Abstract
Hereditary breast and ovarian cancer syndrome (HBOC) represents 5–10% of all patients with breast cancer and is associated with high-risk pathogenic alleles in BRCA1/2 genes, but only for 25% of cases. We aimed to find new pathogenic alleles in a panel of 143 cancer-predisposing genes in 300 Mexican cancer patients with suspicion of HBOC and 27 high-risk patients with a severe family history of cancer, using massive parallel sequencing. We found pathogenic variants in 23 genes, including BRCA1/2. In the group of cancer patients 15% (46/300) had a pathogenic variant; 11% (33/300) harbored variants with unknown clinical significance (VUS) and 74% (221/300) were negative. The high-risk group had 22% (6/27) of patients with pathogenic variants, 4% (1/27) had VUS and 74% (20/27) were negative. The most recurrent mutations were the Mexican founder deletion of exons 9-12 and the variant p.G228fs in BRCA1, each found in 5 of 17 patients with alterations in this gene. Rare VUS with potential impact at the protein level were found in 21 genes. Our results show for the first time in the Mexican population a higher contribution of pathogenic alleles in other susceptibility cancer genes (54%) than in BRCA1/2 (46%), highlighting the high locus heterogeneity of HBOC and the necessity of expanding genetic tests for this disease to include broader gene panels.
1. Introduction
Breast cancer (BC, OMIM#114480) is the most prevalent cancer in the world and accounts for 14.7 million of mortality cases [1]. Approximately, 10% of BC cases have a genetic, inherited etiology referred as Hereditary Breast and Ovarian Cancer (HBOC) with an important impact in genetic counseling and cancer prevention interventions [2].
Pathogenic variants in BRCA1 and BRCA2 genes are the most prevalent in HBOC, collectively contributing to 15–25% of the cases [3]. Pathogenic alleles in these genes frequently have high penetrance and have been found in different populations, including countries from Latin America, such as Mexico, Colombia, Argentina, Chile, Brazil, among others [4,5]. However, locus heterogeneity has been found in patients without mutations in BRCA1 and BRCA2 [6] together with additional pathogenic variants at lower frequency and in genes that confer moderate risk including CHEK2, PALB2, ATM, FANCM, ATR, STK11, RAD51C, BRIP1, CDH1, NF1, NBN and ERCC3 [7,8]. The prevalence of these novel, moderate-risk genes in HBOC patients has recently started to be defined by massive parallel sequencing (MPS). Studies indicate that causal variants have very low frequency in most of the populations studied and are spread in a larger array of genes that remain unexplored (Table 1). Until now, the contribution of pathogenic variants in genes other than BRCA1 and BRCA2 has not been entirely defined and studies in Latin American populations are still scarce.

Table 1.
Summary of gene panel studies in hereditary breast cancer.
In recent years, important efforts to define common susceptibility loci for breast cancer in large cohorts have identified more than 90 SNPs, which predispose to this disease [26]. However, the risk conferred by these common susceptibility loci can explain up to 14% of hereditary breast cancer aggregation in the European population [27]. Additional SNPs remain to be discovered and association studies need to be conducted in other populations to better define the prevalence and clinical relevance of novel pathogenic alleles [28]. The identification of rare or population specific, high/moderate-risk pathogenic alleles could be translated into better molecular diagnosis, personalized risk assessment and treatment [23].
To determine the prevalence of pathogenic variants in cancer predisposing genes in Mexican patients, an understudied mixed population, and the potential benefit for molecular diagnosis with gene panel testing, we performed a germline genetic analysis in 327 patients with a clinical indication of HBOC. We analyzed all cases using a panel of 143 genes associated with different inherited oncologic diseases, by massive parallel sequencing.
2. Results
2.1. Clinical and Epidemiological Description of Breast Cancer Cases
Clinical and pathological characteristics of a total of 300 sequenced cases diagnosed with breast cancer are described in Table 2. Mean age at diagnosis was 41 years (range 23–69, SD: 7.3). Seventy one percent of cases had a family history of cancer, 85% reported at least one pregnancy and the average parity was 3 children (SD: 1.6), 60% never used oral contraceptives and 93% reported not being current alcohol drinkers. Importantly, sixty two percent of all cases were overweight, obese or extremely obese. Mutational status was defined as the presence of a pathogenic or likely pathogenic variant (American College of Medical Genetics and Genomics classification) in any of the 143 genes evaluated [29]. Fifteen percent of this group had a pathogenic or likely pathogenic variant.

Table 2.
Clinical and epidemiological characteristics of 300 women with breast cancer and 27 familial breast cancer risk women.
Age at diagnosis was the only epidemiological characteristic statistically associated with mutational status (p = 0.04). No association was found between stage, histological subtype, hormone receptor status and mutational status in cases. Analysis by individual gene showed no association between presence of a mutation and a clinical or pathological characteristic.
Patients in the older age group (60–69 years) were characterized by presenting with early stage tumors (I/II) and absence of mutations.
2.2. Pathogenic Variants in the Breast Cancer Cases
In the group diagnosed with breast cancer (300 cases), we detected 46 pathogenic or likely pathogenic variants in 46 patients (Figure 1; Table S1), including 22 frameshift changes, 13 stop gain/loss mutations and 4 splicing variants. Fifty-six percent (26/46) of mutations detected were already reported in ClinVar as pathogenic. Twenty six percent (12/46) of the pathogenic variants were recurrent, and no genetic alteration was found in 73.6% (221/300) of the patients (Figure 1).
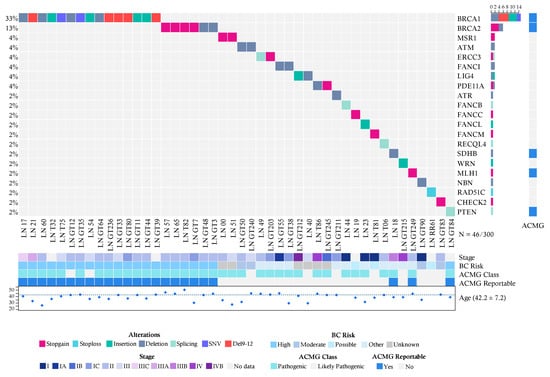
Figure 1.
Allelic distribution of the pathogenic variants in patients with cancer. The grid panel depicts the pathogenic mutations found in each patient color-coded for each type. Right panel: gene reportable by the suggestion of the ACMG (light blue = yes, gray = no). Bottom axis: patient ID. Left axis: relative frequency of mutations per gene. Right axis: mutated gene. Right bar plot: absolute frequency and type of pathogenic mutation per gene. Bottom panel indicates: stage (I-IVB); risk associated with a pathogenic variant; ACMG variant class (pathogenic, likely pathogenic); gene reportable by the suggestion of the ACMG (light blue = yes, gray = no); age distribution.
Notably, BRCA1 (5%, 15/300), BRCA2 (2%, 6/300) and PTEN (0.3%, 1/300) were the only high-risk mutated genes associated with HBOC (Figure 1). No Ashkenazi founder mutations were detected and no pathogenic variants were found in other high-risk HBOC genes such as TP53, CDH1, PALB2 and STK11. Pathogenic alterations in moderate-risk genes were found in ATM (0.6%, 2/300), CHEK2 (0.3% 1/300) and NBN (0.3%) (Table S1) (Figure 1).
2.3. Pathogenic Variants in Familial Breast Cancer Risk Patients without Cancer Diagnosis
In the group of 27 patients without cancer and with suspicion of familial breast cancer risk, we found pathogenic variants in 6 individuals (22%) (Figure 2). The affected genes were BRCA1 (2/27), BRCA2 (1/27), FANCF (1/27), PDE11A (1/27) and POLH (1/27).
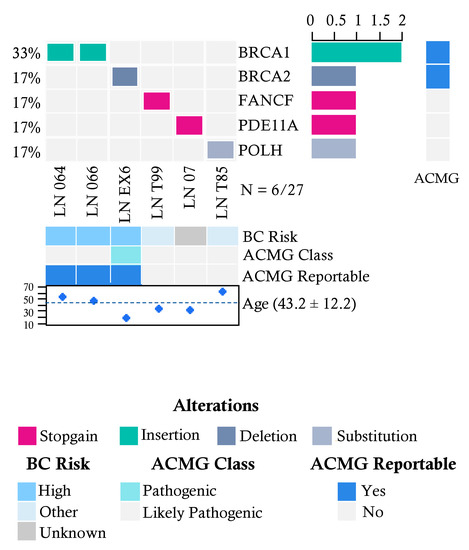
Figure 2.
Allelic distribution of the pathogenic variants in high-risk patients with a severe family history of cancer. The grid panel depicts the pathogenic mutations found in each patient color-coded for each type. Right panel: gene reportable by the suggestion of the ACMG (light blue = yes, gray = no). Bottom axis: patient ID. Left axis: relative frequency of mutations per gene. Right axis: mutated gene. Right bar plot: absolute frequency and type of pathogenic mutation per gene. Bottom panel indicates: risk associated with a pathogenic variant; ACMG variant class (pathogenic, likely pathogenic); gene reportable by the suggestion of the ACMG (light blue = yes, gray = no); age distribution.
2.4. Recurrent Mutations in BRCA1 and BRCA2 in Both Groups
There were 2 recurrent pathogenic alleles in BRCA1, including p.G228fs in five individuals (29%, 5/17), and the Mexican founder mutation in BRCA1 (deletion of exons 9-12), was present in 29% (5/17) (Figure 1, Figure 2 and Figure 3). One recurrent mutation was found in BRCA2: p.R2494X, which was detected in 2 patients (Table S1).
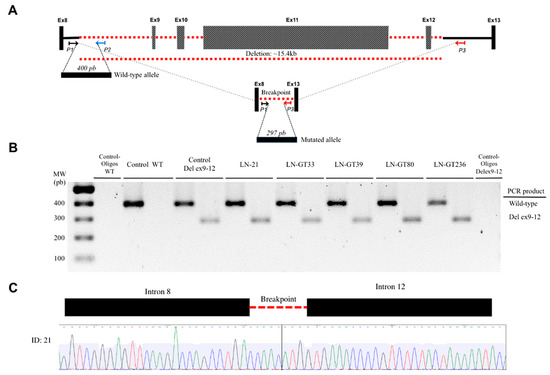
Figure 3.
Detection of BRCA1 deletion of exons 9-12. (A) The locus of exons BRCA1 8 to 13 is indicated. Exons (not to scale) are depicted as boxes and introns as lines, where the discontinuous red line indicates the deleted exons 9-12. The location and orientation of the primers used for amplification of the wild-type (P1, P2) and the mutant alleles (P1, P3) are shown. The PCR products for both amplicons are depicted as horizontal boxes, with their respective number of bp. (B) The resolved PCR products of the patients with the deletion are shown, along with the wild-type and negative controls. (C) The electropherogram of the sequence shows the intron-intron junction in the deletion.
2.5. Pathogenic Variants in Genes with Unknown Risk in Breast Cancer
Deleterious variants in low risk HBOC genes were found in two cancer patients in the genes FANCI (0.6%, 2/300), ERCC3 (0.6%), and in one patient in the genes ATR, FANCB, FANCC, FANCF, FANCL, FANCM, MLH1, RAD51C, POLH, RECQL4, SDHB and WRN (0.3%, 1/300) (Figure 1 and Figure 2). Furthermore, analysis of genes associated with risk of other inherited neoplastic syndromes different to HBOC identified heterozygous pathogenic variants for MSR1 (4%, 2/52), LIG4 (4%, 2/52) and PDE11A (6%, 3/52) in the affected women in both groups (Figure 1 and Figure 2, Table 3).

Table 3.
Syndromes associated with the pathogenic variants detected.
2.6. Description of Variants with Unknown Clinical Significance by Phosphorylation Site Disruption Analysis
We found 38 VUS in 21 genes, 4 of which were found in homozygosity (Table S2). These VUS have MAF < 0.001 in ExAC, and 1000 Genomes databases and not all of them are classified as VUS in ClinVar. To better define the potential effect of VUS in gene functionality, we evaluated the impact of the amino acid change in the context of phosphorylation sites. There was no enrichment in these sites for the occurrence of VUS. However, changes that potentially affect the phosphorylation regulation were found in the AIP and APC genes (Figure S2). The changes affected the FKBP C domain and APC basic domain for AIP and APC, respectively.
3. Discussion
In this work, we evaluated genetic alterations in an expanded panel of 143 genes associated with oncologic inherited diseases, including breast, colon, gastric, among others, by MPS in two groups of high-risk HBOC patients. This is the first study in a Latin American population that analyzes a large cancer risk gene panel by MPS. Overall in all the individuals included in this study, we detected pathogenic variants in 16% (52/327), including 7% (24/327) of variants in BRCA1/2, and 8% (28/327) in genes other than BRCA1/2 (Table 1). These mutations were found in 21 genes previously associated with more than 25 inherited conditions related to cancer (Table 3). Globally, 8% (27/327) of patients had a pathogenic mutation in one of the genes categorized by the American College of Medical Genetics ACMG as a secondary finding with clinical validity and utility to improve medical outcome [30]. Interestingly, half of the pathogenic variants, 50% (26/52), have not been reported before in any Latin-American population, which highlights the current need to expand the evaluation of the genetic diversity of under-studied, mixed populations such as Mexicans and its association to HBOC. These results also confirm the high level of locus heterogeneity that has been described for HBOC [6,23,31] (Table 1).
Age at diagnosis was the only epidemiological or clinical variable associated with the presence of a pathogenic mutation in breast cancer cases, supporting the NCCN criteria for HBOC. Several studies have identified additional life style and genetic risk factors modifying the penetrance in BRCA1/2 mutation carriers [32]. A recent meta-analysis evaluated potentially risk-modification factors for BRCA1 and BRCA2 carriers such as age at first pregnancy, parity, breastfeeding, use of oral contraceptives, smoking and radiation exposure [32]. The loss of at least 10 pounds of body weight before the age of 30 was associated with a reduced risk of BC between 30 to 49 years in BRCA1 mutation carriers [32]. Interestingly, in our analysis around 60% of BC cases were overweight or obese at the time of diagnosis but only 15% of patients had a pathogenic mutation in any of the 143 genes evaluated.
In addition, genetic studies of risk-modifiers focused in the BRCA1 and BRCA2 genes have identified 26 and 16 SNPs associated with BC risk in BRCA1/2 mutation carriers, which have small associated effect sizes (1.05–1.26) per copy of the minor allele [10,33,34]. Given the descriptive focus of our study, the possible combined effect of common low-risk alleles with the detected pathogenic variants was not evaluated. However, these genetic risk-modifiers are thought to account for less than 10% of the genetic variance [10,33,35]. The lack of evidence that associates these modifying factors with pathogenic variants in other genes of high- and medium-penetrance that participate in the development of HBOC is an unsolved concern. These potential allelic interactions could act as genetic modifiers of the risk of pathogenic variants present (especially) in low penetrance genes and might account for the clinical differences in disease presentation and outcome [36].
To our knowledge there is no information on modifiable risk factors for HBOC pathogenic variant carriers in Latin America available to compare the findings from our study. Larger prospective studies on HBOC mutation carriers that incorporate information on a variety of environmental exposures, ancestry and lifestyle factors are required to identify modifying risk factors in Latin America. These studies should include index patients and selected families in diverse representative populations to provide (i) reliable estimates of the allelic frequencies of the pathogenic alleles and modifying variants, (ii) the risk they confer and that may ultimately (iii) facilitate genetic counseling for patients carrying pathogenic variants with demonstrated clinical utility.
An interesting finding was that patients in the older age group (60–69 years) were characterized by presenting with early stage tumors (I/II) and absence of mutations even though they fulfilled the NCNN criteria for HBOC. Given the late presentation and the early stage of the disease, it is possible that these patients may have single pathogenic variants in genes or loci of lower risk not included in our analysis. Another possibility is that these patients may carry a combination of different low-risk loci (not identified in this work) that have additive or epistatic effects, as has been observed in other types of cancer [37,38]. Some reports have previously shown a series of low-risk alleles in genes involved in DNA repair, modification and metabolism related pathways, which act in concert to increase the risk of BC [39,40,41,42]. With the further generalized implementation of WES and WGS population-scale studies these potential multi-allelic interactions will be identified. In addition, a higher frequency of early stage tumors in patients above 60 years with a strong family history of BC might be explained by the increased awareness of this group to comply with current BC screening guidelines [43]. This represents a direct and additional benefit for early detection when identifying high-risk BC cases.
Pathogenic alterations in the HBOC moderate-risk genes ATM, CHEK2 and NBN, were found in a frequency of 0.6%, 0.3%, and 0.3%, respectively. Additionally, we found 7 monoallelic pathogenic variants (2%, 7/327) in 6 genes (besides BRCA1/2) of the interstrand crosslink DNA repair Fanconi anemia pathway (FANCB, FANCC, FANCF, FANCI, FANCL, FANCM) and 1 in RAD51C, a Fanconi-like phenotype gene [44]. The allelic frequency of these variants in the Latin American population spans the 0–0.0015 interval (ExAC). These results confirm findings from other multi-gene panel studies in HBOC patients (Table 1). Although strong evidence regarding the contribution of mutations in some Fanconi anemia genes to HBOC is still limited [45,46], our results provide additional support for this potential association.
Interestingly, we detected pathogenic variants in MSR1, LIG4 and PDE11A, genes not previously associated with HBOC, both in BC patients and high-risk cases. Moreover, the mutation MSR1 p.R293X was found in two unrelated patients. This mutation has been associated with Barrett’s esophagus and esophageal adenocarcinoma in European families [47], and with hereditary prostate cancer [48]; although contradictory results also exist [49,50]. The mutation p.R505fs in the Non-homologous end joining (NHEJ) ligase LIG4, found in one patient, has an allele frequency of 0.0000247 in ExAC. This mutation is located in the ATP dependent DNA ligase C terminal region, and produces a protein lacking both of BRCT-I and BRCT-II domains that are required for chromatin binding [51], abrogating LIG4 function. Recently, germline mutations in LIG4 have been suggested to predispose to diffuse large B-cell lymphomas [52] and to sensitize cell to ionizing radiation, causing immunodeficiency and delay in growth and development in homozygous or compound heterozygous carriers (OMIM#606593) [53]. Given the biochemical function of LIG4 in NHEJ and the low prevalence of its mutations, germline monoallelic mutations could influence HBOC risk, although additional studies are needed to establish this association. In one patient we found a frameshift pathogenic variant p.G57fs in PDE11A, a gene previously associated with different neoplasms including Carney multiple neoplasia complex, prostate cancer and testicular germ cell tumors [54,55].
Overall, 10.8% of patients that were negative for a pathogenic mutation in any of the 143 genes tested had VUS defined following the ACMG criteria. VUS constitute a universal concern in cancer genetics diagnostic settings. The risk conferred by VUS must be addressed by generating more evidence of their allelic frequency in different populations, and by conducting co-segregation analyses, as well as efforts to define their function at protein level using experimental models. Remarkably, we found 2 VUS-AIP p.V49M in homozygosis and APC p.S2535G in heterozygosis—that potentially affect the phosphorylation regulation of the protein. In fact, mutations in the chaperone aryl hydrocarbon receptor-interacting protein (AIP) have been found in familial cases of pituitary adenomas [56]. Experimental in vitro evidence showed that AIP V49M interferes with AIP activity and stability [57]. APC p.S2535G was predicted to disrupt a phosphorylation site in the protein basic domain, which interacts with the microtubules [58]. Neither this amino acid change nor any other change in this position has been reported in COSMIC, OMIM or ClinVar. Consequently, further functional studies are needed to determine the impact of the APC p.S2535G variant.
On the other hand, seventy-four percent of all patients did not harbor alterations in any of the 143 genes studied. Even though we tested the deletion of exons 9-12 in BRCA1, a mutation with a founder effect and the highest frequency reported in Mexican population [59], additional larger rearrangements, undetectable by MPS could account for the lack of mutation detection in this group of patients. The frequency of large genomic rearrangements in BRCA1/2 varies considerably among populations but higher frequencies are related to founder effect variants [60]. In our study, we found that almost one out of three patients (5/17) with BRCA1 pathogenic variants had the Mexican founder mutation (deletion of exons 9-12), which highlights the additional value of evaluating this alteration through a rapid test, such as the one we used. It is also possible that patients who tested negative for any of the genes evaluated may harbor variants in noncoding regions that we did not analyzed. Additional mechanisms of pathogenesis that may play a role in susceptibility to BC might include pathogenic variants affecting splicing mechanisms that disrupt RNA-binding protein (RBBSs) and splicing regulatory (SRBSs) binding sites as well as transcription factor binding site disruption or promoter mutations [31,61].
An additional limitation of this study is the lack of population paired-controls, which could lead to the wrong attribution of common, non-deleterious variants as pathogenic, and which also could eliminate an important amount of common VUS present in this population. It has been described that each person carries up to 100 loss-of-function variants, thirty of which could be in homozygosis, and these are not necessarily disease-causing variants [62,63]. To exclude for non-pathogenic natural variation, we used the largest international databases (ExAC, 1000G, ESP) available in our filtering algorithm. These repositories contain whole genome and exome information from a large number (N = 60,706) of sequenced individuals, with broad ancestral diversity, including 5789 Latinos (2254 males and 3535 females). It has been reported that ExAC is not overrepresented for pathogenic variants, which supports its use to estimate normal variation [64]. We estimate that this strategy may have helped to palliate the effect of lack of paired-controls on our study.
Future germline analyses of cohort studies and population based case-control studies specifically focused on underrepresented populations such as the Latin American region and including women with BC with and without HBOC susceptibility are necessary to validate our results.
Overall, we found 54% of pathogenic variants in genes other than BRCA1 and BRCA2. Consistently, other studies using a panel sequencing approach have found a proportion of 5–64% of pathogenic variants in non-BRCA genes (Table 1). The rate of pathogenic variant detection in these studies tend to be dependent on the total number of genes analyzed, rather than the number of individuals studied. For example, the largest study evaluated a panel of 21 genes in 65,057 patients with BC and found pathogenic variants in 8 non-BRCA genes [10], and our study using a panel of 143 genes in 327 individuals, detected pathogenic variants in 21 non-BRCA cancer-associated genes. Therefore, to further elucidate the wider variation in genes with pathogenic variants that influence HBOC, more studies that investigate larger panels, or ultimately the whole exomes or genomes are needed. Likewise, penetrance and polygenic analyses of rare and common variation will aid to provide more accurate assessment of genetic cancer risk in the clinical setting.
The findings of this work have relevant clinical and public health implications. The frequency of mutations we found in high and intermediate penetrance HBOC associated genes, emphasizes the additional advantage of using a complete gene panel testing instead of single selected mutation approach, increasing the number of identified high-risk individuals who might benefit from personalized prevention or clinical intervention programs. In addition, the implementation of extended gene panels constitutes an efficient development to accelerate de detection of a broader number of high-risk mutation carriers. We detected patients with mutations in BRCA1 and BRCA2 but also in the MLH1, SDHB and PTEN genes that are suggested to be reported by the ACMG given their risk to develop other hereditary conditions [30]. The gradual adoption of clinically informative gene panel testing along with genetic counseling programs will eventually be a key component in the prevention of cancer and other genetic diseases. Lastly, our results highlight the current necessity for the establishment of prospective cohorts in understudied populations, such as the Latin American, to better establish factors that modify penetrance and to identify association relationships of new genetic variants with the disease. These studies could provide enough evidence to direct public health guidelines for risk assessment programs in specific populations including genetic testing recommendations, lifestyle and treatment interventions.
4. Materials and Methods
4.1. Study Population and Data Collection
A total of 327 patients were enrolled based on criteria established in the Genetic/Familial High-Risk Assessment: Breast and Ovarian of the National Comprehensive Cancer Network (NCCN) guidelines, version 2.2015 (https://www.nccn.org/). A transversal series of 300 Mexican female patients diagnosed with primary breast cancer (stages I–IV) were recruited at 4 centers from different states in Mexico: Instituto de Salud del Estado de México, Instituto Estatal de Cancerología de Guerrero, Centro de Investigación Biomédica, Torreón, Coahuila and Hospital de Oncología del CMN Siglo XXI de la Ciudad de México. A second group of 27 individuals without cancer diagnosis who meet NCCN criteria for HBOC susceptibility were additionally included. The protocol was approved by the Ethics Committee of each center (IECG-CEICANCL290515-05GENCMAHER; IECC-2015-01; ISEM-02092015; INSP-CI:1065; INSP-341) and was conducted in accordance with the Declaration of Helsinki. Patients provided written informed consent for participation in this study, and their samples were anonymized and sent to the Laboratorio Nacional en Salud: Diagnóstico Molecular y Efecto Ambiental en Enfermedades Crónico-Degenerativas, Facultad de Estudios Superiores Iztacala, UNAM. Epidemiological and clinical information was obtained from hospital records when available. After the review of their clinical records and age of onset (<45 years), all patients with suspicion of HBOC were invited to participate in this study. After a complete and detailed explanation of the study and written informed consent, a questionnaire of enrollment was used to evaluate the fulfillment of inclusion criteria.
4.2. Sample Preparation and DNA Extraction
For all patients enrolled, 4 mL samples of blood were collected and stored locally at −80 °C. The period between sample collection and freezing never exceeded 36 h. Peripheral blood DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions. DNA concentration was quantified with the Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, USA) and the integrity and purity of the material was verified by agarose gel electrophoresis and spectrophotometry, respectively.
4.3. Library Preparation and Massive Parallel Sequencing
Peripheral Blood DNA was used for library preparation with the GeneRead Cancer Predisposition V2 Kit (Qiagen, Hilden, Germany), which targets 143 genes, which loss of function is a well-known mechanism associated with 88 inherited oncologic diseases based on data from the College of American Pathologists (CAP) guidelines, NCCN guidelines, late-stage clinical trials, The Cancer Genome Atlas (TCGA), and Ingenuity® Knowledge Base. The amplification was divided in 4-pool PCR reactions with a total of 6582 amplicons. Pair-end sequencing was performed with the MiSeq System platform (Illumina, San Diego, USA). Briefly, 40 ± 2.5 ng of DNA was amplified with the GeneRead DNAseq Gene Panel Kit (Qiagen, Hilden, Germany) and purified with Agencourt AMPure XP magnetic Beads (Beckman Coulter, Brea, USA). The amplified fragments were end-repaired, dA-tailed and the adapter GeneRead Adapter 1 Set plex (Qiagen, Hilden, Germany) was ligated using the GeneRead DNA Library I Core Kit. Amplified segments were then size-selected (200–300 bp) using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, USA). New England Biolabs (Ipswich, USA) barcodes were incorporated by PCR amplification in 10 PCR cycles and the products were purified. The libraries were diluted to 4.0 nM and were pooled in batches of 60-80 samples. Library quality was evaluated by DNA quantification with Qubit after size-selection, and by Bioanalyzer (Agilent, Santa Clara, USA) profiling with the High Sensitivity DNA Kit after adaptor-ligated molecules amplification and final library pooling. Pooled barcoded libraries were diluted to 15.0 pM and sequenced with a MiSeq Reagent Kit V2 2 × 150 cycles (Illumina, San Diego, USA) to reach a theoretical average coverage of 100× for each sample.
4.4. Pathogenic Variant Detection
Alignment and variant calling were performed with BWA and GATK (Broad Institute, Cambridge, USA). FastQC files were aligned to the human genome reference hg19 with BWA-MEM; indels were realigned and bases recalibrated. Adaptors were soft-clipped and reads with <20 bp were eliminated. The overall (327) mean sequencing depth of all samples was 70.3× (SD: 21.35) with a range 30–156×, excluding one sample with depth 20× (Figure S1). Variant calling was done with HaplotypeCaller (Broad Institute, Cambridge, USA). Variants were annotated with ANNOVAR and InterVar [65,66]. Mutation description follows Human Genome Variation Society (HGVS) nomenclature (http://www.hgvs.org/). Variant classification followed the five-tier criteria of the American College of Medical Genetics and Genomics (ACMG) [29] and was manually curated. We excluded variants that were synonymous, with depth <5.0× or with mutant allele fraction <20% and those present in homopolymeric tracts >8 bp. All splicing and null variants (stop-gain/loss, frameshift indels) and missense variants defined as pathogenic in ClinVar were considered unequivocally pathogenic (https://www.ncbi.nlm.nih.gov/clinvar). Null variants present at the 3′ extreme end of the gene that were reported as conflicting in ClinVar were classified as unknown clinical significance (VUS). Minor allelic frequency <0.001 in either the ExAC database, 1000 Genomes (1000G) project or the Exome Sequencing Project (ESP6500) was used to capture rare, potentially pathogenic null and missense variants. Low frequency (<0.001) missense variants predicted as deleterious alleles by SIFT or PolyPhen-2 but with no further evidence of pathogenicity in vitro/vivo or clinically were classified as VUS. All filtered variants were manually curated by inspection of the BAM files with the IGV software (Broad Institute). All pathogenic variants were confirmed by two independent assays of Sanger sequencing. Variants in BRCA1 and BRCA2 were further assessed in the Huntsman Cancer Institute Breast Cancer Genes Prior Probabilities site (http://priors.hci.utah.edu/PRIORS/index.php) to evaluate their potential impact. Variants in MLH1, MSH2, MSH6, PMS2 were also investigated in the Leiden Open Variation Database (http://hci-lovd.hci.utah.edu/home.php).
4.5. Detection of Exon 9-12 Deletion in BRCA1
The deletion in exons 9-12 founder mutation was detected by PCR amplification of the mutant and wildtype allele, using specific primers based on the Weitzel et al. method [59]. The PCR products were resolved in 1.5% agarose gels to identify the amplification of the truncated allele and sequenced.
4.6. Phosphorylation Site Disruption Analysis
To evaluate the impact of missense changes in phosphorylation sites, the protein sequences and amino acid changes of all VUS variants (Table S2) were submitted to the ReKINect portal (http://rekinect.science/home). Only mutations predicted to disrupt the phosphorylation site and those which have previous evidence of functional impact in experimental studies were considered.
4.7. Statistical Analyses
Characteristics of cases with confirmed diagnosis of breast cancer were summarized with descriptive statistics. The association between demographic and clinical characteristics on the presence of pathological mutations was assessed using univariate analyses (unadjusted logistic regression model). Age at diagnosis and BMI were included as continuous variables, whereas all other factors were considered to be categorical variables. The logistic regression model utilized all available data (complete and missing). p values of less than 0.05 were considered to indicate statistical significance. All the analyses were conducted using STATA 13.0.
5. Conclusions
Our results show that 16% of the patients with suspicion of HBOC carried a pathogenic mutation in at least one of the 143 genes tested. Fifty-four percent of all pathogenic alterations were not present in BRCA1 and BRCA2, highlighting the locus heterogeneity of this disease. We found 10% of patients with VUS, which require further studies to establish their significance. The genetic information derived from this study could guide the treatment, appropriate follow-up and prophylactic measures in these families and our findings emphasize the benefit of gene panel sequencing service for candidate patients. Although currently clinical guidelines for patients with the pathogenic mutations detected in several of these genes are lacking, the detection of these variants together with a suggestive family history may warrant for a post-test management change, including close follow-up and monitoring. Future efforts will collectively provide enough evidence of the clinical impact of these variants and will foster the development of consensus population-specific guidelines for clinical management.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6694/10/10/361/s1, Figure S1: Mean sequence depth of all samples analyzed; Figure S2: Phosphorylation site disruption in AIP and APC; Table S1: Pathogenic genetic alterations detected in 327 patients; Table S2: Variants with unknown clinical significance detected in 327 patients.
Author Contributions
Conceptualization, F.V.P., R.G., J.O. and S.P.; Data curation, R.Q.U., C.E.D.V., G.T.M., M.D. and F.V.P.; Formal analysis, R.Q.U., C.E.D.V., M.D., R.M.Á.G., L.A.H., I.R., L.I.T, Y.I.C., C.F., J.O., S.P. and F.V.P.; Funding acquisition, L.A.H., L.I.T. and F.V.P.; Investigation, M.P.R.C., M.S.T., A.F.M., O.M.G., L.G.E., J.O., S.P. and F.V.P.; Methodology, R.Q.U., C.E.D.V., M.P.R.C., M.S.T., A.F.M., O.M.G., L.G.E., H.M.G., E.A.R.J., L.E.R.C., R.M.Á.G. and V.F.O.; Project administration, R.Q.U. and F.V.P.; Resources, R.G., G.T.M., I.D.E., H.O.D.L., F.R.L., V.J., V.H.G.B., P.R.F., P.K.E.S., J.H.S.C. and F.V.P.; Software, R.Q.U., L.E.R.C., J.O. and S.P.; Supervision, C.E.D.V., R.G., M.P.R.C., M.S.T., A.F.M., O.M.G. and L.G.E.; Validation, R.Q.U., C.E.D.V., H.M.G., E.A.R.J., R.M.Á.G. and V.F.O.; Visualization, R.Q.U.; Writing—original draft, S.P. and F.V.P.; Writing—review & editing, R.Q.U., C.E.D.V., R.G., G.T.M., M.D., I.D.E., H.O.D.L., F.R.L., V.J., V.H.G.B., P.R.F., P.K.E.S., J.H.S.C., C.F.M.C., L.A.H., I.R., L.I.T., Y.I.C., C.F., J.O., S.P. and F.V.P.
Funding
This work was supported by the National Autonomous University of Mexico (UNAM: PAPIIT IA204215) and by the National Council for Science and Technology (CONACyT: 285879, 264410, 271685). Rosalía Quezada-Urban received a scholarship from CONACyT and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Acknowledgments
We are thankful with M. C. Laura Margarita Márquez Valdemar for her support with the Sanger sequencing assays used for validation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ervik, M.L.F.; Ferlay, J.; Mery, L.; Soerjomataram, I.; Bray, F. Cancer Today. Lyon: International Agency for Research on Cancer. 2016. Available online: http://gco.iarc.fr/today (accessed on 24 November 2016).
- Ford, D.; Easton, D.F.; Stratton, M.; Narod, S.; Goldgar, D.; Devilee, P.; Bishop, D.T.; Weber, B.; Lenoir, G.; Chang-Claude, J.; et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am. J. Hum. Genet. 1998, 62, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Kast, K.; Rhiem, K.; Wappenschmidt, B.; Hahnen, E.; Hauke, J.; Bluemcke, B.; Zarghooniet, V.; Herold, N.; Ditsch, N.; Kiechle, M.; et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J. Med. Genet. 2016, 53, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Torres-Mejia, G.; Royer, R.; Llacuachaqui, M.; Akbari, M.R.; Giuliano, A.R.; Martinez-Matsushita, L.; Angeles-Llerenas, A.; Ortega-Olvera, C.; Ziv, E.; Lazcano-Ponce, E.; et al. Recurrent BRCA1 and BRCA2 mutations in Mexican women with breast cancer. Cancer Epidemiol. Biomark. Prev. 2015, 24, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Dutil, J.; Golubeva, V.A.; Pacheco-Torres, A.L.; Diaz-Zabala, H.J.; Matta, J.L.; Monteiro, A.N. The spectrum of BRCA1 and BRCA2 alleles in Latin America and the Caribbean: A clinical perspective. Breast Cancer Res. Treat. 2015, 154, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, F.C.; van Overeem Hansen, T.; Sorensen, C.S. Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer 2016, 16, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Xie, M.; Wendl, M.C.; Wang, J.; McLellan, M.D.; Leiserson, M.D.; Huang, K.-L.; Wyczalkowski, M.A.; Jayasinghe, R.; Banerjee, T.; et al. Patterns and functional implications of rare germline variants across 12 cancer types. Nat. Commun. 2015, 6, 10086. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Hart, S.N.; Vijai, J.; Schrader, K.A.; Slavin, T.P.; Thomas, T.; Wubbenhorst, B.; Ravichandran, V.; Moore, R.M.; Hu, C.; et al. Evaluation of ACMG-Guideline-Based Variant Classification of Cancer Susceptibility and Non-Cancer-Associated Genes in Families Affected by Breast Cancer. Am. J. Hum. Genet. 2016, 98, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Lee, S.T.; Nam, E.J.; Han, J.W.; Lee, J.Y.; Kim, J.; Kim, T.I.; Park, H.S. Variants of cancer susceptibility genes in Korean BRCA1/2 mutation-negative patients with high risk for hereditary breast cancer. BMC Cancer 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Shimelis, H.; Hu, C.; Hart, S.N.; Polley, E.C.; Na, J.; Hallberg, E.; Moore, R.; Thomas, A.; Lilyquist, J.; et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017, 3, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Kraus, C.; Hoyer, J.; Vasileiou, G.; Wunderle, M.; Lux, M.P.; Fasching, P.A.; Krumbiegel, M.; Uebe, S.; Reuter, M.; Beckmann, M.W.; et al. Gene panel sequencing in familial breast/ovarian cancer patients identifies multiple novel mutations also in genes others than BRCA1/2. Int. J. Cancer 2017, 140, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Lolas Hamameh, S.; Renbaum, P.; Kamal, L.; Dweik, D.; Salahat, M.; Jaraysa, T.; Rayyan, A.A.; Casadei, S.; Mandell, J.B.; Gulsuner, S.; et al. Genomic analysis of inherited breast cancer among Palestinian women: Genetic heterogeneity and a founder mutation in TP53. Int. J. Cancer 2017, 141, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Tedaldi, G.; Tebaldi, M.; Zampiga, V.; Danesi, R.; Arcangeli, V.; Ravegnani, M.; Cangini, I.; Pirini, F.; Petracci, E.; Rocca, A.; et al. Multiple-gene panel analysis in a case series of 255 women with hereditary breast and ovarian cancer. Oncotarget 2017, 8, 47064–47075. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Shao, K.; Qin, Q.; Wang, X.; Song, S.; Wang, X. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered. Cancer Clin. Pract. 2017, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Cock-Rada, A.M.; Ossa, C.A.; Garcia, H.I.; Gomez, L.R. A multi-gene panel study in hereditary breast and ovarian cancer in Colombia. Fam. Cancer 2017, 17, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Susswein, L.R.; Marshall, M.L.; Nusbaum, R.; Vogel Postula, K.J.; Weissman, S.M.; Yackowski, L.; Vaccari, E.M.; Bissonnette, J.; Booker, J.K.; Cremona, M.L.; et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet. Med. 2016, 18, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.; Shin, V.Y.; Au, C.H.; Law, F.B.; Ho, D.N.; Ip, B.K.; Wong, A.T.C.; Lau, S.S.; To, R.M.Y.; Choy, G.; et al. Detection of Germline Mutation in Hereditary Breast and/or Ovarian Cancers by Next-Generation Sequencing on a Four-Gene Panel. J. Mol. Diagn. 2016, 18, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Meeks, H.; Feng, B.J.; Healey, S.; Thorne, H.; Makunin, I.; Campbell, I.; Southey, M.; Mitchell, G.; Clouston, D.; et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J. Med. Genet. 2016, 53, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Mannan, A.U.; Singh, J.; Lakshmikeshava, R.; Thota, N.; Singh, S.; Sowmya, T.S.; Mishra, A.; Sinha, A.; Deshwal, S.; Soni, M.R.; et al. Detection of high frequency of mutations in a breast and/or ovarian cancer cohort: Implications of embracing a multi-gene panel in molecular diagnosis in India. J. Hum. Genet 2016, 61, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.H.; Kuo, W.H.; Huang, A.C.; Lu, Y.S.; Lin, C.H.; Kuo, S.H.; Wang, M.-Y.; Liu, C.-Y.; Cheng, F.T.-F.; Yeh, M.-H.; et al. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget 2016, 7, 8310–8320. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Battelli, C.; Allen, B.; Kaldate, R.; Bhatnagar, S.; Bowles, K.; Timms, K.; Garber, J.E.; Herold, C.; Ellisen, L.; et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015, 121, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, S.E.; Kobayashi, Y.; Anderson, M.J.; Yang, S.; Desmond, A.J.; Mills, M.A.; Nilsen, G.B.; Jacobs, K.B.; Monzon, F.A.; Kurian, A.W.; et al. A Systematic Comparison of Traditional and Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Genes in More Than 1000 Patients. J. Mol. Diagn. 2015, 17, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Desmond, A.; Kurian, A.W.; Gabree, M.; Mills, M.A.; Anderson, M.J.; Kobayashi, Y.; Horick, N.; Yang, S.; Shannon, K.M.; Tung, N.; et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 2015, 1, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.; Faust, U.; Sturm, M.; Hackmann, K.; Grundmann, K.; Harmuth, F.; Bosse, K.; Kehrer, M.; Benkert, T.; Klink, B.; et al. HBOC multi-gene panel testing: Comparison of two sequencing centers. Breast Cancer Res. Treat. 2015, 152, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, Y.; Nakagomi, H.; Sakamoto, I.; Amemiya, K.; Oyama, T.; Mochizuki, H.; Omata, M. Multigene panel analysis identified germline mutations of DNA repair genes in breast and ovarian cancer. Mol. Genet. Genomic Med. 2015, 3, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.N.; Nathanson, K.L. Common breast cancer risk variants in the post-COGS era: A comprehensive review. Breast Cancer Res. 2013, 15, 212. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Beesley, J.; Lindstrom, S.; Canisius, S.; Dennis, J.; Lush, M.J.; Maranian, M.J.; Bolla, M.K.; Wang, Q.; Shah, M.; et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015, 47, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.P.; Beesley, J.; Amin Al Olama, A.; Michailidou, K.; Tyrer, J.; Kote-Jarai, Z.; Lawrenson, K.; Lindstrom, S.; Ramus, S.J.; Thompson, D.J.; et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov. 2016, 6, 1052–1067. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Kalia, S.S.; Adelman, K.; Bale, S.J.; Chung, W.K.; Eng, C.; Evans, J.P.; Herman, G.E.; Hufnagel, S.B.; Klein, T.E.; Korf, B.R.; et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017, 19, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Caminsky, N.G.; Mucaki, E.J.; Perri, A.M.; Lu, R.; Knoll, J.H.; Rogan, P.K. Prioritizing Variants in Complete Hereditary Breast and Ovarian Cancer Genes in Patients Lacking Known BRCA Mutations. Hum. Mutat. 2016, 37, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Friebel, T.M.; Domchek, S.M.; Rebbeck, T.R. Modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: Systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, M.M.; Kuchenbaecker, K.B.; Vijai, J.; Klein, R.J.; Kirchhoff, T.; McGuffog, L.; Barrowdale, D.; Dunning, A.M.; Lee, A.; Dennis, J.; et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013, 9, e1003173. [Google Scholar] [CrossRef] [PubMed]
- Milne, R.L.; Antoniou, A.C. Modifiers of breast and ovarian cancer risks for BRCA1 and BRCA2 mutation carriers. Endocr. Relat. Cancer 2016, 23, T69–T84. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Neuhausen, S.L.; Robson, M.; Barrowdale, D.; McGuffog, L.; Mulligan, A.M.; Andrulis, I.L.; Spurdle, A.B.; Schmidt, M.K.; Schmutzler, R.K.; et al. Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. 2014, 16, 3416. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Barrowdale, D.; Andrulis, I.L.; Domchek, S.M.; Eccles, D.; Nevanlinna, H.; et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: Results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol. Biomark. Prev. 2012, 21, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Ciampa, J.; Yeager, M.; Amundadottir, L.; Jacobs, K.; Kraft, P.; Chung, C.; Wacholder, S.; Yu, K.; Wheeler, W.; Thun, M.J.; et al. Large-scale exploration of gene-gene interactions in prostate cancer using a multistage genome-wide association study. Cancer Res. 2011, 71, 3287–3295. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, Z.; Song, Z.; Chen, J.; Shi, Y. Genome-wide two-locus interaction analysis identifies multiple epistatic SNP pairs that confer risk of prostate cancer: A cross-population study. Int. J. Cancer 2017, 140, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Cherdyntseva, N.V.; Denisov, E.V.; Litviakov, N.V.; Maksimov, V.N.; Malinovskaya, E.A.; Babyshkina, N.N.; Slonimskaya, E.M.; Voevoda, M.I.; Choinzonov, E.L. Crosstalk between the FGFR2 and TP53 genes in breast cancer: Data from an association study and epistatic interaction analysis. DNA Cell Biol. 2012, 31, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Naushad, S.M.; Pavani, A.; Digumarti, R.R.; Gottumukkala, S.R.; Kutala, V.K. Epistatic interactions between loci of one-carbon metabolism modulate susceptibility to breast cancer. Mol. Biol. Rep. 2011, 38, 4893–4901. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.; Furriol, J.; Tormo, E.; Ballester, S.; Lluch, A.; Eroles, P. Epistatic interaction of Arg72Pro TP53 and -710 C/T VEGFR1 polymorphisms in breast cancer: Predisposition and survival. Mol. Cell. Biochem. 2013, 379, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, Y.; Mackey, J.R.; Lai, R.; Franco-Villalobos, C.; Lupichuk, S.; Robson, P.J.; Kopciuk, K.; Cass, C.E.; Yasui, Y.; Damaraju, S. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS ONE 2014, 8, e64896. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Cuevas, S.; Guisa-Hohenstein, F.; Labastida-Almendaro, S. First breast cancer mammography screening program in Mexico: Initial results 2005–2006. Breast J. 2009, 15, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Berwick, M.; Satagopan, J.M.; Ben-Porat, L.; Carlson, A.; Mah, K.; Henry, R.; Diotti, R.; Milton, K.; Pujara, K.; Landers, T.; et al. Genetic heterogeneity among Fanconi anemia heterozygotes and risk of cancer. Cancer Res. 2007, 67, 9591–9596. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.J.; Fernandez, V.; Osorio, A.; Barroso, A.; Fernandez, F.; Urioste, M.; Benitez, J. Mutational analysis of FANCL, FANCM and the recently identified FANCI suggests that among the 13 known Fanconi Anemia genes, only FANCD1/BRCA2 plays a major role in high-risk breast cancer predisposition. Carcinogenesis 2009, 30, 1898–1902. [Google Scholar] [CrossRef] [PubMed]
- Orloff, M.; Peterson, C.; He, X.; Ganapathi, S.; Heald, B.; Yang, Y.R.; Bebek, G.; Romigh, T.; Song, J.H.; Wu, W.; et al. Germline mutations in MSR1, ASCC1, and CTHRC1 in patients with Barrett esophagus and esophageal adenocarcinoma. JAMA 2011, 306, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zheng, S.L.; Komiya, A.; Mychaleckyj, J.C.; Isaacs, S.D.; Hu, J.J.; Sterling, D.; Lange, E.M.; Hawkins, G.A.; Turner, A.; et al. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat. Genet. 2002, 32, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Hope, Q.; Bullock, S.; Evans, C.; Meitz, J.; Hamel, N.; Edwards, S.M.; Severi, G.; Dearnaley, D.; Jhavar, S.; Southgate, C.; et al. Macrophage scavenger receptor 1 999C>T (R293X) mutation and risk of prostate cancer. Cancer Epidemiol. Biomark. Prev. 2005, 14, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; McDonnell, S.K.; Cunningham, J.M.; Hebbring, S.; Jacobsen, S.J.; Cerhan, J.R.; Slager, S.L.; Blute, M.L.; Schaid, D.J.; Thibodeau, S.N. No association of germline alteration of MSR1 with prostate cancer risk. Nat. Genet. 2003, 35, 128–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, X.; Kamdar, R.P.; Wanotayan, R.; Sharma, M.K.; Adachi, N.; Matsumoto, Y. C-Terminal region of DNA ligase IV drives XRCC4/DNA ligase IV complex to chromatin. Biochem. Biophys. Res. Commun. 2013, 439, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Leeksma, O.C.; de Miranda, N.F.; Veelken, H. Germline mutations predisposing to diffuse large B.-cell lymphoma. Blood Cancer J. 2017, 7, e532. [Google Scholar] [CrossRef] [PubMed]
- O’Driscoll, M.; Cerosaletti, K.M.; Girard, P.M.; Dai, Y.; Stumm, M.; Kysela, B.; Hirsch, B.; Gennery, A.; Palmer, S.E.; Seidel, J.; et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell 2001, 8, 1175–1185. [Google Scholar] [CrossRef]
- Faucz, F.R.; Horvath, A.; Rothenbuhler, A.; Almeida, M.Q.; Libe, R.; Raffin-Sanson, M.L.; Bertherat, J.; Carraro, D.M.; Soares, F.A.; Molina, G.d.C.; et al. Phosphodiesterase 11A (PDE11A) genetic variants may increase susceptibility to prostatic cancer. J. Clin. Endocrinol. Metab. 2011, 96. [Google Scholar] [CrossRef] [PubMed]
- Horvath, A.; Korde, L.; Greene, M.H.; Libe, R.; Osorio, P.; Faucz, F.R.; Raffin-Sanson, M.L.; Tsang, K.M.; Drori-Herishanu, L.; Patronas, Y.; et al. Functional phosphodiesterase 11A mutations may modify the risk of familial and bilateral testicular germ cell tumors. Cancer Res. 2009, 69, 5301–5306. [Google Scholar] [CrossRef] [PubMed]
- Vierimaa, O.; Georgitsi, M.; Lehtonen, R.; Vahteristo, P.; Kokko, A.; Raitila, A.; Tuppurainen, K.; Ebeling, T.M.L.; Salmela, P.I.; Paschke, R.; et al. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science 2006, 312, 1228–1230. [Google Scholar] [CrossRef] [PubMed]
- Formosa, R.; Vassallo, J. Aryl Hydrocarbon Receptor-Interacting Protein (AIP) N-Terminus Gene Mutations Identified in Pituitary Adenoma Patients Alter Protein Stability and Function. Horm. Cancer 2017, 8, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Deka, J.; Kuhlmann, J.; Muller, O. A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur. J. Biochem. 1998, 253, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Weitzel, J.N.; Lagos, V.I.; Herzog, J.S.; Judkins, T.; Hendrickson, B.; Ho, J.S.; Ricker, C.N.; Lowstuter, K.J.; Blazer, K.R.; Tomlinson, G.; et al. Evidence for common ancestral origin of a recurring BRCA1 genomic rearrangement identified in high-risk Hispanic families. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1615–1620. [Google Scholar] [CrossRef] [PubMed]
- Ewald, I.P.; Ribeiro, P.L.; Palmero, E.I.; Cossio, S.L.; Giugliani, R.; Ashton-Prolla, P. Genomic rearrangements in BRCA1 and BRCA2: A. literature review. Genet. Mol. Biol. 2009, 32, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yao, F.; Luan, H.; Wang, Y.; Dong, X.; Zhou, W.; Wang, Q. Three novel functional polymorphisms in the promoter of FGFR2 gene and breast cancer risk: A HuGE review and meta-analysis. Breast Cancer Res. Treat. 2012, 136, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Cassa, C.A.; Tong, M.Y.; Jordan, D.M. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum. Mutat. 2013, 34, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; Balasubramanian, S.; Frankish, A.; Huang, N.; Morris, J.; Walter, K.; Walter, K.; Jostins, L.; Habegger, L.; Pickrell, J.K.; et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 2012, 335, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Gardner, S.A.; Hovhannisyan, H.; Natalizio, A.; Weymouth, K.S.; Chen, W.; Thibodeau, I.; Bogdanova, E.; Letovsky, S.; Willis, A.; et al. Exploring the landscape of pathogenic genetic variation in the ExAC population database: Insights of relevance to variant classification. Genet. Med. 2016, 18, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017, 100, 267–280. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).