haltiwanger, R.S. O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. J. Biol. Chem. 2017, 292, 15964–15973. [Google Scholar] [CrossRef] [PubMed]
- Stahl, M.; Uemura, K.; Ge, C.; Shi, S.; Tashima, Y.; Stanley, P. Roles of Pofut1 and Fucose in Mammalian Notch Signaling. J. Biol. Chem. 2008, 283, 13638–13651. [Google Scholar] [CrossRef] [PubMed]
- Weng, A.P. Activating Mutations of NOTCH1 in Human T Cell Acute Lymphoblastic Leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Göthert, J.R.; Brake, R.L.; Smeets, M.; Dührsen, U.; Begley, C.G.; Izon, D.J. NOTCH1 pathway activation is an early hallmark of SCL T leukemogenesis. Blood 2007, 110, 3753–3762. [Google Scholar] [CrossRef] [PubMed]
- Bolós, V.; Mira, E.; Martínez-Poveda, B.; Luxán, G.; Cañamero, M.; Martínez-A, C.; Mañes, S.; de la Pompa, J.L. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. 2013, 15, R54. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.-W.; Hsieh, R.-H.; Huang, K.-H.; Fen-Yau Li, A.; Chi, C.-W.; Wang, T.-Y.; Tseng, M.-J.; Wu, K.-J.; Yeh, T.-S. Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression. Carcinogenesis 2012, 33, 1459–1467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.; Ji, Z.-Z.; Zheng, P.-S. Notch1 regulates the growth of human colon cancers. Cancer 2010, 116, 5207–5218. [Google Scholar] [CrossRef] [PubMed]
- Kroes, R.A.; Dawson, G.; Moskal, J.R. Focused microarray analysis of glyco-gene expression in human glioblastomas. J. Neurochem. 2007, 103, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Ogawara, K.; Kimura, R.; Shimizu, F.; Baba, T.; Minakawa, Y.; Higo, M.; Kasamatsu, A.; Endo-Sakamoto, Y.; Shiiba, M.; et al. Protein O-fucosyltransferase 1: A potential diagnostic marker and therapeutic target for human oral cancer. Int. J. Oncol. 2013, 43, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Dong, P.; Liu, L.; Gao, Q.; Duan, M.; Zhang, S.; Chen, S.; Xue, R.; Wang, X. Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochem. Biophys. Res. Commun. 2016, 473, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wang, Z.; Huang, B.; Zhang, J.; Ge, Y.; Fan, Q.; Wang, Z. Bioinformatics insight into glycosyltransferase gene expression in gastric cancer: POFUT1 is a potential biomarker. Biochem. Biophys. Res. Commun. 2017, 483, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Kallioniemi, A.; Kallioniemi, O.; Sudar, D.; Rutovitz, D.; Gray, J.; Waldman, F.; Pinkel, D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Noguchi, T.; Kawahara, K.; Kashima, K.; Daa, T.; Yokoyama, S. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: Gain of 20q and loss of 18q are associated with tumor progression. Mod. Pathol. 2004, 17, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, R.N.; Selan, C.; Wall, M.; Baker, E.; Nandurkar, H.; Campbell, L.J. The paradox of 20q11.21 amplification in a subset of cases of myeloid malignancy with chromosome 20 deletion. Genes Chromosom. Cancer 2010, 49, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Postma, C.; Hermsen, M.; Coffa, J.; Baak, J.; Mueller, J.; Mueller, E.; Bethke, B.; Schouten, J.; Stolte, M.; Meijer, G. Chromosomal instability in flat adenomas and carcinomas of the colon. J. Pathol. 2005, 205, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.W.M.; Tiirikainen, M.; Cheng, I.; Lum-Jones, A.; Seifried, A.; Church, J.M.; Gryfe, R.; Weisenberger, D.J.; Lindor, N.M.; Gallinger, S.; et al. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, CpG island methylator phenotype-negative colon cancer. Genes Chromosom. Cancer 2013, 52, 450–466. [Google Scholar] [CrossRef] [PubMed]
- Loriol, C.; Audfray, A.; Dupuy, F.; Germot, A.; Maftah, A. The two N-glycans present on bovine Pofut1 are differently involved in its solubility and activity. FEBS J. 2007, 274, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Schuster-Gossler, K.; Harris, B.; Johnson, K.R.; Serth, J.; Gossler, A. Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC Dev. Biol. 2009, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Haltiwanger, R.S. Role of glycosylation of Notch in development. Semin. Cell Dev. Biol. 2010, 21, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lee, G.F.; Kelley, R.F.; Spellman, M.W. Identification of a GDP-L-fucose: Polypeptide fucosyltransferase and enzymatic addition of O-linked fucose to EGF domains. Glycobiology 1996, 6, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Al-Shareffi, E.; Haltiwanger, R.S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Tian, L.; Yu, Y.; Li, F.; Wang, X.; Li, C.; Deng, S.; Yu, X.; Cai, X.; Zuo, Z.; et al. Overexpression of Pofut1 and activated Notch1 may be associated with poor prognosis in breast cancer. Biochem. Biophys. Res. Commun. 2017, 491, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Lefort, N.; Feyeux, M.; Bas, C.; Féraud, O.; Bennaceur-Griscelli, A.; Tachdjian, G.; Peschanski, M.; Perrier, A.L. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat. Biotechnol. 2008, 26, 1364–1366. [Google Scholar] [CrossRef] [PubMed]
- Birkenkamp-Demtroder, K. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut 2005, 54, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Frattini, M. Different Genetic Features Associated with Colon and Rectal Carcinogenesis. Clin. Cancer Res. 2004, 10, 4015–4021. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, R.; Granato, D.C.; Carnielli, C.M.; Cervigne, N.K.; Oliveria, C.E.; Martinez, C.A.R.; Yokoo, S.; Fonseca, F.P.; Lopes, M.; Santos-Silva, A.R.; et al. Agrin and Perlecan Mediate Tumorigenic Processes in Oral Squamous Cell Carcinoma. PLoS ONE 2014, 9, e115004. [Google Scholar] [CrossRef] [PubMed]
- Hugen, N.; Brown, G.; Glynne-Jones, R.; de Wilt, J.H.W.; Nagtegaal, I.D. Advances in the care of patients with mucinous colorectal cancer. Nat. Rev. Clin. Oncol. 2016, 13, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Li, P.; Lu, F.; Liu, N.; Dai, J.; Ye, J.; Qu, X.; Sun, X.; Ma, D.; Park, J.; et al. Notch1 is required for hypoxia-induced proliferation, invasion and chemoresistance of T-cell acute lymphoblastic leukemia cells. J. Hematol. Oncol. 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, Y.; Wang, S.; Liu, Y.; Zheng, L.; Yang, J.; Huang, W.; Ye, Y.; Luo, W.; Xiao, D. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci. Rep. 2015, 4, 3963. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, M.; Andrici, J.; Sioson, L.; Sheen, A.; Clarkson, A.; Gill, A.J. Loss of Hes1 expression is associated with poor prognosis in colorectal adenocarcinoma. Hum. Pathol. 2016, 57, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Reedjik, M.; Odorcic, S.; Zhang, H.; Chetty, R.; Tennert, C.; Dickson, B.C.; Lockwood, G.; Gallinger, S.; Egan, S.E. Activation of Notch signaling in human colon adenocarcinoma. Int. J. Oncol. 1992. [Google Scholar] [CrossRef]
- Maier, M.M.; Gessler, M. Comparative Analysis of the Human and Mouse Hey1 Promoter: Hey Genes Are New Notch Target Genes. Biochem. Biophys. Res. Commun. 2000, 275, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Jarriault, S.; Brou, C.; Logeat, F.; Schroeter, E.H.; Kopan, R.; Israel, A. Signalling downstream of activated mammalian Notch. Nature 1995, 377, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Candy, P.A.; Phillips, M.R.; Redfern, A.D.; Colley, S.M.; Davidson, J.A.; Stuart, L.M.; Wood, B.A.; Zeps, N.; Leedman, P.J. Notch-induced transcription factors are predictive of survival and 5-fluorouracil response in colorectal cancer patients. Br. J. Cancer 2013, 109, 1023–1030. [Google Scholar] [CrossRef] [PubMed]
- Hulleman, E.; Quarto, M.; Vernell, R.; Masserdotti, G.; Colli, E.; Kros, J.M.; Levi, D.; Gaetani, P.; Tunici, P.; Finocchiaro, G.; et al. A role for the transcription factor HEY1 in glioblastoma. J. Cell. Mol. Med. 2008, 13, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Kakuda, S.; Haltiwanger, R.S. Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands. Dev. Cell 2017, 40, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Okajima, T.; Xu, A.; Lei, L.; Irvine, K.D. Chaperone Activity of Protein O-fucosyltransferase 1 Promotes Notch Receptor Folding. Science 2005, 307, 1599–1603. [Google Scholar] [CrossRef] [PubMed]
- Ajima, R.; Suzuki, E.; Saga, Y. Pofut1 point-mutations that disrupt O-fucosyltransferase activity destabilize the protein and abolish Notch1 signaling during mouse somitogenesis. PLoS ONE 2017, 12, e0187248. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Gao, X.-M.; Winbanks, C.E.; Boey, E.J.H.; Tham, Y.K.; Kiriazis, H.; Gregorevic, P.; Obad, S.; Kauppinen, S.; Du, X.-J.; et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. USA 2012, 109, 17615–17620. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Clinical Parameters | N | POFUT1 | p Value | |
|---|---|---|---|---|
| High (%) | Low (%) | |||
| Gender | ||||
| Female | 290 | 203 (70.0) | 87 (30.0) | 0.10206 |
| Male | 331 | 251 (75.8) | 80 (24.2) | |
| Age (years) | ||||
| ≤60 | 193 | 146 (75.6) | 47 (24.4) | 0.38296 |
| >60 | 426 | 308 (72.3) | 118 (27.7) | |
| Tumor Issue Site | ||||
| Colon | 454 | 313 (68.9) | 141 (31.1) | 0.00010 |
| Rectum | 163 | 138 (84.6) | 25 (15.4) | |
| Pathologic Stage | ||||
| Stage I | 105 | 83 (79.0) | 22 (21.0) | 0.00019 |
| Stage II | 229 | 145 (63.3) | 84 (36.7) | |
| Stage III | 179 | 134 (74.9) | 45 (25.1) | |
| Stage IV | 88 | 75 (85.2) | 13 (14.8) | |
| Pathology T Stage | ||||
| T1 | 20 | 17 (85.0) | 3 (15.0) | 0.16581 |
| T2 | 105 | 84 (80.0) | 21 (20.0) | |
| T3 | 423 | 300 (70.9) | 123 (29.1) | |
| T4 | 70 | 50 (71.4) | 20 (28.6) | |
| Pathology N Stage | ||||
| N0 | 352 | 245 (69.6) | 107 (30.4) | 0.05423 |
| N1 | 150 | 120 (80.0) | 30 (20.0) | |
| N2 | 115 | 85 (73.9) | 30 (26.1) | |
| Pathology M Stage | ||||
| M0 | 460 | 325 (70.7) | 135 (29.3) | 0.01087 |
| M1 | 87 | 73 (83.9) | 14 (16.1) | |
| Histological Type | ||||
| Colon adenocarcinoma | 388 | 291 (75.0) | 97 (25.0) | 0.00001 |
| Colon mucinous adenocarcinoma | 62 | 19 (30.6) | 43 (69.4) | |
| Rectal adenocarcinoma | 147 | 130 (88.4) | 17 (11.6) | |
| Rectal mucinous adenocarcinoma | 13 | 6 (46.2) | 7 (53.8) | |
| Clinical Parameters | N | NOTCH1 | p Value | |
|---|---|---|---|---|
| High (%) | Low (%) | |||
| Gender | ||||
| Female | 290 | 214 (73.8) | 76 (26.2) | 0.74724 |
| Male | 331 | 248 (74.9) | 83 (25.1) | |
| Age (years) | ||||
| ≤60 | 193 | 141 (73.1) | 52 (26.9) | 0.58601 |
| >60 | 426 | 320 (75.1) | 106 (24.9) | |
| Tumor Issue Site | ||||
| Colon | 454 | 322 (70.9) | 132 (29.1) | 0.00099 |
| Rectum | 163 | 137 (84.0) | 26 (16.0) | |
| Pathologic Stage | ||||
| Stage I | 105 | 80 (76.2) | 25 (23.8) | 0.59717 |
| Stage II | 229 | 163 (71.2) | 66 (28.8) | |
| Stage III | 179 | 135 (75.4) | 44 (24.6) | |
| Stage IV | 88 | 68 (77.3) | 20 (22.7) | |
| Pathology T Stage | ||||
| T1 | 20 | 11 (55.0) | 9 (45.0) | 0.16062 |
| T2 | 105 | 79 (75.2) | 26 (24.8) | |
| T3 | 423 | 313 (74.0) | 110 (26.0) | |
| T4 | 70 | 56 (80.0) | 14 (20.0) | |
| Pathology N Stage | ||||
| N0 | 352 | 257 (73.0) | 95 (27.0) | 0.66047 |
| N1 | 150 | 114 (76.0) | 36 (24.0) | |
| N2 | 115 | 88 (76.5) | 27 (23.5) | |
| Pathology M Stage | ||||
| M0 | 460 | 335 (72.8) | 125 (27.2) | 0.41731 |
| M1 | 87 | 67 (77.0) | 20 (23.0) | |
| Histological Type | ||||
| Colon adenocarcinoma | 388 | 284 (73.2) | 104 (26.8) | 0.00173 |
| Colon mucinous adenocarcinoma | 62 | 37 (59.7) | 25 (40.3) | |
| Rectal adenocarcinoma | 147 | 124 (84.4) | 23 (15.6) | |
| Rectal mucinous adenocarcinoma | 13 | 10 (76.9) | 3 (23.1) | |
| Clinical Parameters | N | POFUT1/NOTCH1 | p Value | |||
|---|---|---|---|---|---|---|
| High/High (%) | Low/Low (%) | High/Low (%) | Low/High (%) | |||
| Gender | ||||||
| Female | 290 | 169 (58.3) | 42 (14.5) | 34 (11.7) | 45 (15.5) | 0.4020 |
| Male | 331 | 205 (61.9) | 37 (11.2) | 46 (13.9) | 43 (13.0) | |
| Age (years) | ||||||
| ≤60 | 193 | 116 (60.1) | 22 (11.4) | 30 (15.5) | 25 (13.0) | 0.56050 |
| >60 | 426 | 258 (60.6) | 56 (13.1) | 50 (11.7) | 62 (14.6) | |
| Tumor Issue Site | ||||||
| Colon | 454 | 248 (54.6) | 67 (14.8) | 65 (14.3) | 74 (16.3) | 0.00004 |
| Rectum | 163 | 124 (76.1) | 12 (7.4) | 14 (8.6) | 13 (8.0) | |
| Pathologic Stage | ||||||
| Stage I | 105 | 68 (64.8) | 10 (9.5) | 15 (14.3) | 12 (11.4) | 0.00498 |
| Stage II | 229 | 123 (53.7) | 44 (19.2) | 22 (9.6) | 40 (17.5) | |
| Stage III | 179 | 107 (59.8) | 17 (9.5) | 27 (15.1) | 28 (15.6) | |
| Stage IV | 88 | 62 (70.5) | 7 (8) | 13 (14.8) | 6 (6.8) | |
| Pathology T Stage | ||||||
| T1 | 20 | 10 (50.0) | 2 (10.0) | 7 (35.0) | 1 (5.0) | 0.09107 |
| T2 | 105 | 68 (64.8) | 10 (9.5) | 16 (15.2) | 11 (10.5) | |
| T3 | 423 | 250 (59.1) | 60 (14.2) | 50 (11.8) | 63 (14.9) | |
| T4 | 70 | 43 (61.4) | 7 (10.0) | 7 (10.0) | 13 (18.6) | |
| Pathology N Stage | ||||||
| N0 | 352 | 204 (58.0) | 54 (15.3) | 41 (11.6) | 53 (15.1) | 0.42789 |
| N1 | 150 | 97 (64.7) | 13 (8.7) | 23 (15.3) | 17 (11.3) | |
| N2 | 115 | 70 (60.9) | 12 (10.4) | 15 (13.0) | 18 (15.7) | |
| Pathology M Stage | ||||||
| M0 | 460 | 266 (57.8) | 66 (14.3) | 59 (12.8) | 69 (15.0) | 0.07716 |
| M1 | 87 | 61 (70.1) | 8 (9.2) | 12 (13.8) | 6 (6.9) | |
| Histological Type | ||||||
| Colon adenocarcinoma | 388 | 235 (60.6) | 48 (12.4) | 56 (14.4) | 49 (12.6) | 1.5335 × 10−15 |
| Colon mucinous adenocarcinoma | 62 | 12 (19.4) | 18 (29.0) | 7 (11.3) | 25 (40.3) | |
| Rectal adenocarcinoma | 147 | 116 (78.9) | 9 (6.1) | 14 (9.5) | 8 (5.4) | |
| Rectal mucinous adenocarcinoma | 13 | 6 (46.2) | 3 (23.1) | 0 (0) | 4 (30.8) | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chabanais, J.; Labrousse, F.; Chaunavel, A.; Germot, A.; Maftah, A. POFUT1 as a Promising Novel Biomarker of Colorectal Cancer. Cancers 2018, 10, 411. https://doi.org/10.3390/cancers10110411
Chabanais J, Labrousse F, Chaunavel A, Germot A, Maftah A. POFUT1 as a Promising Novel Biomarker of Colorectal Cancer. Cancers. 2018; 10(11):411. https://doi.org/10.3390/cancers10110411
Chicago/Turabian StyleChabanais, Julien, François Labrousse, Alain Chaunavel, Agnès Germot, and Abderrahman Maftah. 2018. "POFUT1 as a Promising Novel Biomarker of Colorectal Cancer" Cancers 10, no. 11: 411. https://doi.org/10.3390/cancers10110411
APA StyleChabanais, J., Labrousse, F., Chaunavel, A., Germot, A., & Maftah, A. (2018). POFUT1 as a Promising Novel Biomarker of Colorectal Cancer. Cancers, 10(11), 411. https://doi.org/10.3390/cancers10110411





