Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis
Abstract
1. Introduction
2. Results
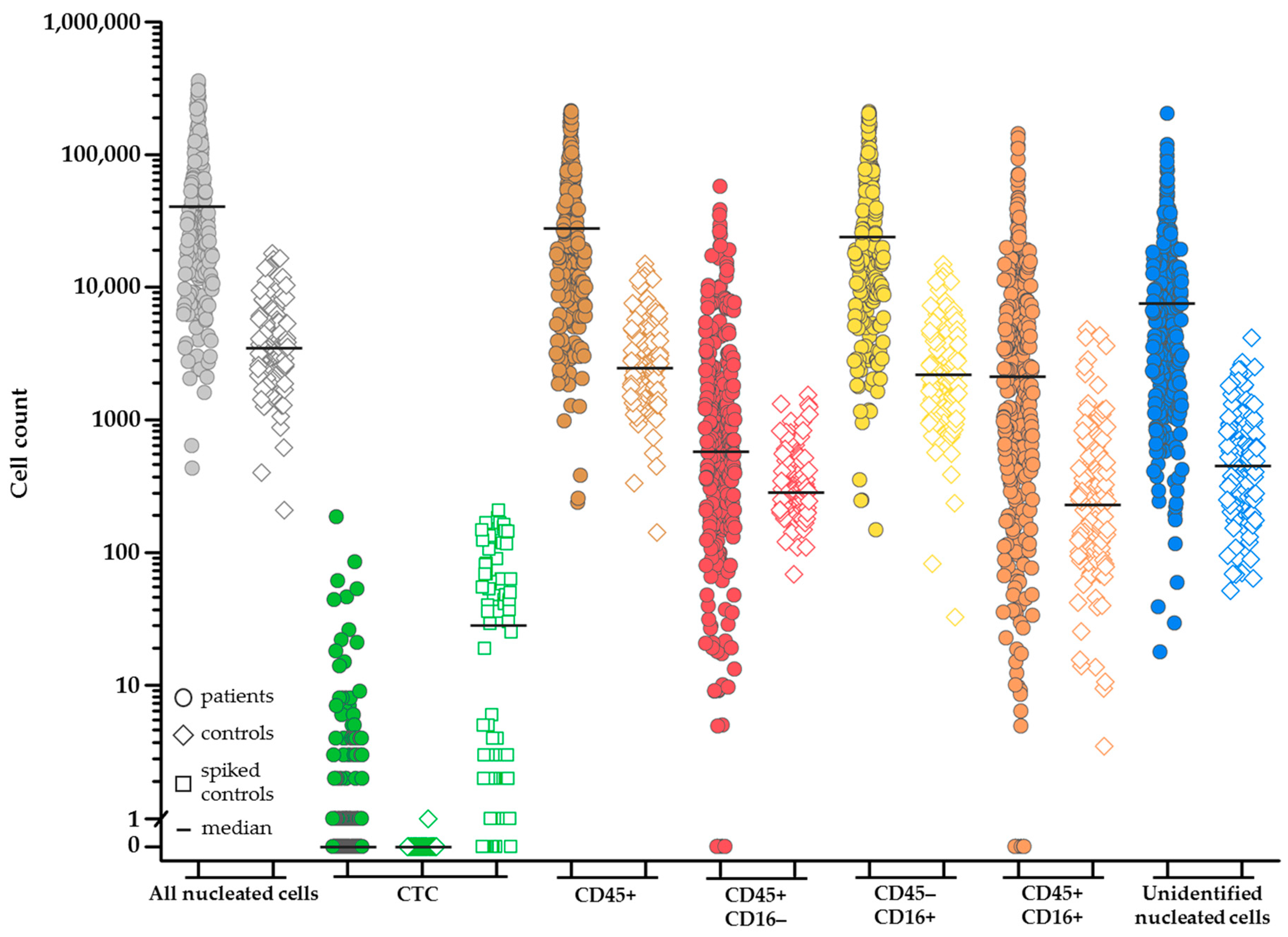
2.1. Enumeration of Nucleated Cells in the EpCAM-Enriched Cells
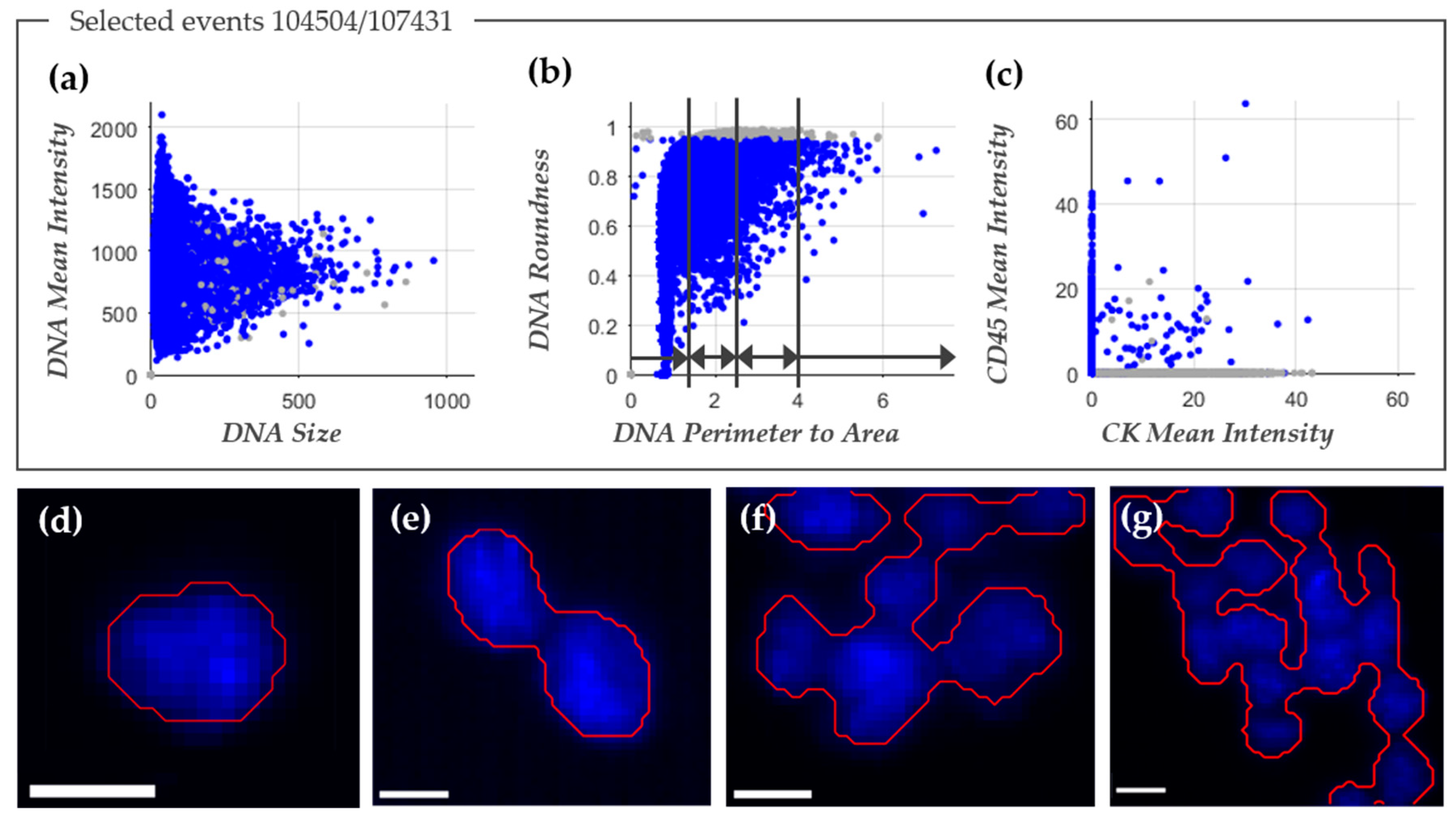
2.2. Improved Image Analysis
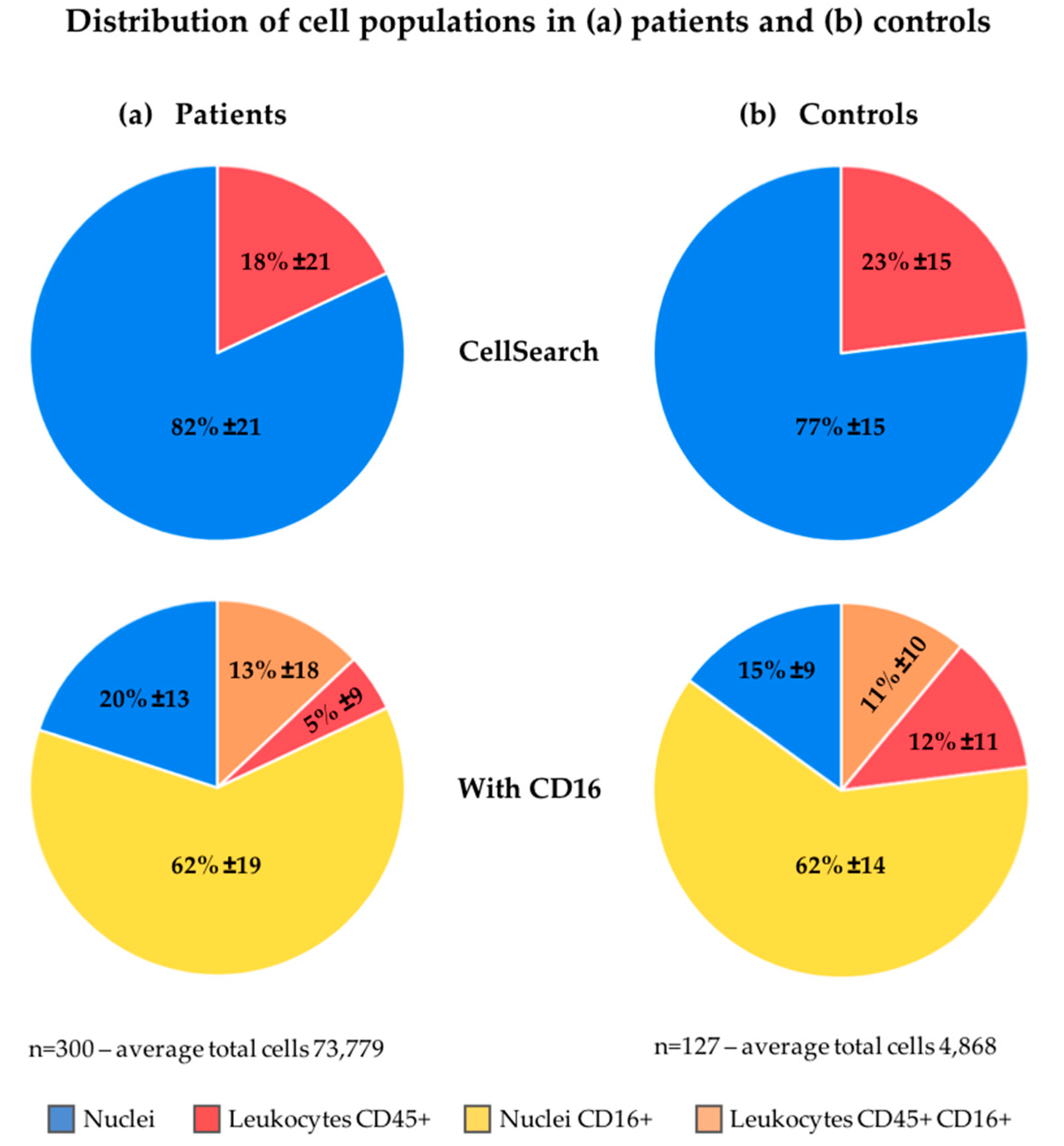
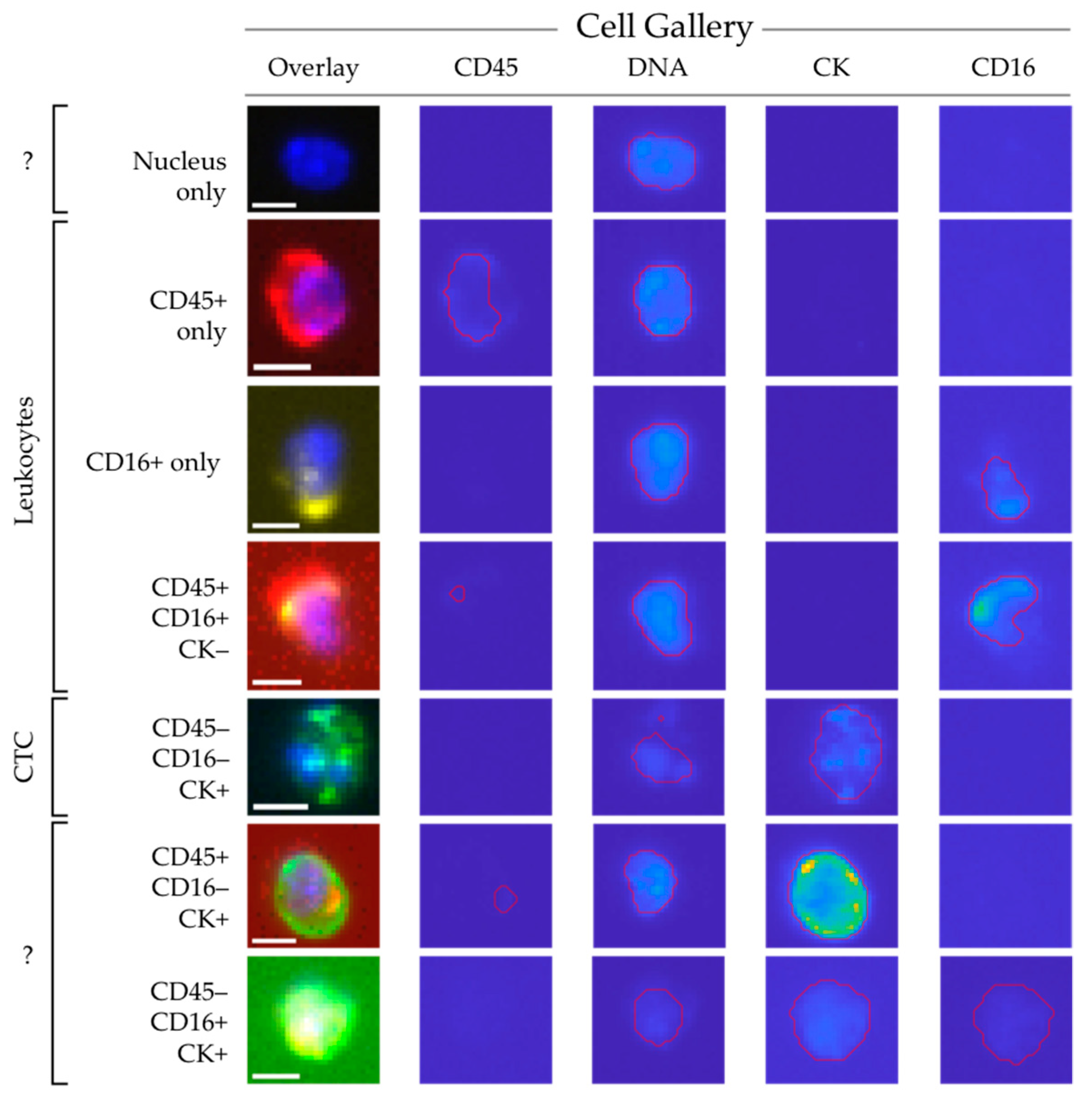
2.3. Assignment of Nucleated Cells to a Cell Lineage
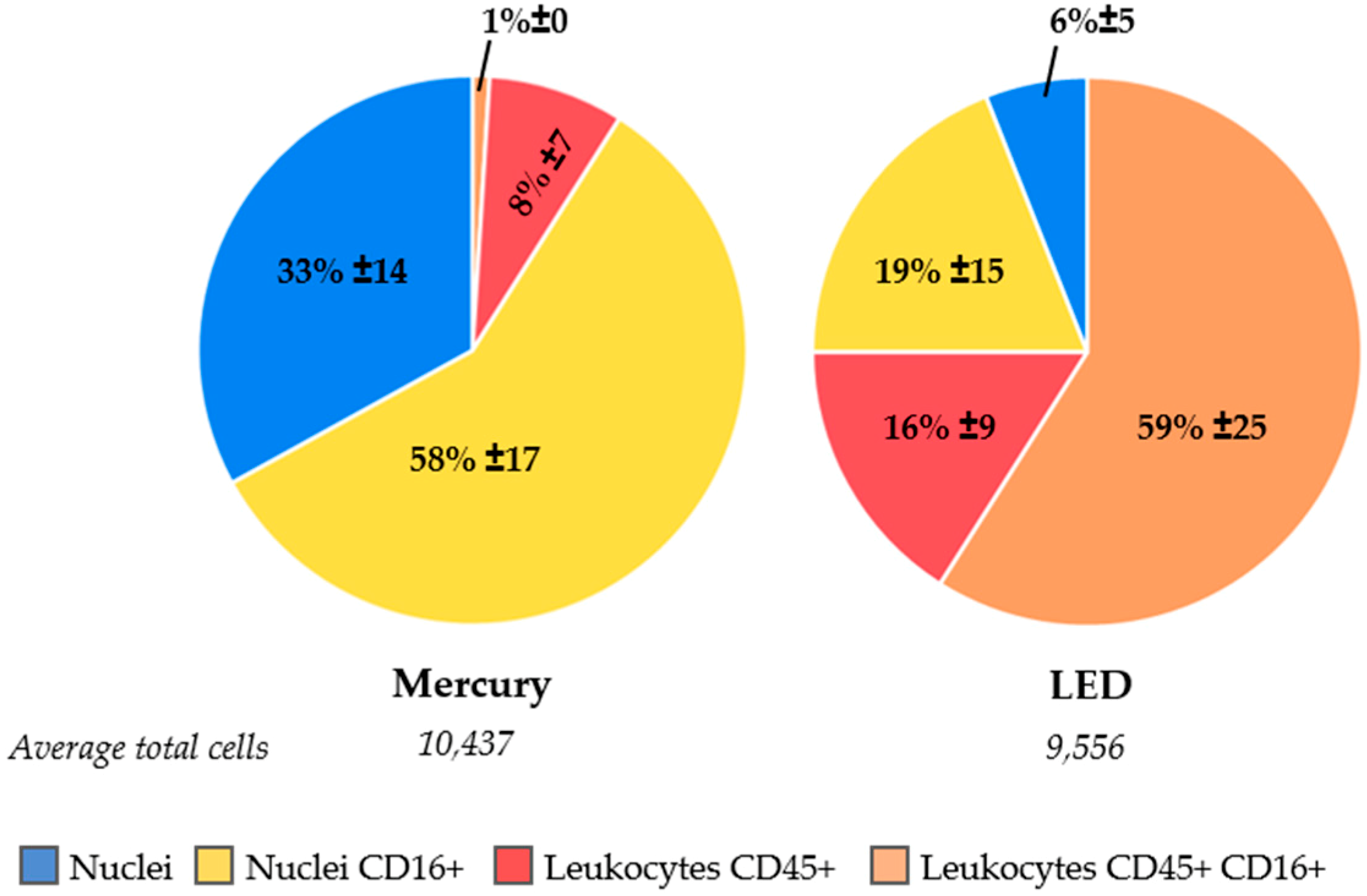
2.4. Increasing Leukocyte Identification by Adding CD16 Immunostaining
2.5. LED as a Light Source to Improve Excitation Efficiency of CD45-APC and CD16-PerCP
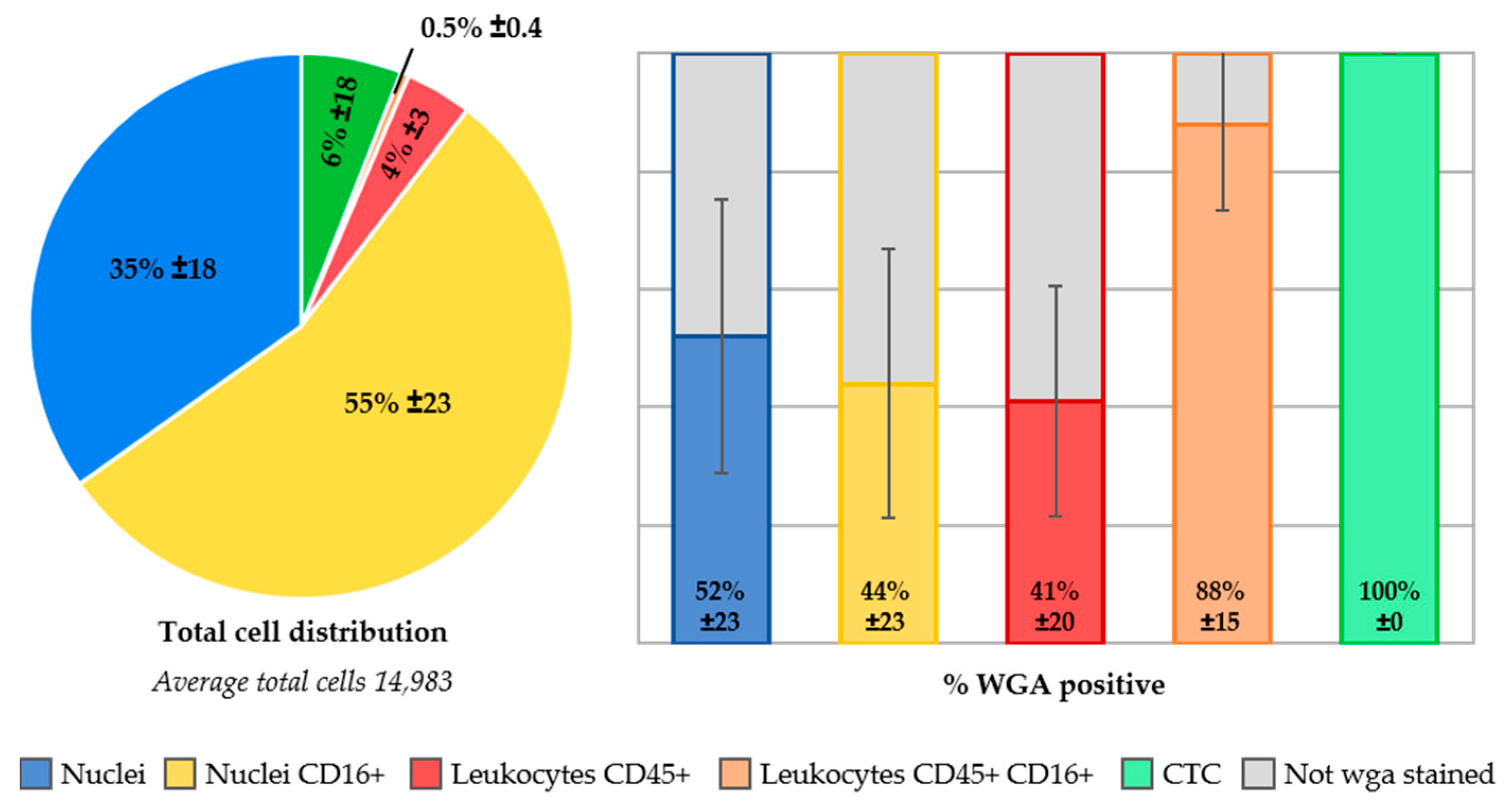
2.6. Identification of Unstained Nuclei by Adding Wheat Germ Agglutinin Immunostaining
3. Discussion
- Hematopoietic cells without sufficient expression of CD45 and CD16: This suggests they would need additional CD markers for identification or an improved labeling method that amplifies low signals, which might yield increased detection of very dim stained cells and separate the fluorescent signal of densely packed cells [11,26,27,28]. Early myeloid cells have recently been observed to surround the tumor in high numbers, and as these cells do not yet express CD16, this remains a possibility [29];
- CTC with no or low expression of the CK antigens detected by the C11 and A53.B/A2 clones: Since these clones only recognize a subset of the CK present in a cell, it might be beneficial to include antibody clones that recognize all CK. Previously, we have shown that adding several CK clones to the CellSearch antibody cocktail improved the detection of CTC positive patients by 11% [30]. Also, it might be possible that EpCAM+/CK- CTC are present, remaining undetected because of the downregulation of epithelial markers through epithelial-to-mesenchymal transition [31,32,33]. In order to detect these cells, antibodies specific to this process could be added to the CellSearch immunostaining [34,35,36];
- Cells of other origin: Such as circulating stromal, endothelial, or stem cells [37]. Detection of these cells would also require the addition of other antibodies to the assay.
4. Materials and Methods
4.1. Cancer Patients, Patients with Benign Disease, and Healthy Volunteers
4.2. Cell Lines and Spiking
4.3. Processing Blood with CellSearch
4.4. Image Acquisition
4.5. Image Analysis with ACCEPT
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Racila, E.; Euhus, D.; Weiss, A.J.; Rao, C.; McConnell, J.; Terstappen, L.W.M.M.; Uhr, J.W. Detection and characterization of carcinoma cells in the blood. Proc. Natl. Acad. Sci. USA 1998, 95, 4589–4594. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Hiltermann, T.J.N.; Pore, M.M.; Van den Berg, A.; Timens, W.; Boezen, H.M.; Liesker, J.J.W.; Schouwink, J.H.; Wijnands, W.J.A.; Kerner, G.S.M.A.; Kruyt, F.A.E.; et al. Circulating tumor cells in small-cell lung cancer: A predictive and prognostic factor. Ann. Oncol. 2012, 23, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and Prognostic Significance of Circulating Tumor Cells in Patients With Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Hiraiwa, K.; Takeuchi, H.; Hasegawa, H.; Saikawa, Y.; Suda, K.; Ando, T.; Kumagai, K.; Irino, T.; Yoshikawa, T.; Matsuda, S.; et al. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann. Surg. Oncol. 2008, 15, 3092–3100. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Bui, T.; Connelly, M.; Doyle, G.; Karydis, I.; Middleton, M.R.; Clack, G.; Malone, M.; Coumans, F.A.W.; Terstappen, L.W.M.M. Circulating melanoma cells and survival in metastatic melanoma. Int. J. Oncol. 2011, 38, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, P.; Gradilone, A.; De Berardinis, E.; Busetto, G.M.; Raimondi, C.; Gandini, O.; Nicolazzo, C.; Petracca, A.; Vincenzi, B.; Farcomeni, A.; et al. Prognostic value of circulating tumor cells in nonmuscle invasive bladder cancer: A CellSearch analysis. Ann. Oncol. 2012, 23, 2352–2356. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Van Dalum, G. The Role and Definition of Cancer Cells in Blood. Ph.D. Thesis, University of Twente, Enschede, The Netherlands, 2015. [Google Scholar]
- Lumencore, I. SOLA Light Engine. Available online: http://lumencor.com (accessed on 21 August 2018).
- Chazotte, B. Labeling Membrane Glycoproteins or Glycolipids with Fluorescent Wheat Germ Agglutinin. Cold Spring Harb. Protoc. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Legant, W.R.; Shao, L.; Grimm, J.B.; Brown, T.A.; Milkie, D.E.; Avants, B.B.; Lavis, L.D.; Betzig, E. High-density three-dimensional localization microscopy across large volumes. Nat. Methods 2016, 13, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Solórzano, C.; Bouquelet, S.; Pereyra, M.A.; Blanco-Favela, F.; Slomianny, M.C.; Chavez, R.; Lascurain, R.; Zenteno, E.; Agundis, C. Isolation and characterization of the potential receptor for wheat germ agglutinin from human neutrophils. Glycoconj. J. 2006, 23, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Sharon, N.; Lis, H. Lectins; Springer: New York, NY, USA, 2007; ISBN 978-1-4020-6953-6. [Google Scholar]
- Emde, B.; Heinen, A.; Gödecke, A.; Bottermann, K. Wheat germ agglutinin staining as a suitable method for detection and quantification of fibrosis in cardiac tissue after myocardial infarction. Eur. J. Histochem. 2014, 58, 2448. [Google Scholar] [CrossRef] [PubMed]
- Adair, W.L.; Kornfeld, S. Isolation of the receptors for wheat germ agglutinin and the Ricinus communis lectins from human erythrocytes using affinity chromatography. J. Biol. Chem. 1974, 249, 4696–4704. [Google Scholar]
- Zeune, L.L.; Van Dalum, G.; Terstappen, L.W.M.M.; Van Gils, S.A.; Brune, C. Multiscale Segmentation via Bregman Distances and Nonlinear Spectral Analysis. SIAM J. Imaging Sci. 2017, 10, 111–146. [Google Scholar] [CrossRef]
- Zeune, L.L.; De Wit, S.; Berghuis, A.M.S.; IJzerman, M.J.; Terstappen, L.W.M.M.; Brune, C. How to agree on a CTC: Evaluating the Consensus in Circulating Tumor Cell Scoring. Cytom. Part A 2018. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 21 August 2018).
- Zeune, L.L.; De Wit, S.; Van Dalum, G.; Nanou, A.; Andree, K.C.; Swennenhuis, J.F.; Terstappen, L.W.M.M.; Brune, C. Deep Learning to Identify Circulating Tumor Cells by ACCEPT. In Proceedings of the ISMRC, Montpellier, France, 3–5 May 2018. [Google Scholar]
- Scholtens, T.M. Automated Classification and Enhanced Characterization of Circulating Tumor Cells by Image Cytometry. Ph.D. Thesis, University of Twente, Enschede, The Netherlands, 2012. [Google Scholar]
- Coumans, F.A.W.; Terstappen, L.W.M.M. Detection and Characterization of Circulating Tumor Cells by the CellSearch Approach. Methods Mol. Biol. 2015, 1347, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Jamur, M.C.; Oliver, C. Permeabilization of Cell Membranes. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: New York, NY, USA, 2010; Volume 588, pp. 63–66. [Google Scholar]
- Faget, L.; Hnasko, T.S. Tyramide Signal Amplification for Immunofluorescent Enhancement. In Methods in Molecular Biology (Clifton, N.J.); Humana Press: New York, NY, USA, 2015; Volume 1318, pp. 161–172. [Google Scholar]
- Zhou, S.; Yuan, L.; Hua, X.; Xu, L.; Liu, S. Signal amplification strategies for DNA and protein detection based on polymeric nanocomposites and polymerization: A review. Anal. Chim. Acta 2015, 877, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Gustafsdottir, S.M.; Schallmeiner, E.; Fredriksson, S.; Gullberg, M.; Söderberg, O.; Jarvius, M.; Jarvius, J.; Howell, M.; Landegren, U. Proximity ligation assays for sensitive and specific protein analyses. Anal. Biochem. 2005, 345, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Spataro, C.; Zagato, E.; Di Mitri, D.; Gil, V.; Crespo, M.; De Bernardis, G.; Losa, M.; Mirenda, M.; Pasquini, E.; et al. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature 2018, 559, 363–369. [Google Scholar] [CrossRef] [PubMed]
- De Wit, S.; Van Dalum, G.; Lenferink, A.T.M.; Tibbe, A.G.J.; Hiltermann, T.J.N.; Groen, H.J.M.; Van Rijn, C.J.M.; Terstappen, L.W.M.M. The detection of EpCAM+ and EpCAM- circulating tumor cells. Sci. Rep. 2015, 5, 12270. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.J.; Ortega, F.G.; Alvarez-Cubero, M.J.; Nadal, R.; Sanchez-Rovira, P.; Salido, M.; Rodríguez, M.; García-Puche, J.L.; Delgado-Rodriguez, M.; Solé, F.; et al. EMT and EGFR in CTCs cytokeratin negative non-metastatic breast cancer. Oncotarget 2014, 5, 7486–7497. [Google Scholar] [CrossRef] [PubMed]
- Willipinski-Stapelfeldt, B.; Riethdorf, S.; Assmann, V.; Woelfle, U.; Rau, T.; Sauter, G.; Heukeshoven, J.; Pantel, K. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin. Cancer Res. 2005, 11, 8006–8014. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Hou, J.M.; Sloane, R.; Lancashire, L.; Priest, L.; Nonaka, D.; Ward, T.H.; Backen, A.; Clack, G.; Hughes, A.; et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J. Thorac. Oncol. 2012, 7, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13, R59. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating Tumor Cells from Patients with Advanced Prostate and Breast Cancer Display Both Epithelial and Mesenchymal Markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, S.; Liu, Z.; Huang, J.; Pu, X.; Li, J.; Yang, D.; Deng, H.; Yang, N.; Xu, J. Classification of Circulating Tumor Cells by Epithelial-Mesenchymal Transition Markers. PLoS ONE 2015, 10, e0123976. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, P.; Huang, P.H.; Xie, Y.; Mai, J.D.; Wang, L.; Nguyen, N.T.; Huang, T.J. Rare cell isolation and analysis in microfluidics. Lab Chip 2014, 14, 626–645. [Google Scholar] [CrossRef] [PubMed]
- Van Dalum, G.; Stam, G.J.; Scholten, L.F.A.; Mastboom, W.J.B.; Vermes, I.; Tibbe, A.G.J.; De Groot, M.R.; Terstappen, L.W.M.M. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int. J. Oncol. 2015, 46, 1361–1368. [Google Scholar] [CrossRef] [PubMed]
- Zeune, L.L.; Van Dalum, G.; Decraene, C.; Proudhon, C.; Fehm, T.; Neubauer, H.; Rack, B.; Alunni-Fabbroni, M.; Terstappen, L.W.M.M.; Van Gils, S.A.; Brune, C. Quantifying HER-2 expression on circulating tumor cells by ACCEPT. PLoS ONE 2017, 12, e0186562. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
| Analysis Type | Regression Coefficient | 95% Confidence Interval | p-Value |
|---|---|---|---|
| Crude analysis | 17.0 | 14.1–20.6 | <0.001 |
| Corrected analysis for age | 19.2 | 15.3–24.2 | <0.001 |
| Corrected analysis for age, and patient variables (sample age, CTC count and treatment) | 12.4 | 9.4–16.4 | <0.001 |
| Cell Population | Mean Intensity Value | Overlay Nucleus | Size (µm2) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DAPI | CD45 | CK | CD16 1 | wga 1 | with CK | with CD45 | with CD16 | CK | ||
| CTC | CK+/CD45- | >0 | 0 | ≥50 | 0 | >0 | >0.4 | ND | ND | >9 |
| Leukocytes | CD45+/CD16- | >0 | >0 | 0 | 0 | >0 | ND | >0 | ND | ND |
| CD45+/CD16+ | >0 | >0 | 0 | >0 | >0 | ND | >0 | >0 | ND | |
| CD45-/CD16+ | >0 | 0 | 0 | >0 | >0 | ND | ND | >0 | ND | |
| Nucleus | Bare nucleus | >0 | 0 | 0 | 0 | 0 | ND | ND | ND | ND |
| Unstained cell | >0 | 0 | 0 | 0 | >0 | ND | ND | ND | ND | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Wit, S.; Zeune, L.L.; Hiltermann, T.J.N.; Groen, H.J.M.; Dalum, G.V.; Terstappen, L.W.M.M. Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis. Cancers 2018, 10, 377. https://doi.org/10.3390/cancers10100377
De Wit S, Zeune LL, Hiltermann TJN, Groen HJM, Dalum GV, Terstappen LWMM. Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis. Cancers. 2018; 10(10):377. https://doi.org/10.3390/cancers10100377
Chicago/Turabian StyleDe Wit, Sanne, Leonie L. Zeune, T. Jeroen N. Hiltermann, Harry J. M. Groen, Guus Van Dalum, and Leon W. M. M. Terstappen. 2018. "Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis" Cancers 10, no. 10: 377. https://doi.org/10.3390/cancers10100377
APA StyleDe Wit, S., Zeune, L. L., Hiltermann, T. J. N., Groen, H. J. M., Dalum, G. V., & Terstappen, L. W. M. M. (2018). Classification of Cells in CTC-Enriched Samples by Advanced Image Analysis. Cancers, 10(10), 377. https://doi.org/10.3390/cancers10100377






