Nanowire-Based Biosensors: From Growth to Applications
Abstract
1. Introduction
2. Working Principle and Fabrication Process
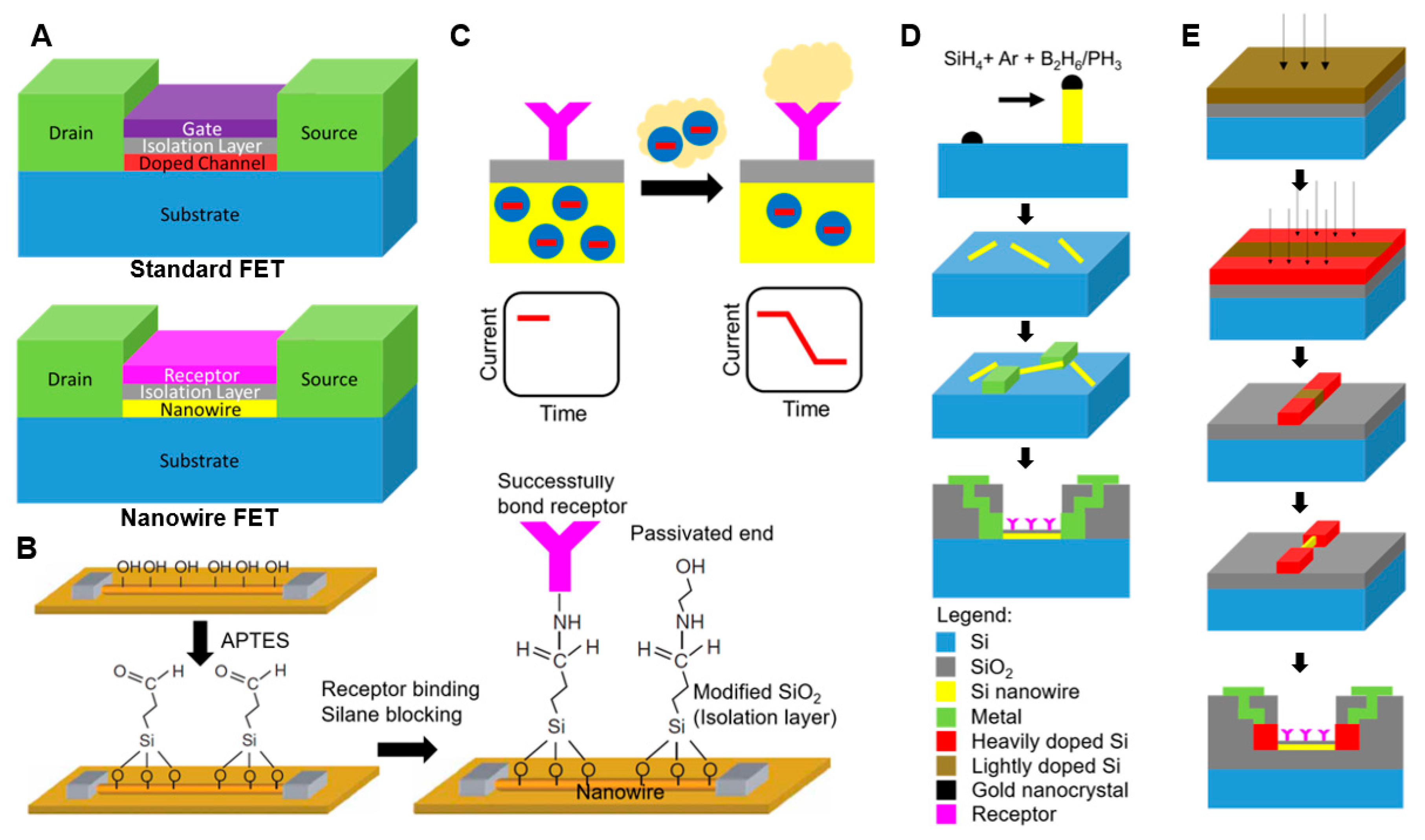
2.1. Fundamentals of Nanowire Field-Effect Transistors
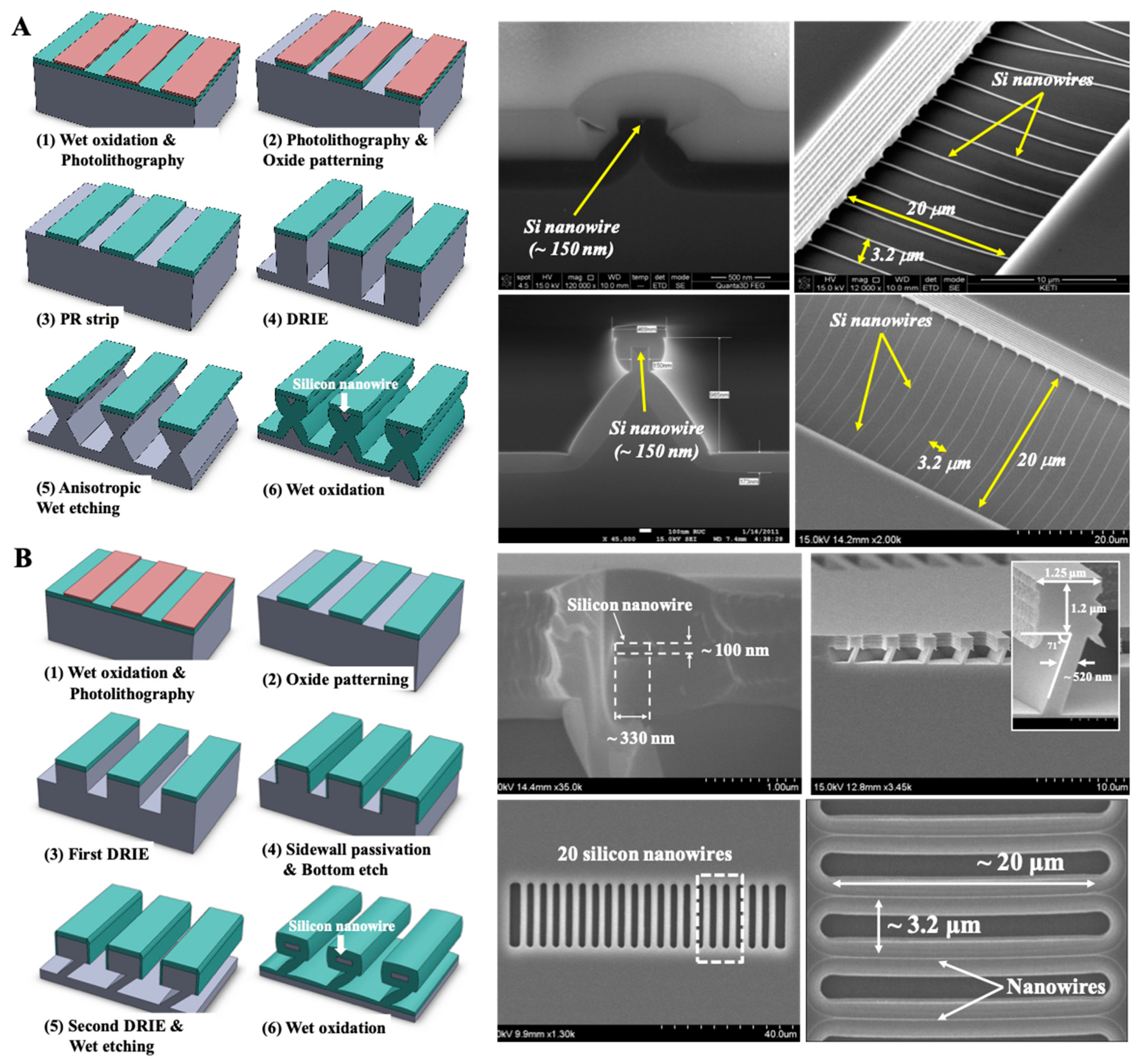
2.2. Fabrication Process
3. Detection of Various Biological Agents
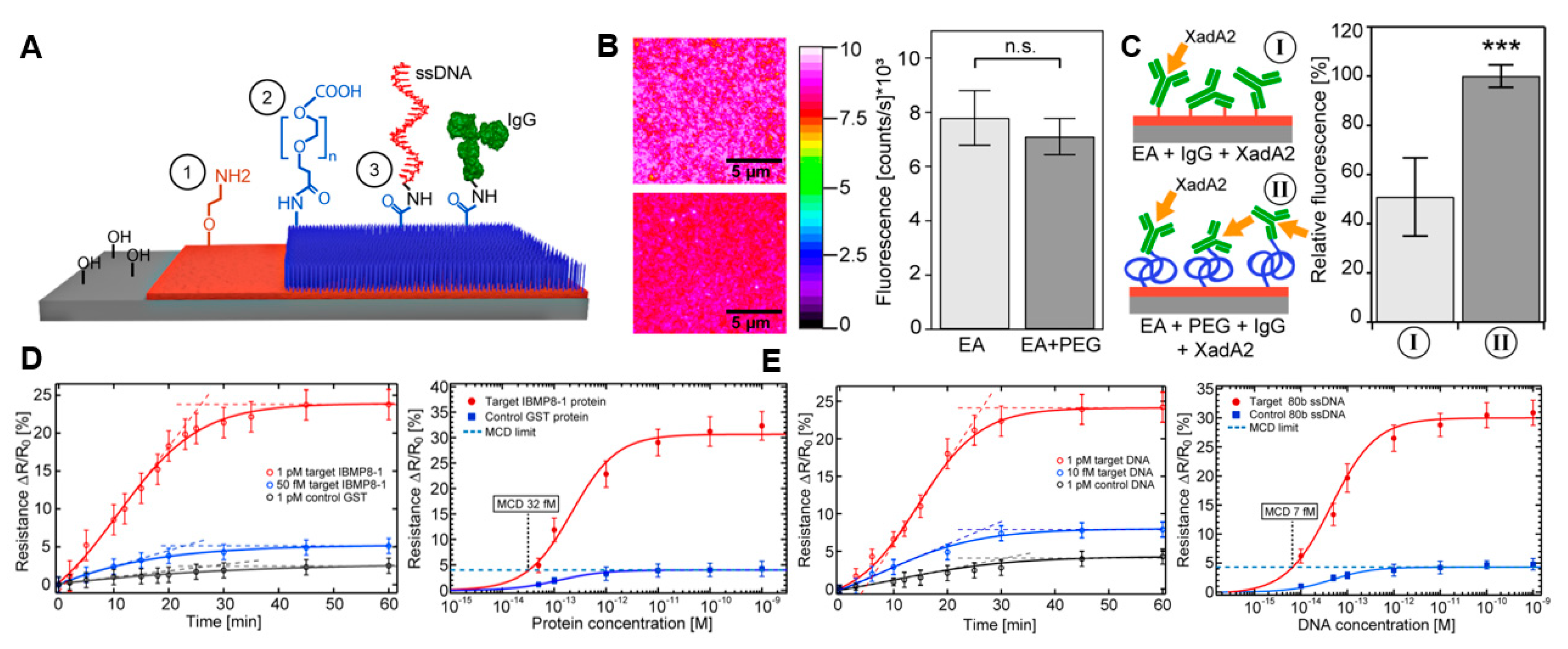
3.1. Proteins
3.2. DNA and RNA
3.3. Viruses
4. Recent Advances in Nanowire Biosensors
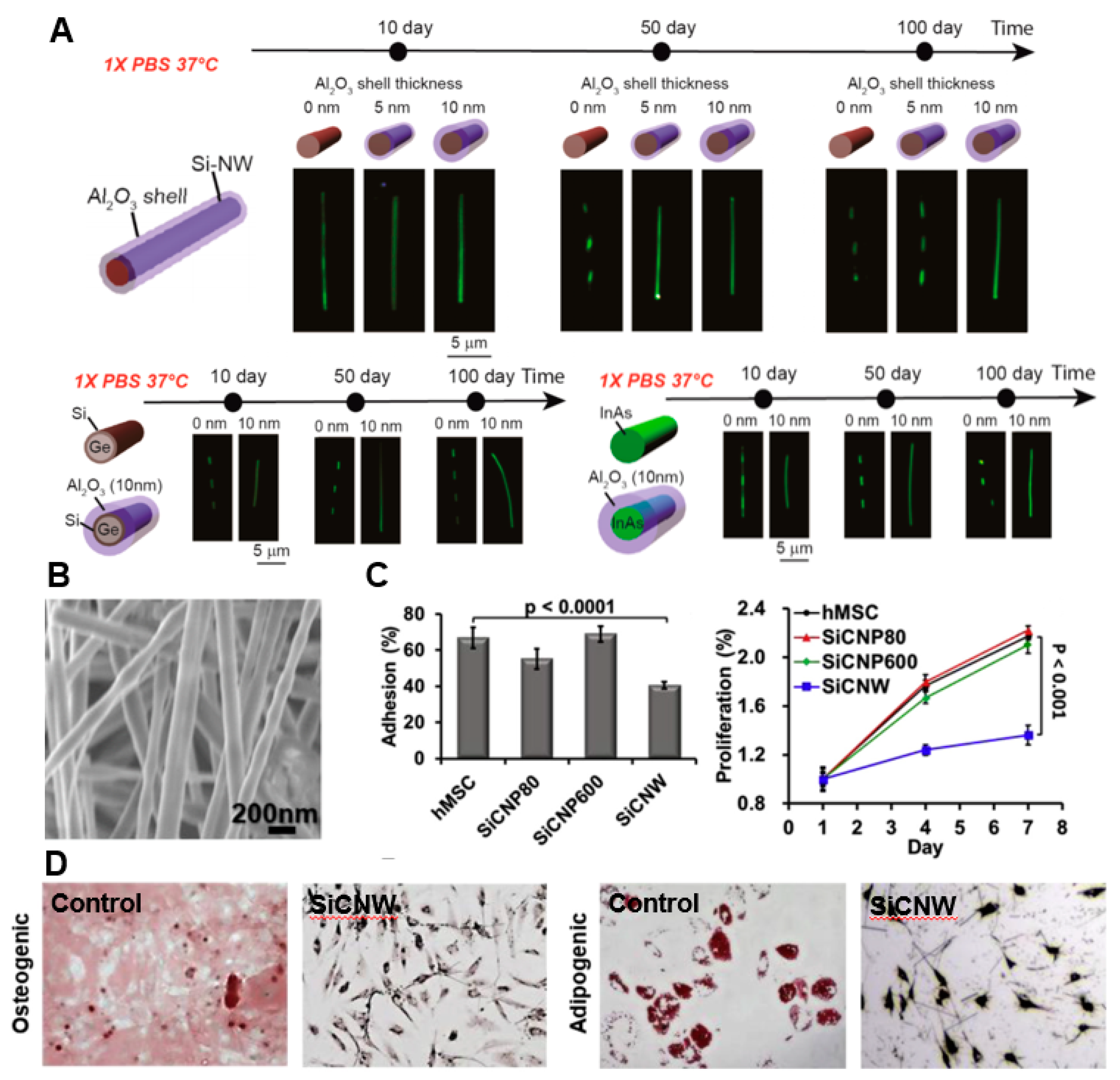
4.1. In Vivo Sensing
4.2. Integration with Paper-Based Devices
4.3. Self-Powering
4.4. Signal Processing and Data Analysis
5. Summary and Perspective
Funding
Conflicts of Interest
References
- Etzioni, R.; Urban, N.; Ramsey, S.; McIntosh, M.; Schwartz, S.; Reid, B.; Radich, J.; Anderson, G.; Hartwell, L. The case for early detection. Nat. Rev. Cancer 2003, 3, 243–252. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Wulfkuhle, J.D.; Liotta, L.A.; Petricoin, E.F. Proteomic applications for the early detection of cancer. Nat. Rev. Cancer 2003, 3, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Adam, T.; Hashim, U. Highly sensitive silicon nanowire biosensor with novel liquid gate control for detection of specific single-stranded DNA molecules. Biosens. Bioelectron. 2015, 67, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Park, C.; Kwon, D.; Kim, D.; Meyyappan, M.; Jeon, S.; Lee, J.-S. Silicon nanowire biosensors for detection of cardiac troponin I (cTnI) with high sensitivity. Biosens. Bioelectron. 2016, 77, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Patolsky, F.; Zheng, G.; Lieber, C.M. Nanowire sensors for medicine and the life sciences. Nanomedicine 2006, 1, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Poole; Owens, F.J. Introduction to Nanotechnology; Wiley: Hoboken, NJ, USA, 2003. [Google Scholar]
- Wang, Y.; Herron, N. Nanometer-Sized Semiconductor Clusters: Materials Synthesis, Quantum Size Effects, and Photophysical Properties. J. Phys. Chem. 1991, 95, 525–532. [Google Scholar] [CrossRef]
- Lee, W.; Ko, H.; Cho, D.D.; Koo, K.; Seo, J. A Review of Nanotechnology for Highly Sensitive Photodetectors for Vision Sensors of Insect-like Robots. Sens. Mater. 2015, 27, 465–474. [Google Scholar] [CrossRef]
- Kurkina, T.; Balasubramanian, K. Towards in vitro molecular diagnostics using nanostructures. Cell. Mol. Life Sci. 2012, 69, 373–388. [Google Scholar] [CrossRef]
- Alivisatos, P. The use of nanocrystals in biological detection. Nat. Biotechnol. 2004, 22, 47–52. [Google Scholar] [CrossRef]
- Ballou, B.; Lagerholm, B.C.; Ernst, L.A.; Bruchez, M.P.; Waggoner, A.S. Noninvasive Imaging of Quantum Dots in Mice. Bioconjugate Chem. 2004, 15, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Jaffer, F.A.; Weissleder, R. Seeing Within: Molecular Imaging of the Cardiovascular System. Circ. Res. 2004, 94, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.M.; Josephson, L.; Weissleder, R. Use of magnetic nanoparticles as nanosensors to probe for molecular interactions. ChemBioChem 2004, 5, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2015, 36, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wei, Q.; Park, H.; Lieber, C.M. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science 2001, 293, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Seker, F.; Meeker, K.; Kuech, T.F.; Ellis, A.B. Surface chemistry of prototypical bulk II-VI and III-V semiconductors and implications for chemical sensing. Chem. Rev. 2000, 100, 2505–2536. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.F.; Schultz, J.S. Handbook of Chemical and Biological Sensors; Institute of Physics Pub.: Washington, DC, USA, 1996. [Google Scholar]
- Cui, Y.; Zhong, Z.H.; Wang, D.L.; Wang, W.U.; Lieber, C.M. High performance silicon nanowire field effect transistors. Nano Lett. 2003, 3, 149–152. [Google Scholar] [CrossRef]
- Lieber, C.M. The incredible shrinking circuit—Researchers have built nanotransistors and nanowires. Now they just need to find a way to put them all together. Sci. Am. 2001, 285, 58–64. [Google Scholar] [CrossRef]
- Ajayan, P.M.; Tour, J.M. Nanotube composites. Nature 2007, 447, 1066–1068. [Google Scholar] [CrossRef]
- Zhang, G.J.; Ning, Y. Silicon nanowire biosensor and its applications in disease diagnostics: A review. Anal. Chim. Acta 2012, 749, 1–15. [Google Scholar] [CrossRef]
- Kaushik, A.; Kumar, R.; Huey, E.; Bhansali, S.; Nair, N.; Nair, M. Silica nanowires: Growth, integration, and sensing applications. Microchim. Acta 2014, 181, 1759–1780. [Google Scholar] [CrossRef]
- Wang, Z.; Lee, S.; Koo, K.; Kim, K. Nanowire-Based Sensors for Biological and Medical Applications. IEEE Trans. Nanobiosci. 2016, 15, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Koo, K.; Ko, H.; Seo, J.-M.; Cho, D.D. Review of High-resolution Retinal Prosthetic System for Vision Rehabilitation: Our Perspective Based on 18 Years of Research. Sens. Mater. 2018, 30, 1393. [Google Scholar] [CrossRef]
- Sze, S.M. Physics of Semiconductor Devices, 2nd ed.; Wiley: New York, NY, USA, 1981. [Google Scholar]
- Bergveld, P. Development, operation, and application of the ion-sensitive field-effect transistor as a tool for electrophysiology. IEEE Trans. Biomed. Eng. 1972, 19, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.M. Nanoscale Science and Technology: Building a Big Future from Small Things. MRS Bull. 2003, 28, 486–491. [Google Scholar] [CrossRef]
- Wu, Y.; Xiang, J.; Yang, C.; Lu, W.; Lieber, C.M. Single-crystal metallic nanowires and metal/semiconductor nanowire heterostructures. Nature 2004, 430, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Lu, W.; Jin, S.; Lieber, C.M. Synthesis and fabrication of high-performance n-type silicon nanowire transistors. Adv. Mater. 2004, 16, 1890–1893. [Google Scholar] [CrossRef]
- Patolsky, F.; Zheng, G.; Lieber, C.M. Fabrication of silicon nanowire devices for ultrasensitive, label-free, real-time detection of biological and chemical species. Nat. Protoc. 2006, 1, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Patolsky, F.; Zheng, G.; Hayden, O.; Lakadamyali, M.; Zhuang, X.; Lieber, C.M. Electrical detection of single viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 14017–14022. [Google Scholar] [CrossRef]
- Zheng, G.; Patolsky, F.; Cui, Y.; Wang, W.U.; Lieber, C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005, 23, 1294–1301. [Google Scholar] [CrossRef]
- McAlpine, M.C.; Ahmad, H.; Wang, D.; Heath, J.R. Highly ordered nanowire arrays on plastic substrates for ultrasensitive flexible chemical sensors. Nat. Mater. 2007, 6, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Poghossian, A.; Cherstvy, A.; Ingebrandt, S.; Offenhäusser, A.; Schöning, M.J. Possibilities and limitations of label-free detection of DNA hybridization with field-effect-based devices. Sens. Actuators B Chem. 2005, 111–112, 470–480. [Google Scholar] [CrossRef]
- Ghatak, K.P.; Bhattacharya, S. Debye Screening Length Effects of Nanostructured Materials; Springer International Publishing: New York, NY, USA, 2014; ISBN 9783319013381. [Google Scholar]
- Poghossian, A.; Weil, M.; Cherstvy, A.G.; Schöning, M.J. Electrical monitoring of polyelectrolyte multilayer formation by means of capacitive field-effect devices. Anal. Bioanal. Chem. 2013, 405, 6425–6436. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, G.-J. Silicon nanowire-transistor biosensor for study of molecule-molecule interactions. Rev. Anal. Chem. 2014, 33, 95–110. [Google Scholar] [CrossRef]
- Stern, E.; Wagner, R.; Sigworth, F.J.; Breaker, R.; Fahmy, T.M.; Reed, M.A. Importance of the debye screening length on nanowire field effect transistor sensors. Nano Lett. 2007, 7, 3405–3409. [Google Scholar] [CrossRef] [PubMed]
- Baraban, L.; Zörgiebel, F.; Pahlke, C.; Baek, E. Nanowire Field Effect Transistors: Principles and Applications; Springer International Publishing: New York, NY, USA, 2014; ISBN 978-1-4614-8123-2. [Google Scholar]
- Li, C.P.; Lee, C.S.; Ma, X.L.; Wang, N.; Zhang, R.Q.; Lee, S.T. Growth Direction and Cross-Sectional Study of Silicon Nanowires. Adv. Mater. 2003, 15, 607–609. [Google Scholar] [CrossRef]
- Wagner, R.S.; Ellis, W.C. Vapor-Liquid-Solid Mechanism of Single Crystal Growth. Appl. Phys. Lett. 1964, 4, 89. [Google Scholar] [CrossRef]
- Whang, D.; Jin, S.; Wu, Y.; Lieber, C.M. Large-scale hierarchical organization of nanowire arrays for integrated nanosystems. Nano Lett. 2003, 3, 1255–1259. [Google Scholar] [CrossRef]
- Yu, G.; Cao, A.; Lieber, C.M. Large-area blown bubble films of aligned nanowires and carbon nanotubes. Nat. Nanotechnol. 2007, 2, 372–377. [Google Scholar] [CrossRef]
- Huang, Y.; Duan, X.; Wei, Q.; Lieber, C.M. Directed assembly of one-dimensional nanostructures into functional networks. Science 2001, 291, 630–633. [Google Scholar] [CrossRef]
- Fan, Z.; Ho, J.C.; Jacobson, Z.A.; Yerushalmi, R.; Alley, R.L.; Razavi, H.; Javey, A. Wafer-scale assembly of highly ordered semiconductor nanowire arrays by contact printing. Nano Lett. 2008, 8, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bhiladvala, R.B.; Morrow, T.J.; Sioss, J.A.; Lew, K.-K.; Redwing, J.M.; Keating, C.D.; Mayer, T.S. Bottom-up assembly of large-area nanowire resonator arrays. Nat. Nanotechnol. 2008, 3, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Freer, E.M.; Grachev, O.; Duan, X.; Martin, S.; Stumbo, D.P. High-yield self-limiting single-nanowire assembly with dielectrophoresis. Nat. Nanotechnol. 2010, 5, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Vu, X.T.; Eschermann, J.F.; Stockmann, R.; Ghoshmoulick, R.; Offenhäusser, A.; Ingebrandt, S. Top-down processed silicon nanowire transistor arrays for biosensing. Phys. Status Solidi Appl. Mater. Sci. 2009, 206, 426–434. [Google Scholar] [CrossRef]
- Vu, X.T.; GhoshMoulick, R.; Eschermann, J.F.; Stockmann, R.; Offenhäusser, A.; Ingebrandt, S. Fabrication and application of silicon nanowire transistor arrays for biomolecular detection. Sens. Actuators B Chem. 2010, 144, 354–360. [Google Scholar] [CrossRef]
- Lee, K.-N.; Jung, S.-W.; Shin, K.-S.; Kim, W.-H.; Lee, M.-H.; Seong, W.-K. Fabrication of Suspended Silicon Nanowire Arrays. Small 2008, 4, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jung, S.W.; Park, S.; Ahn, J.; Hong, S.J.; Yoo, H.J.; Lee, M.H.; Cho, D.I. Fabrication and Evaluation of Silicon Nanowire Photodetectors on Flexible Substrate for Retinal Prosthetic System. Sens. Mater. 2012, 24, 205–220. [Google Scholar] [CrossRef]
- Lee, S.; Jung, S.W.; Ahn, J.; Yoo, H.J.; Oh, S.J. Microelectrode array with integrated nanowire FET switches for high-resolution retinal prosthetic systems. J. Micromech. Microeng. 2014, 24, 075018. [Google Scholar] [CrossRef]
- Wang, Z.; Samanipour, R.; Kim, K. Organ-on-a-Chip Platforms for Drug Screening and Tissue Engineering. In Biomedical Engineering: Frontier Research and Converging Technologies; Springer International Publishing: New York, NY, USA, 2015; pp. 209–233. [Google Scholar]
- Wang, Z.; Samanipour, R.; Koo, K.; Kim, K. Organ-on-a-Chip Platforms for Drug Delivery and Cell Characterization: A Review. Sens. Mater. 2015, 27, 487–506. [Google Scholar]
- Chua, J.H.; Chee, R.E.; Agarwal, A.; She, M.W.; Zhang, G.J. Label-free electrical detection of cardiac biomarker with complementary metal-oxide semiconductor-compatible silicon nanowire sensor arrays. Anal. Chem. 2009, 81, 6266–6271. [Google Scholar] [CrossRef] [PubMed]
- Puppo, F.; Doucey, M.A.; Delaloye, J.F.; Moh, T.S.Y.; Pandraud, G.; Sarro, P.M.; De Micheli, G.; Carrara, S. SiNW-FET in-Air Biosensors for High Sensitive and Specific Detection in Breast Tumor Extract. IEEE Sens. J. 2016, 16, 3374–3381. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Gu, J.; Wang, H.; Zhu, Z.; Hong, X.; Zhang, Z.; Lu, Q.; Qiu, J.; Wang, X.; et al. A Conductive Nanowire-Mesh Biosensor for Ultrasensitive Detection of Serum C-Reactive Protein in Melanoma. Adv. Funct. Mater. 2018, 1802482, 1–9. [Google Scholar] [CrossRef]
- Kim, A.; Ah, C.S.; Yu, H.Y.; Yang, J.H.; Baek, I.B.; Ahn, C.G.; Park, C.W.; Jun, M.S.; Lee, S. Ultrasensitive, label-free, and real-time immunodetection using silicon field-effect transistors. Appl. Phys. Lett. 2007, 91, 29–32. [Google Scholar] [CrossRef]
- Stern, E.; Vacic, A.; Rajan, N.K.; Criscione, J.M.; Park, J.; Fahmy, T.M.; Reed, M.A. Label-free biomarker detection from whole blood. Nat. Nanotechnol. 2010, 5, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Davis, J.J. Electrical biosensors and the label free detection of protein disease biomarkers. Chem. Soc. Rev. 2013, 42, 5944–5962. [Google Scholar] [CrossRef] [PubMed]
- Janissen, R.; Sahoo, P.K.; Santos, C.A.; Da Silva, A.M.; Von Zuben, A.A.G.; Souto, D.E.P.; Costa, A.D.T.; Celedon, P.; Zanchin, N.I.T.; Almeida, D.B.; et al. InP Nanowire Biosensor with Tailored Biofunctionalization: Ultrasensitive and Highly Selective Disease Biomarker Detection. Nano Lett. 2017, 17, 5938–5949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, G.; Chua, J.H.; Chee, R.; Wong, E.H.; Buddharaju, K.D.; Singh, N.; Gao, Z.; Balasubramanian, N. DNA Sensing by Silicon Nanowire: Charge Layer Distance Dependence. Nano 2008, 8, 1066–1070. [Google Scholar] [CrossRef]
- Wu, C.C.; Ko, F.H.; Yang, Y.S.; Hsia, D.L.; Lee, B.S.; Su, T.S. Label-free biosensing of a gene mutation using a silicon nanowire field-effect transistor. Biosens. Bioelectron. 2009, 25, 820–825. [Google Scholar] [CrossRef]
- Hahm, J.; Lieber, C.M. Direct Ultrasensitive Electrical Detection of DNA and DNA Sequence Variations Using Nanowire Nanosensors. Nano Lett. 2004, 4, 51–54. [Google Scholar] [CrossRef]
- Höök, F.; Ray, A.; Nordén, B.; Kasemo, B. Characterization of PNA and DNA immobilization and subsequent hybridization with DNA using acoustic-shear-wave attenuation measurements. Langmuir 2001, 17, 8305–8312. [Google Scholar] [CrossRef]
- Kilså Jensen, K.; Ørum, H.; Nielsen, P.E.; Nordén, B. Kinetics for hybridization of peptide nucleic acids (PNA) with DNA and RNA studied with the BIAcore technique. Biochemistry 1997, 36, 5072–5077. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Musick, M.D.; Nicewarner, S.R.; Salinas, F.G.; Benkovic, S.J.; Natan, M.J.; Keating, C.D. Colloidal Au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J. Am. Chem. Soc. 2000, 122, 9071–9077. [Google Scholar] [CrossRef]
- Zhang, G.J.; Huang, M.J.; Ang, J.J.; Liu, E.T.; Desai, K.V. Self-assembled monolayer-assisted silicon nanowire biosensor for detection of protein-DNA interactions in nuclear extracts from breast cancer cell. Biosens. Bioelectron. 2011, 26, 3233–3239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Huang, M.J.; Ang, J.J.; Yao, Q.; Ning, Y. Label-free detection of carbohydrate-protein interactions using nanoscale field-effect transistor biosensors. Anal. Chem. 2013, 85, 4392–4397. [Google Scholar] [CrossRef] [PubMed]
- Lu, N.; Gao, A.; Dai, P.; Song, S.; Fan, C.; Wang, Y.; Li, T. CMOS-compatible silicon nanowire field-effect transistors for ultrasensitive and label-free microRNAs sensing. Small 2014, 10, 2022–2028. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Gao, Z.F.; Luo, H.Q.; Li, N.B. Sensitive detection of HIV gene by coupling exonuclease III-assisted target recycling and guanine nanowire amplification. Sens. Actuators B Chem. 2017, 238, 1017–1023. [Google Scholar] [CrossRef]
- Gao, A.; Yang, X.; Tong, J.; Zhou, L.; Wang, Y.; Zhao, J.; Mao, H.; Li, T. Multiplexed detection of lung cancer biomarkers in patients serum with CMOS-compatible silicon nanowire arrays. Biosens. Bioelectron. 2017, 91, 482–488. [Google Scholar] [CrossRef]
- Yasui, T.; Yanagida, T.; Ito, S.; Konakade, Y.; Takeshita, D.; Naganawa, T.; Nagashima, K.; Shimada, T.; Kaji, N.; Nakamura, Y.; et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci. Adv. 2017, 3, e1701133. [Google Scholar] [CrossRef]
- Doucey, M.A.; Carrara, S. Nanowire Sensors in Cancer. Trends Biotechnol. 2018, 1–14. [Google Scholar] [CrossRef]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS--beginning to understand a new virus. Nat. Rev. Microbiol. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- Atlas, R.M. Bioterrorism and biodefence research: Changing the focus of microbiology. Nat. Rev. Microbiol. 2003, 1, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-J.; Zhang, L.; Huang, M.J.; Luo, Z.H.H.; Tay, G.K.I.; Lim, E.-J.A.; Kang, T.G.; Chen, Y. Silicon nanowire biosensor for highly sensitive and rapid detection of Dengue virus. Sens. Actuators B Chem. 2010, 146, 138–144. [Google Scholar] [CrossRef]
- Shen, F.; Wang, J.; Xu, Z.; Wu, Y.; Chen, Q.; Li, X.; Jie, X.; Li, L.; Yao, M.; Guo, X.; et al. Rapid flu diagnosis using silicon nanowire sensor. Nano Lett. 2012, 12, 3722–3730. [Google Scholar] [CrossRef] [PubMed]
- Kao, L.T.H.; Shankar, L.; Kang, T.G.; Zhang, G.; Tay, G.K.I.; Rafei, S.R.M.; Lee, C.W.H. Multiplexed detection and differentiation of the DNA strains for influenza A (H1N1 2009) using a silicon-based microfluidic system. Biosens. Bioelectron. 2011, 26, 2006–2011. [Google Scholar] [CrossRef]
- Inci, F.; Tokel, O.; Wang, S.; Gurkan, U.A.; Tasoglu, S.; Kuritzkes, D.R.; Demirci, U. Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano 2013, 7, 4733–4745. [Google Scholar] [CrossRef] [PubMed]
- Ibarlucea, B.; Akbar, T.F.; Kim, K.; Rim, T.; Baek, C.K.; Ascoli, A.; Tetzlaff, R.; Baraban, L.; Cuniberti, G. Ultrasensitive detection of Ebola matrix protein in a memristor mode. Nano Res. 2018, 11, 1057–1068. [Google Scholar] [CrossRef]
- Zhou, W.; Dai, X.; Lieber, C.M. Advances in nanowire bioelectronics. Rep. Prog. Phys. 2017, 80, 016701. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Liu, J.; Fu, T.M.; Dai, X.; Zhou, W.; Lieber, C.M. Three-dimensional macroporous nanoelectronic networks as minimally invasive brain probes. Nat. Mater. 2015, 14, 1286–1292. [Google Scholar] [CrossRef]
- Cui, Y.; Lauhon, L.J.; Gudiksen, M.S.; Wang, J.; Lieber, C.M. Diameter-controlled synthesis of single-crystal silicon nanowires. Appl. Phys. Lett. 2001, 78, 2214–2216. [Google Scholar] [CrossRef]
- Ahn, S.; Spuhler, P.S.; Chiari, M.; Cabodi, M.; Selim Ünlü, M. Quantification of surface etching by common buffers and implications on the accuracy of label-free biological assays. Biosens. Bioelectron. 2012, 36, 222–229. [Google Scholar] [CrossRef]
- Tian, B.; Liu, J.; Dvir, T.; Jin, L.; Tsui, J.H.; Qing, Q.; Suo, Z.; Langer, R.; Kohane, D.S.; Lieber, C.M. Macroporous nanowire nanoelectronic scaffolds for synthetic tissues. Nat. Mater. 2012, 11, 986–994. [Google Scholar] [CrossRef]
- Hwang, S.-W.; Tao, H.; Kim, D.-H.; Cheng, H.; Song, J.-K.; Rill, E.; Brenckle, M.A.; Panilaitis, B.; Won, S.M.; Kim, Y.-S.; et al. A Physically Transient Form of Silicon Electronics. Science 2012, 337, 1640–1644. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Dai, X.; Fu, T.; Xie, C.; Liu, J.; Lieber, C.M. Long Term Stability of Nanowire Nanoelectronics in Physiological Environments. Nano Lett. 2014, 14, 1614–1619. [Google Scholar] [CrossRef]
- Peled, A.; Pevzner, A.; Peretz Soroka, H.; Patolsky, F. Morphological and chemical stability of silicon nanostructures and their molecular overlayers under physiological conditions: Towards long-term implantable nanoelectronic biosensors. J. Nanobiotechnol. 2014, 12, 7. [Google Scholar] [CrossRef]
- Liu, J.; Fu, T.-M.; Cheng, Z.; Hong, G.; Zhou, T.; Jin, L.; Duvvuri, M.; Jiang, Z.; Kruskal, P.; Xie, C.; et al. Syringe-injectable electronics. Nat. Nanotechnol. 2015, 10, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Cacchioli, A.; Ravanetti, F.; Alinovi, R.; Pinelli, S.; Rossi, F.; Negri, M.; Bedogni, E.; Campanini, M.; Galetti, M.; Goldoni, M.; et al. Cytocompatibility and cellular internalization mechanisms of SiC/SiO2 nanowires. Nano Lett. 2014, 14, 4368–4375. [Google Scholar] [CrossRef]
- Poudineh, M.; Wang, Z.; Labib, M.; Ahmadi, M.; Zhang, L.; Das, J.; Ahmed, S.U.; Angers, S.; Kelley, S.O. Three-Dimensional Nanostructured Architectures Enable Efficient Neural Differentiation of Mesenchymal Stem Cells via Mechanotransduction. Nano Lett. 2018, 18, 7188–7193. [Google Scholar] [CrossRef]
- Chen, F.; Li, G.; Zhao, E.R.; Li, J.; Hableel, G.; Lemaster, J.E.; Bai, Y.; Sen, G.L.; Jokerst, J.V. Cellular toxicity of silicon carbide nanomaterials as a function of morphology. Biomaterials 2018, 179, 60–70. [Google Scholar] [CrossRef]
- Alaraby, M.; Hernández, A.; Marcos, R. Systematic in vivo study of NiO nanowires and nanospheres: Biodegradation, uptake and biological impacts. Nanotoxicology 2018, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Wang, Y.H.; Liu, X. Flexible physical sensors made from Paper substrates integrated with zinc oxide nanostructures. Flex. Print. Electron. 2017, 2, 034001. [Google Scholar] [CrossRef]
- Li, X.; Zhao, C.; Liu, X. A paper-based microfluidic biosensor integrating zinc oxide nanowires for electrochemical glucose detection. Microsyst. Nanoeng. 2015, 1, 15014. [Google Scholar] [CrossRef]
- Kalia, S.; Boufi, S.; Celli, A.; Kango, S. Nanofibrillated cellulose: Surface modification and potential applications. Colloid Polym. Sci. 2014, 292, 5–31. [Google Scholar] [CrossRef]
- Missoum, K.; Belgacem, M.N.; Bras, J. Nanofibrillated cellulose surface modification: A review. Materials 2013, 6, 1745–1766. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Song, J. Piezoelectric Nanogenerators Based on Zinc Oxide Nanowire Arrays. Science 2006, 312, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Qin, Y.; Xu, C.; Wei, Y.; Yang, R.; Wang, Z.L. Self-powered nanowire devices. Nat. Nanotechnol. 2010, 5, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, Y.; Xu, C.; Lin, L.; Snyder, R.L.; Wang, Z.L. Self-Powered System with Wireless Data Transmission. Nano Lett. 2011, 11, 2572–2577. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, J.; Zhao, S.; Yang, W.; Yuan, Z.; Yin, Y.; Du, X.; Li, N.W.; Zhang, X.; Li, X.; et al. Self-powered nanofiber-based screen-print triboelectric sensors for respiratory monitoring. Nano Res. 2018, 11, 3771–3779. [Google Scholar] [CrossRef]
- Chen, B.D.; Tang, W.; He, C.; Jiang, T.; Xu, L.; Zhu, L.P.; Gu, G.Q.; Chen, J.; Shao, J.J.; Luo, J.J.; et al. Ultrafine Capillary-Tube Triboelectric Nanogenerator as Active Sensor for Microliquid Biological and Chemical Sensing. Adv. Mater. Technol. 2018, 3, 1–10. [Google Scholar] [CrossRef]
- Yan, Y.; Zheng, W.; Su, L.; Mao, L. Carbon-nanotube-based glucose/O2 biofuel cells. Adv. Mater. 2006, 18, 2639–2643. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y.; Chu, M.; Deng, W.; Tan, Y.; Ma, M.; Su, X.; Xie, Q.; Yao, S. Facile fabrication of network film electrodes with ultrathin Au nanowires for nonenzymatic glucose sensing and glucose/O2 fuel cell. Biosens. Bioelectron. 2014, 52, 105–110. [Google Scholar] [CrossRef]
- Haddad, R.; Thery, J.; Gauthier-Manuel, B.; Elouarzaki, K.; Holzinger, M.; Le Goff, A.; Gautier, G.; El Mansouri, J.; Martinent, A.; Cosnier, S. High performance miniature glucose/O2 fuel cell based on porous silicon anion exchange membrane. Electrochem. Commun. 2015, 54, 10–13. [Google Scholar] [CrossRef]
- Zhao, M.; Gao, Y.; Sun, J.; Gao, F. Mediatorless Glucose Biosensor and Direct Electron Transfer Type Glucose/Air Biofuel Cell Enabled with Carbon Nanodots. Anal. Chem. 2015, 87, 2615–2622. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.J.; Liu, Y.; Yang, R.; Wang, Z.L. Hybrid Nanogenerator for Concurrently Biomedical and Biochemical Energy. ACS Nano 2010, 4, 3647–3652. [Google Scholar] [CrossRef] [PubMed]
- Potyrailo, R.A. Multivariable Sensors for Ubiquitous Monitoring of Gases in the Era of Internet of Things and Industrial Internet. Chem. Rev. 2016, 116, 11877–11923. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Li, X.; Gu, Z.; Kurup, P.U. Recognition of explosive precursors using nanowire sensor array and decision tree learning. IEEE Sens. J. 2012, 12, 2384–2391. [Google Scholar] [CrossRef]
- Shehada, N.; Cancilla, J.C.; Torrecilla, J.S.; Pariente, E.S.; Brönstrup, G.; Christiansen, S.; Johnson, D.W.; Leja, M.; Davies, M.P.A.; Liran, O.; et al. Silicon Nanowire Sensors Enable Diagnosis of Patients via Exhaled Breath. ACS Nano 2016, 10, 7047–7057. [Google Scholar] [CrossRef] [PubMed]
- Nakhleh, M.K.; Amal, H.; Jeries, R.; Broza, Y.Y.; Aboud, M.; Gharra, A.; Ivgi, H.; Khatib, S.; Badarneh, S.; Har-Shai, L.; et al. Diagnosis and Classification of 17 Diseases from 1404 Subjects via Pattern Analysis of Exhaled Molecules. ACS Nano 2017, 11, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.; Schrittwieser, J.; Simonyan, K.; Antonoglou, I.; Huang, A.; Guez, A.; Hubert, T.; Baker, L.; Lai, M.; Bolton, A.; et al. Mastering the game of Go without human knowledge. Nature 2017, 550, 354–359. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Wu, F.; Cao, X.; Li, Z.; Alharbi, M.; Abbas, A.N.; Amer, M.R.; Zhou, C. Highly Sensitive and Wearable In2O3 Nanoribbon Transistor Biosensors with Integrated On-Chip Gate for Glucose Monitoring in Body Fluids. ACS Nano 2018, 12, 1170–1178. [Google Scholar] [CrossRef]
- Kim, J.; Sempionatto, J.R.; Imani, S.; Hartel, M.C.; Barfidokht, A.; Tang, G.; Campbell, A.S.; Mercier, P.P.; Wang, J. Simultaneous Monitoring of Sweat and Interstitial Fluid Using a Single Wearable Biosensor Platform. Adv. Sci. 2018, 5, 1800880. [Google Scholar] [CrossRef]
- Mishra, R.K.; Hubble, L.J.; Martín, A.; Kumar, R.; Barfidokht, A.; Kim, J.; Musameh, M.M.; Kyratzis, I.L.; Wang, J. Wearable Flexible and Stretchable Glove Biosensor for On-Site Detection of Organophosphorus Chemical Threats. ACS Sens. 2017, 2, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Barfidokht, A.; Karajic, A.; Sempionatto, J.R.; Wang, J.; Wang, J. Wearable potentiometric tattoo biosensor for on-body detection of G-type nerve agents simulants. Sens. Actuators B Chem. 2018, 273, 966–972. [Google Scholar] [CrossRef]
- Kim, J.; Jeerapan, I.; Sempionatto, J.R.; Barfidokht, A.; Mishra, R.K.; Campbell, A.S.; Hubble, L.J.; Wang, J. Wearable Bioelectronics: Enzyme-Based Body-Worn Electronic Devices. Acc. Chem. Res. 2018, 51, 2820–2828. [Google Scholar] [CrossRef] [PubMed]
| Features | Application | Reference |
|---|---|---|
| Top-down fabrication process using SCS wafer | Photodiode and FET for the retinal prosthetic systems | [52,53] |
| High sensitivity using PEG cross-linker | Detection of protein and DNA | [62] |
| Long-term stability using Al2O3 shell coating | In vivo sensing | [89] |
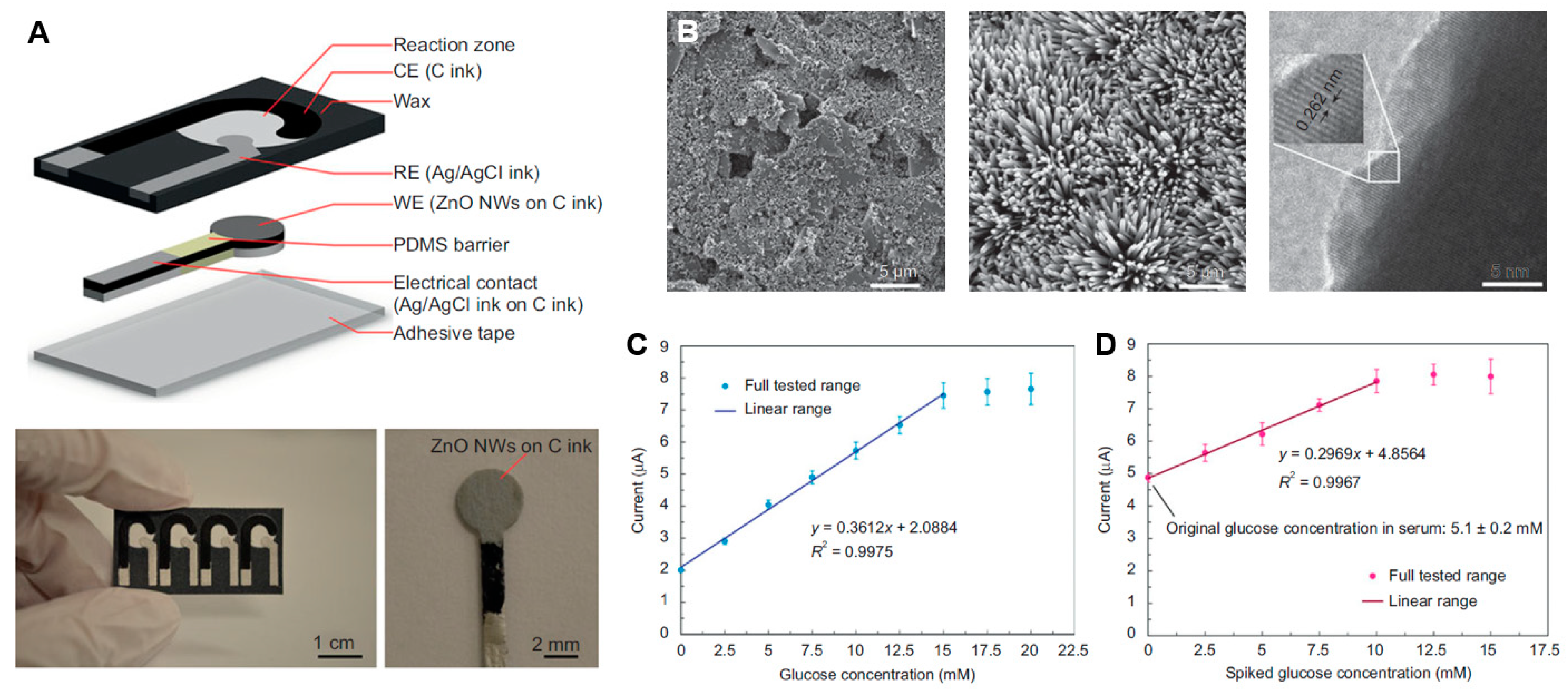
| Integrating nanowires with disposable device | Glucose detection | [97] |
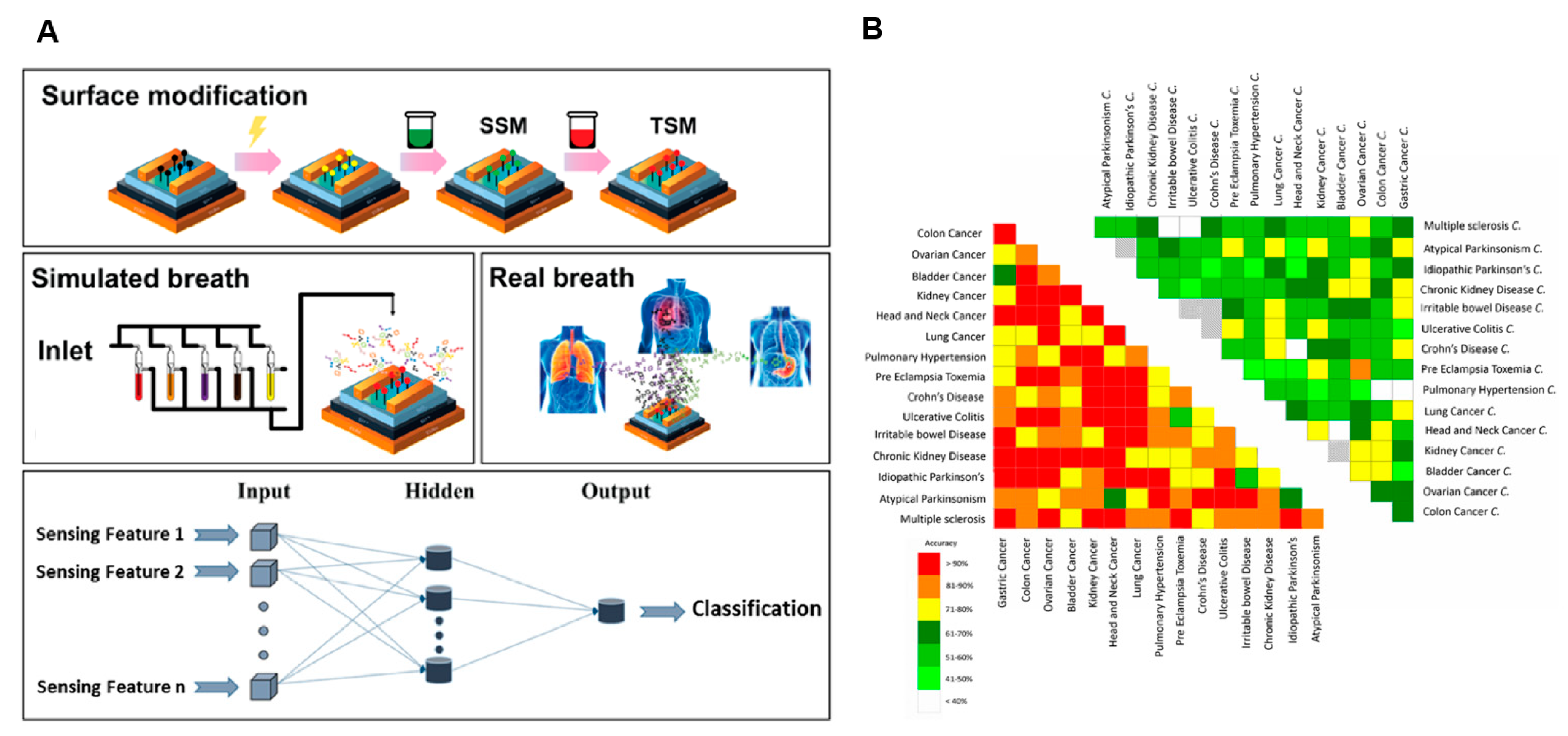
| Multivariable detection using machine learning | Multiple disease diagnosis | [112,113] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambhorkar, P.; Wang, Z.; Ko, H.; Lee, S.; Koo, K.-i.; Kim, K.; Cho, D.-i. Nanowire-Based Biosensors: From Growth to Applications. Micromachines 2018, 9, 679. https://doi.org/10.3390/mi9120679
Ambhorkar P, Wang Z, Ko H, Lee S, Koo K-i, Kim K, Cho D-i. Nanowire-Based Biosensors: From Growth to Applications. Micromachines. 2018; 9(12):679. https://doi.org/10.3390/mi9120679
Chicago/Turabian StyleAmbhorkar, Pranav, Zongjie Wang, Hyuongho Ko, Sangmin Lee, Kyo-in Koo, Keekyoung Kim, and Dong-il (Dan) Cho. 2018. "Nanowire-Based Biosensors: From Growth to Applications" Micromachines 9, no. 12: 679. https://doi.org/10.3390/mi9120679
APA StyleAmbhorkar, P., Wang, Z., Ko, H., Lee, S., Koo, K.-i., Kim, K., & Cho, D.-i. (2018). Nanowire-Based Biosensors: From Growth to Applications. Micromachines, 9(12), 679. https://doi.org/10.3390/mi9120679









