Abstract
Targeting exosome for liquid biopsy has gained significant attention for its diagnostic and therapeutic potential. For detecting neuronal disease diagnosis such as Alzheimer’s disease (AD), the main technique for identifying AD still relies on positron-emission tomography (PET) imaging to detect the presence of amyloid-β (Aβ). While the detection of Aβ in cerebrospinal fluid has also been suggested as a marker for AD, the lack of quantitative measurements has compromised existing assays. In cerebrospinal fluid, in addition to Aβ, T-Tau, and P-Tau, alpha-synuclein has been considered a biomarker of neurodegeneration. This review suggests that and explains how the exosome can be used as a neuronal diagnostic component. To this end, we summarize current progress in exosome preparation/isolation and quantification techniques and comment on the outlooks for neuronal exosome-based diagnostic techniques.
1. Introduction
Alzheimer’s disease (AD) and Parkinson’s disease (PD) are the most prevalent age-related neurodegenerative disorders that result from synaptic degeneration and nerve cell death [1]. Dementia is a general mental disorder, characterized by memory impairment and loss of judgment, and around 50% of all patients with dementia have comorbid AD.
AD currently affects around 5.5 million adults in the United States and this figure is expected to increase to 13.8 million by 2050 [2]. It has been estimated that new AD diagnoses were made every 66 s in 2016, an estimate which has reached one in every 33 s in 2050 [3]. In addition, it is estimated that, in 2014, 29.3 per 100,000 people of the population died from AD in the United States. Furthermore, more than 15 million families and caregivers in the United States dedicated about 18.2 billion hours of care to patients with AD or dementia in 2016, which has been estimated to have cost over $230 billion [3]. In addition, the total cost of dementia-related health care, long-term care, and hospice services was estimated to have been $259 billion in 2017, and the social costs of dementia due to AD are only increasing [3].
The early stage detection and diagnosis of AD plays a key role in patient care, since early detection allows patients to take preventative measures before irreversible brain damage occurs [4,5,6]. The Mini-Mental State Examination (MMSE) and the Logical Memory (LM) test are used to measure cognitive status in patients with AD [7]. Generally, biomarkers, such as amyloid-β (Aβ), deposition in the brain can be assessed using neuroimaging methods (i.e., MRI and PET) or cerebrospinal fluid (CSF) analysis [8,9,10,11].
The pathogenesis of AD has been reported to be potentially driven by excessive production and deposition of Aβ protein. Such abnormal Aβ levels can typically be detected in biofluids, such as CSF. It has been reported that continuously monitoring Aβ levels could facilitate early AD diagnosis and treatment before the onset of AD symptoms [11,12]. Nakamura et al. recently demonstrated blood-based clinical validation using plasma-based Aβ had both cost–benefit and scalability advantages [13].
With regard to clinical utility, much effort has been made to validate methods for monitoring neurodegenerative disorders. The exosome has been highlighted for its potential in early stage diagnostics. Nanoscale exosomes (30–150 nm) have been studied to assess proteomic and genetic information about disease diagnostics [14,15]. In particular, neuronal exosomes, as shown in Figure 1, are good candidates for detecting neurodegenerative biomarkers [16,17,18]. Among neurodegenerative biomarkers, Aβ and tau have been considered protein markers of AD and α-synuclein a biomarker of PD (Figure 1).
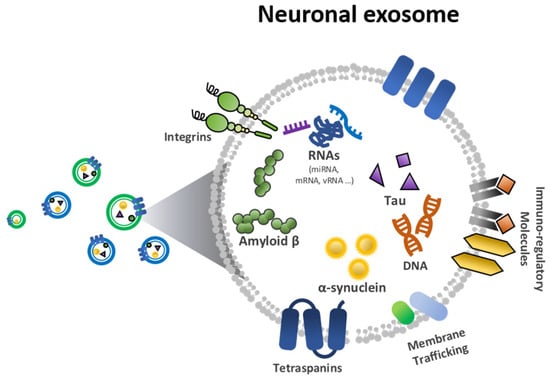
Figure 1.
Biological elements in a neuronal exosome, including amyloid-β (Aβ), tau and α-synuclein.
Typically, neuronal exosomes are collected from human CSF, and the protein markers from a neuronal exosome, i.e., Aβ, tau and α-synuclein, can be extracted from this CSF (Figure 2) [19,20,21,22,23,24,25]. While sample protocols from CSF is well-established, it does present several risks, including pain and leaks that can cause infection, nerve damage, and headaches. The invasive nature of CSF collection means that blood-based detection might be the simplest and most powerful method [26,27]. Since neuronal exosomes can penetrate the blood-brain-barrier, they can be identified in the blood samples, as shown in Figure 2 [28,29].
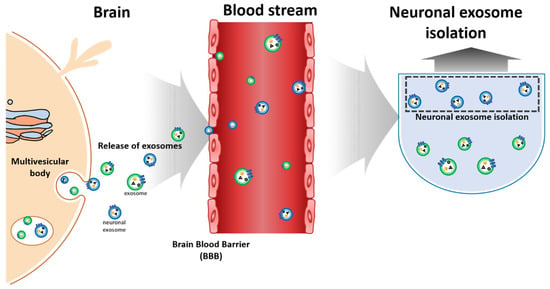
Figure 2.
Schemes of neuronal exosomes isolation/detection from blood.
To diagnose AD and PD using neuronal exosomes in the blood, exosomes must be isolated from the blood. Because of the paucity of biomarker studies in peripheral blood, it is difficult to link biomarker levels with brain pathology due to the relative uncertainty of their tissue of origin [30]. AD and PD diagnoses are thus limited by the unclear origin of exosomes in blood. In contrast, neuronal exosomes can be used to assess for biomarkers of AD and PD since they are known to originate from brain tissue [31]. Neuronal exosomes are thus not only possible, but advantageous, to use instead of exosomes for the identification of AD and PD biomarkers. There are many ways to implement this, and several studies have investigated how best to increase the purity and yield of exosome separations [32,33]. The next step is to separate neuronal exosomes from the isolated exosomes. While many techniques on how to separate exosomes from the blood have been reported, only one technique has been reported concerning the separation of neuronal exosomes from isolated exosomes, using a specific protein presented on the surface of the neuronal exosome [34]. Generally, the amount of exosome in the blood is not enough for a general-purpose ELISA; thus, new techniques that increase the yield and purity of both exosomes and neuronal exosomes are required [35].
2. Exosome Preparation/Isolation Methods
2.1. Differential Ultracentrifugation
Differential ultracentrifugation is a typical method for exosome separation that uses sequential speed steps of ultracentrifugation (Figure 3). By controlling sequential speeds, the crude homogenate is first centrifuged to sediment cells, dead cells, and cell debris and the resulting pellet is discarded. A classical way to improve exosome purification after differential ultracentrifugation is to isolate the exosome again by ultracentrifugation; in this way, a higher purity is possible, but with a reduced amount of exosome [36,37]. Centrifugation is considered the standard method for separating exosomes from biological fluids/media; however, the major drawback is that the properties of the separated exosomes may depend on the speed of rotation. Moreover, high speeds of rotation can lead to protein aggregation, making results of the Bradford assay less reliable [38]. The efficiency of separation is low and expensive equipment is required to operate differential ultracentrifugation.
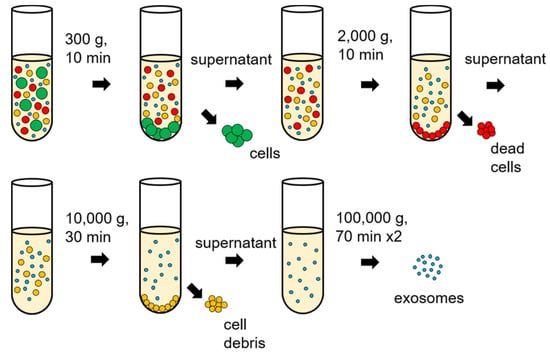
Figure 3.
Differential ultracentrifugation process for exosome isolations.
2.2. Density Gradient Ultracentrifugation
Density gradient ultracentrifugation has recently emerged as a technique for sorting exosomes from biological samples. We showed the density gradient ultracentrifugation process in Figure 4. Density gradient ultracentrifugation is a combination of centrifugation and density gradient and used gradient media, such as sucrose, Nycodenz (iohexol), and iodixanol. Exosomes are isolated via ultracentrifugation to the layer in which the density of the gradient media is equal to that of the exosomes [38,39]. This has the advantage of yielding higher purity because the separations are made according to density rather than size. Other advantages include less protein aggregation, less contamination, and good morphological properties. However, to date, this approach has been limited in its reproducibility, since it is very sensitive to the ultracentrifugation time. Moreover, it requires expensive equipment and stacking and selectively separating layers remains difficult.
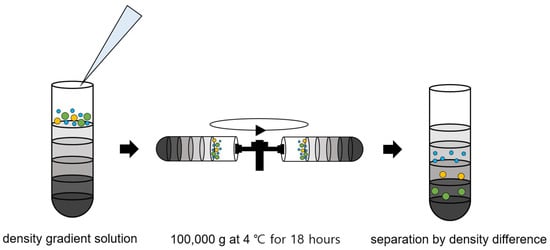
Figure 4.
Density gradient ultracentrifugation process for exosome isolation.
2.3. Size Exclusion
Figure 5 showed size exclusion methods, including filtration, size exclusion chromatography and the combination of filtration and ultracentrifugation. Exosomes can be purified by size exclusion chromatography (SEC), also referred to as gel permeation chromatography. First, the column is packed with porous polymeric beads with a constant pore size and the separated exosome for diagnosis [40,41,42]. Using an ultrafiltration membrane, exosomes can be separated by size [33,43,44,45,46]. The disadvantage of this method is that the separated exosomes also contain elements of similar size to the exosome such as high density lipoprotein (HDL) or other proteins. Another drawback to this approach is that it is time-consuming and can damage the exosome [47].
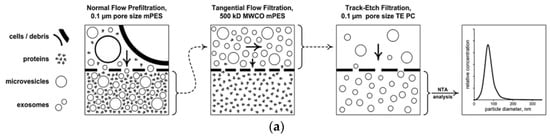
Figure 5.
Size exclusion methods. (a) filtration [42], (b) size exclusion chromatography [46], and (c) combination of filtration and ultracentrifugation [45]. Reprinted with permission from [42,45,46].
2.4. Polymer-Based Exosome Isolation
The polymer-based exosome isolation technique was developed by using polyethylene glycol (PEG) and PEG-dextran-based aqueous two-phase system (ATPS), as shown in Figure 6. For exosome isolation based on PEG, we mixed a biological fluid and a polymer with a precipitating solution, incubated it, and then separated it at low-speed centrifugation (Figure 6a). Generally, Exoquick™ (System Biosciences, USA) is a well-known method for polyethylene glycol (PEG) polymer-based technique [48].
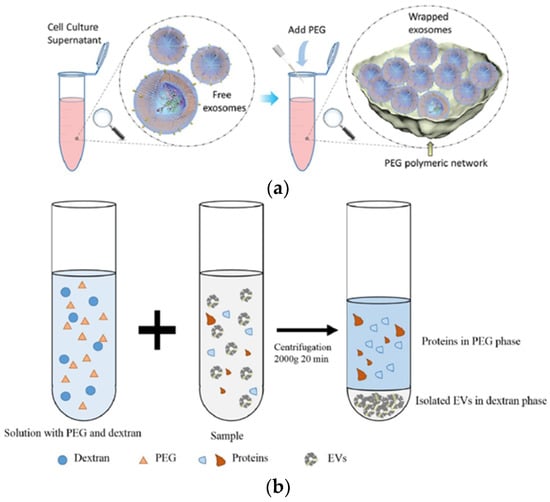
Figure 6.
Polymer based exosome isolation of (a) polyethylene glycol (PEG) [52] and (b) aqueous two-phase system (ATPS) [37]. Reprinted with permission from [37,52].
The PEG-dextran-based aqueous two-phase system (ATPS), as shown in Figure 6b, has been suggested for separating particles by a quick and simple process [37,49,50]. Polymer-based exosome precipitation does not require an expensive ultracentrifuge or a large sample volume. The disadvantage of this technique is the low purity of the separated exosome by the inclusion of lipoprotein. In addition, the polymer material can remain in the down-stream analysis [47,51,52].
2.5. Immunological Exosome Isolation Techniques
Immunological methods can selectively isolate exosomes from biological fluids by immobilizing specific capture antibodies that bind to specific proteins on the exosome surface (e.g., CD9, CD81, CD63, HLA-G, and Rab5b) (Figure 7) [36,53,54]. Specific capture antibodies detect CD9, CD63, and CD81 which are known as exosome markers that can be used to isolate exosomes.
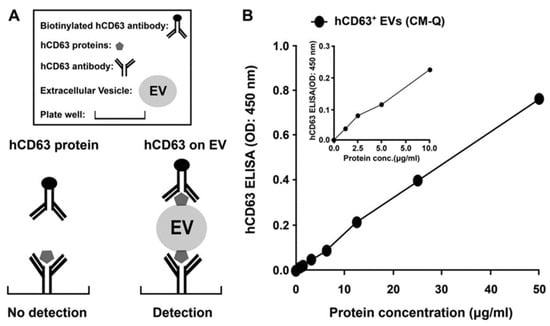
Figure 7.
Immunological exosome isolation techniques using enzyme-linked immunosorbent assay (ELISA) [54]. Reprinted with permission from [54].
Generally, magnetic beads with capture antibodies have been studied [36,53], and enzyme-linked immunosorbent assay (ELISA)-based techniques have also been widely used [55]. ELISA results are expressed as absorbance values which can be used to determine the yield and specificity of the exosome. This approach has several advantages, including its high yield and purity and the selective separation and measurement. The major drawback of this method is that it makes it difficult to handle large volumes, and the isolated antigens lose their activity of the surface functional group [47]. Another major issue is that separation is difficult to achieve in low concentrations because of non-specific binding issues [56].
2.6. Exosome Isolation Using Microfluidic Platform
Microfluidic-based exosome isolation techniques include microstructure-, acoustic wave-, immunological-, and size-based separation (Figure 8). Microfluidics are generally used to separate exosomes by size using micro- or nanostructures (Figure 8a) [57]. Several approaches based on an acoustic wave have been suggested for separating exosomes (Figure 8b) [58]. One interesting way to collect the exosome is through an immunoaffinity technique with a microfluidic channel (Figure 8c) [59,60,61]. In addition, the nanoshearing microfluidics technique with the immunoaffinity method has been reported to increase the reactivity in antibody-antigen interaction. (Figure 8d) [62].
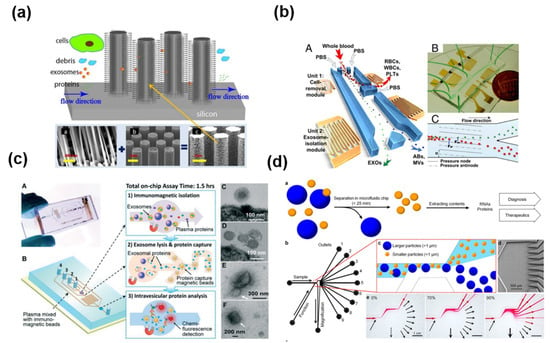
Figure 8.
Exosome isolation using microfluidic platform using (a) microstructure [57], (b) acoustic wave [58], (c) immunological separation [60], and (d) size based separation [62]. Reprinted with permission from [57,58,60,62].
As mentioned above, microfluidics represent a reliable method to separate exosomes according to size, immunoaffinity, and electrical properties. The microfluidic technique is quick, cheap, portable, and easy to automate. Disadvantages include the lack of standardization for clinical samples, limitations of verifying methods, and the small sample volume that can be processed [22].
3. Neuronal Exosome-Based Diagnostic Techniques for AD and PD
3.1. Exosome Quantification Techniques
Several exosome quantification techniques have been proposed, as shown in Figure 9.
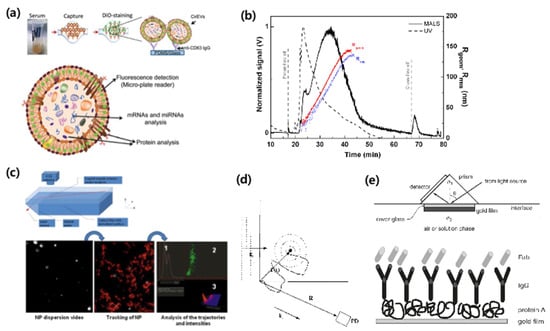
Figure 9.
Exosome quantification methods with (a) immunoaffinity capture (IAC) [59], (b) asymmetrical flow field-flow fractionation (AF4) [63], (c) nanoparticle tracking analysis (NTA) [64] (d) dynamic light scattering (DLS) [65], and (e) surface plasmon resonance (SPR) [66]. Reprinted with permission from [59,63,64,66].
Immunoaffinity capture (IAC) is the exosome capturing technology via immunoaffinity using an indirect isolation method (Figure 9a) [55,59,67]. In general, using a sandwich assay, IAC enables quantification of exosomes by analyzing color, fluorescence, and electrochemical signals. While it has a high selectivity, its drawbacks are its high cost and long running time.
Asymmetrical flow field-flow fractionation (AF4) is a technique for separating and quantifying molecules using field-flow fraction and diffusion (Figure 9b) [63,67]. This method can separate materials, such as nanoparticles, polymers, proteins, and viruses. Although AF4 is advantageous for its short running time and low cost, it also has a low selectivity.
Nanoparticle tracking analysis (NTA) involves the separation and quantification of particles according to their size (Figure 9c). When the exosome is released from body fluids, the rate of Brownian motion differs according to exosome size. NTA uses the rate of Brownian motion to analyze particles using direct, real-time visualization. This technique also tracks the concentration and size of exosomes using a light-scattering technique [64,68,69]. This means that the measurement cost is relatively low. However, NTA also has a long running time and low selectivity.
Dynamic light scattering (DLS) is a technique that determines particle size by light scattered by particles that exhibit Brownian motion (Figure 9d) [65]. Therefore, DLS can measure exosomes in suspension [65,70,71]. Boyd et al. analyzed the main differences between NTA and DLS, showing give different results, but these are all consistent considering the exact nature of each measure and their physical conditions [72]. A major disadvantage of DLS is that it has a limited selectivity and a long running time. It is also limited in its ability to resolve mixtures of exosomes, because it is biased towards the detection of larger particles [71].
Surface plasmon resonance (SPR) is an immunoaffinity-based assay that captures exosomes with receptors on an SPR sensor surface. When the exosome binds to receptors, its optical signals change, and its resonance can then be quantified through a light source [66,73,74]. SPR has a relatively high selectivity and a short running time, but is costly.
3.2. Neuronal Disease-Related Proteins and a New Sensing Platform
Biomarkers of neuronal disease have recently under intense investigation. Aβ, tau, and α-synuclein in neuronal exosomes are great candidates for neuronal disease markers. Specifically, Aβ peptide 42 (Aβ 42), Aβ 40, tau, and tau phosphorylated at threonine 181 (Thr181P) have been extensively studied as biomarkers for AD [75]. However, since they exist in extremely low levels in neuronal exosomes, we described highly sensitive and sensitive sensing platforms below.
To detect Aβ, Rushworth et al. have developed a method to detect Aβ using electrical impedimetric biosensors (Figure 10a) [76] Rama et al detected Aβ using a cyclic voltammetric immunosensor [77]. Oh et al. developed a biosensor based on a carbon nanotube-based field effect transistor (FET) to measure Aβ in human serum (Figure 10b) [78]. Recently, our group and collaborators developed a microelectrode-based impedimetric biosensor that detects plasma-based Aβ which has high selectivity and sensitivity (Figure 10c) [11,12].
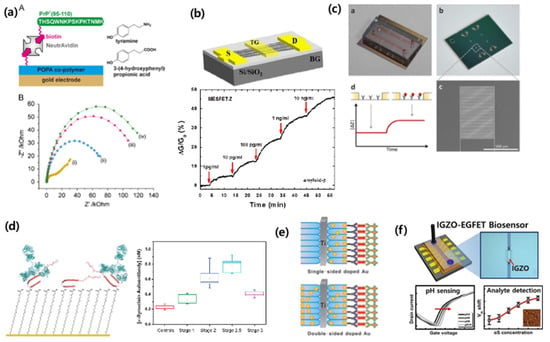
Figure 10.
Biomarker detection in exosome for Alzheimer’s disease (AD) and Parkinson’s disease (PD). (a) Impedimetric Aβ peptide detection [76]. (b) Carbon nanotube-based field effect transistor (FET) for Aβ detection [78]. (c) Impediemtric microelectrodes for Aβ detection in mouse plasma [11]. (d,e) Electrochemical detection of α-synuclein [79,80]. (f) electrolyter-gated FET for α-synuclein detection [81]. Reprinted with permission from [11,76,78,79,80,81].
The levels of tau and phosphorylated tau (p-tau) have been reported to be higher in patients with AD [82]. It has been reported that the combined detection of tau, p-tau, Aβ 40, and Aβ 42 CSF levels is more effective than standalone markers in predicting brain Aβ deposition [83].
α-synuclein is known to be associated with PD and rheumatoid dementia. Bryan et al. [79] reported an electrochemical detection-based method for the detection of α-synuclein in human blood (Figure 10d,e) [79]. MasaroeÌk [80] also used electrochemical detection to detect α-synuclein, and Yaruien et al. [84] used nanotubes sensors based on photoelectrochemistry to detect α-synuclein. Our group and collaborators also conducted an amorphous indium gallium zinc oxide (IGZO)-based electrolyte-gated field-effect transistor method to detect α-synuclein, and found that this method had a high selectivity and sensitivity (Figure 10f) [81].
Aβ and tau levels in plasma and serum are much lower than those found in CSF [85]. Considering the lack of sensing ability of commercial assays, detection platforms with a high sensitivity are urgently needed. Using a detection technology with great sensitivity would allow neurodegenerative diseases to be detected from blood-based exosomes.
3.3. Detection Techniques using Neuronal Disease-Related Proteins from the Neuronal Exosome
Exosomes have been exploited as a novel source of disease biomarkers, and it has been proposed that exosomes may mediate neurological disease [86]. Exosomal transfer of amyloid into the extra-cellular space could be an important pathway in the development of AD [23]. It has been reported that the neuronal exosome contains Aβ, α-synuclein tau, and microRNA [25,81,87]. Researchers have assessed amyloidopathy and tauopathy and their effects on neurodegenerative disorders.
Aβ deposition in senile plaques and cerebral vessels is a neuropathological feature of AD [88]. Amyloid plaque formation primarily results from Aβ peptides, which are considered to play a pivotal role in AD pathogenesis [89]. Thus, techniques for the detection of Aβ have been intensively studied for the early detection of AD [90].
α-synuclein is a 14-kD amino-acid protein, and the aggregation and dysfunction of α-synuclein are common in neurodegenerative disorders; α-synuclein is closely related to PD and dementia with Lewy bodies [91]. In addition to a neuronal exosome, α-synuclein is an abundant protein in the brain that is easily found in the heart, muscle, and other tissues. It is mainly found in the pre-synaptic terminals of the brain, and interacts with phospholipids and proteins [91].
Tau protein is critically important in axonal maintenance and axonal transport [92]. Total tau (T-tau) and p-tau levels in the brain, blood, and CSF are known to be related to neurodegenerative disorders, and reflect neuronal degeneration; T-tau and p-tau levels are considered to be useful biomarkers for assessing neurodegenerative disorders [93].
ELISA is representative method for Aβ [22,34,87], α-synuclein [25], and tau [22]. Real-time polymerase chain reaction (PCR) has been used to analyze microRNA from neuronal exosomes for the diagnosis of amyotrophic lateral sclerosis (ALS) [22,94,95]. Western blot analysis of α-synuclein has been used to uncover the pathology of PD and rheumatoid dementia [96].
CSF is one of the main biological samples used for exosome detection. Street et al. have reported that ultracentrifugation, Western blot analysis, and transmission electron microscopy can be used to identify and perform proteomic profiling of exosomes in human CSF [97]. Chiasserini et al. showed that exosome in human CSF contain prionogenic proteins such as the amyloid precursor protein and the prion protein [98]. Saman et al have suggested that exosome-mediated secretion of p-tau plays a significant role in the abnormal processing of tau and in the genesis of elevated CSF tau in early AD [99]. Rajendran et al. reported that amyloid peptides are released in association with exosomes [23]. The authors showed that exosomal proteins accumulate in the plaques of human AD brains.
As mentioned above, CSF collection is an elaborate and complicated process; therefore, the detection of exosomes from human blood is very important in neuronal diagnosis. Hamlett et al. reported that neuronal exosome levels of Aβ 1–42, p-T181-tau, and p-S396-tau were significantly elevated in individuals with Down syndrome compared with age-matched controls [22]. Levels of AD-related proteins were quantified from neuronally derived blood exosomes to identify biomarkers for prediction and staging of mild cognitive impairment and AD [100]. Stern et al. found that tau-positive exosomes in plasma may be a potential biomarker for chronic traumatic encephalopathy [101]. Shi et al. demonstrated the presence of α-synuclein-containing L1 cell adhesion molecule (L1CAM) exosomes in the blood plasma and quantified α-synuclein within these exosomes [24].
4. Conclusions and Perspectives
Many reports have suggested that neuronal exosome-based diagnoses can be made from a whole blood; however, the use of exosomes in neuronal disease diagnoses is still in early stages of development. To address these challenges of neuronal exosomal diagnoses, an advanced two-step method for exosome separation and detection is needed.
The two-step method for exosome separation is used to selectively isolate and capture neuronally derived blood exosomes, as shown in Figure 11.
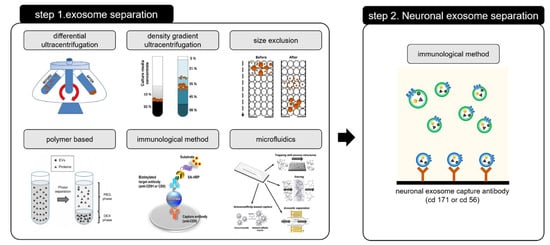
Figure 11.
Two-step process requiring for isolating the neuronal exosome. Step 1 for exosome separation [46,49,102] and Step 2 for neuronal exosome separation. Reprinted with permission from [46,49,102].
The exosome is first separated from the blood, after which the neuronal exosome is isolated from the separated one. Isolating the neuronal exosome from the blood-based exosome is mainly done by using a specific antigen (CD 56, CD 171) which is bound to the neuronal exosome surface [30,103]. The two-step process for isolating the neuronal exosome is expensive and time consuming. In addition, the loss of the amount of exosome extracted during the two steps results in low efficiency and low yield. Time and cost of exosome separation can be reduced by preventing non-specific binding and improving the reactivity by increased antibody-antigen interactions, making separation simpler. Another approach is to separate/preconcentrate the exosome using a microfluidic platform. We have shown that the use of a microfluidic paper-based analytical device chip with preconcentrating function, yields a 5-fold increase in preconcentrating factors under blood-based biofluids (serum) [104,105]. Based on this technique, our group is now trying to separate and concentrate exosomes simultaneously.
Recent studies have reported that exosomes are related to neurodegenerative disorders; however, one current challenge for the application of exosomes is their isolation method. Nonetheless, exosome-based neuronal diagnostic technologies have a huge potential in the development of clinical devices.
Funding
This research was funded by the National Research Foundation of Korea, Grant (NRF-2018R1D1A1A09084044), National Research Foundation of Korea (NRF) and Korean Government (MSIP) (No. NRF-2016R1A2B4010269).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mattson, M.P.; Pedersen, W.A.; Duan, W.; Culmsee, C.; Camandola, S. Cellular and Molecular Mechanisms Underlying Perturbed Energy Metabolism and Neuronal Degeneration in Alzheimer’s and Parkinson’s Diseases. Ann. N. Y. Acad. Sci. 1999, 893, 154–175. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.A.; Greenlund, S.F.; McGuire, L.C.; Lu, H.; Croft, J.B. Deaths from Alzheimer’s Disease—United States, 1999–2014. MMWR. Morb. Mortal. Wkly. Rep. 2017, 66, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2017 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2017, 13, 325–373. [Google Scholar] [CrossRef]
- Liu, S.; Liu, S.; Cai, W.; Pujol, S.; Kikinis, R.; Feng, D. Early diagnosis of Alzheimer’s disease with deep learning. In Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014; pp. 1015–1018. [Google Scholar]
- Small, G.W. Early diagnosis of Alzheimer’s disease: Update on combining genetic and brain-imaging measures. Dialogues Clin. Neurosci. 2000, 2, 241–246. [Google Scholar] [PubMed]
- Mueller, S.G.; Weiner, M.W.; Thal, L.J.; Petersen, R.C.; Jack, C.R.; Jagust, W.; Trojanowski, J.Q.; Toga, A.W.; Beckett, L. Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s Dement. 2005, 1, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.R.; Bing-Canar, H.; Alosco, M.L.; Steinberg, E.G.; Martin, B.; Chaisson, C.; Kowall, N.; Tripodis, Y.; Stern, R.A. Mini Mental State Examination and Logical Memory scores for entry into Alzheimer’s disease trials. Alzheimer’s Res. Ther. 2016, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Bernstein, M.A.; Fox, N.C.; Thompson, P.; Alexander, G.; Harvey, D.; Borowski, B.; Britson, P.J.; Whitwell, J.L.; Ward, C.; et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 2008, 27, 685–691. [Google Scholar] [CrossRef] [PubMed]
- McGeer, P.L.; Kamo, H.; Harrop, R.; McGeer, E.G.; Martin, W.R.W.; Pate, B.D.; Li, D.K.B. Comparison of PET, MRI, and CT with pathology in a proven case of Alzheimer’s disease. Neurology 1986, 36, 1569. [Google Scholar] [CrossRef] [PubMed]
- Frisoni, G.B.; Boccardi, M.; Barkhof, F.; Blennow, K.; Cappa, S.; Chiotis, K.; Démonet, J.-F.; Garibotto, V.; Giannakopoulos, P.; Gietl, A.; et al. Strategic roadmap for an early diagnosis of Alzheimer’s disease based on biomarkers. Lancet Neurol. 2017, 16, 661–676. [Google Scholar] [CrossRef]
- Yoo, Y.K.; Kim, J.; Kim, G.; Kim, Y.S.; Kim, H.Y.; Lee, S.; Cho, W.W.; Kim, S.; Lee, S.-M.; Lee, B.C.; et al. A highly sensitive plasma-based amyloid-β detection system through medium-changing and noise cancellation system for early diagnosis of the Alzheimer’s disease. Sci. Rep. 2017, 7, 8882. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.; Yoon, D.; Kim, G.; Kim, J.; Il Han, S.; Lee, J.; Chae, M.-S.; Lee, S.-M.; Lee, K.; Seon Hwang, K.; et al. An Enhanced Platform to Analyse Low-Affinity Amyloid β Protein by Integration of Electrical Detection and Preconcentrator. Sci. Rep. 2017, 7, 14303. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Kaneko, N.; Villemagne, V.L.; Kato, T.; Doecke, J.; Doré, V.; Fowler, C.; Li, Q.-X.; Martins, R.; Rowe, C.; et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature 2018, 554, 249. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Carpenter, E.; Issadore, D. Detection and isolation of circulating exosomes and microvesicles for cancer monitoring and diagnostics using micro-/nano-based devices. Analyst 2016, 141, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflamm. 2014, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Tsilioni, I.; Panagiotidou, S.; Theoharides, T.C. Exosomes in Neurologic and Psychiatric Disorders. Clin. Ther. 2014, 36, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Frühbeis, C.; Fröhlich, D.; Krämer-Albers, E.M. Emerging Roles of Exosomes in Neuron–Glia Communication. Front. Physiolol. 2012, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, P.R.; Zheng, Y.; Fischer, R.; Heidasch, R.; Gardiner, C.; Evetts, S.; Hu, M.; Wade-Martins, R.; Turner, M.R.; Morris, J.; et al. Identification of distinct circulating exosomes in Parkinson’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zheng, T.; Zhang, B. Exosomes in Parkinson’s Disease. Neurosci. Bull. 2017, 33, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Kapogiannis, D.; Schwartz, J.B.; Lobach, I.V.; Goetzl, L.; Abner, E.L.; Jicha, G.A.; Karydas, A.M.; Boxer, A.; Miller, B.L. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016, 30, 4141–4148. [Google Scholar] [CrossRef] [PubMed]
- Hamlett, E.D.; Goetzl, E.J.; Ledreux, A.; Vasilevko, V.; Boger, H.A.; LaRosa, A.; Clark, D.; Carroll, S.L.; Carmona-Iragui, M.; Fortea, J.; et al. Neuronal exosomes reveal Alzheimer’s disease biomarkers in Down syndrome. Alzheimer’s Dement. 2017, 13, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Liu, C.; Cook, T.J.; Bullock, K.M.; Zhao, Y.; Ginghina, C.; Li, Y.; Aro, P.; Dator, R.; He, C.; et al. Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014, 128, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Stuendl, A.; Kunadt, M.; Kruse, N.; Bartels, C.; Moebius, W.; Danzer, K.M.; Mollenhauer, B.; Schneider, A. Induction of α-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies. Brain 2016, 139, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Grant, R.; Condon, B.; Hart, I.; Teasdale, G.M. Changes in intracranial CSF volume after lumbar puncture and their relationship to post-LP headache. J. Neurol. Neurosurg. Psychiatry 1991, 54, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Eng, R.K.; Seligman, S.J. Lumbar puncture—Induced meningitis. JAMA 1981, 245, 1456–1459. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.J.; O’Loughlin, A.J.; Lakhal, S. Exosomes and the blood–brain barrier: Implications for neurological diseases. Ther. Deliv. 2011, 2, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Mustapic, M.; Eitan, E.; Werner Jr, J.K.; Berkowitz, S.T.; Lazaropoulos, M.P.; Tran, J.; Goetzl, E.J.; Kapogiannis, D. Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Front. Neurosci. 2017, 11, 278. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, L.; Merabova, N.; Darbinian, N.; Martirosyan, D.; Poletto, E.; Fugarolas, K.; Menkiti, O. Diagnostic Potential of Neural Exosome Cargo as Biomarkers for Acute Brain Injury. Ann. Clin. Transl. Neurol. 2017, 5, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome Isolation for Proteomic Analyses and RNA Profiling. In Serum/Plasma Proteomics: Methods and Protocols; Simpson, R.J., Greening, D.W., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 235–246. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Carlson, O.D.; Mustapic, M.; Kapogiannis, D. Low neural exosomal levels of cellular survival factors in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Ziaei, P.; Berkman, C.E.; Norton, M.G. Review: Isolation and Detection of Tumor-Derived Extracellular Vesicles. ACS Appl. Nano Mater. 2018, 1, 2004–2020. [Google Scholar] [CrossRef]
- Clotilde, T.; Sebastian, A.; Graça, R.; Aled, C. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30. [Google Scholar] [CrossRef]
- Kang, H.; Kim, J.; Park, J. Methods to isolate extracellular vesicles for diagnosis. Micro Nano Syst. Lett. 2017, 5, 15. [Google Scholar] [CrossRef]
- Brasaemle, D.L.; Wolins, N.E. Isolation of Lipid Droplets from Cells by Density Gradient Centrifugation. Curr. Protoc. Cell Biol. 2016, 72, 3.15.1–3.15.13. [Google Scholar] [PubMed]
- Iwai, K.; Minamisawa, T.; Suga, K.; Yajima, Y.; Shiba, K. Isolation of human salivary extracellular vesicles by iodixanol density gradient ultracentrifugation and their characterizations. J. Extracell. Vesicles 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Merchant, M.L.; Powell, D.W.; Wilkey, D.W.; Cummins, T.D.; Deegens, J.K.; Rood, I.M.; McAfee, K.J.; Fleischer, C.; Klein, E.; Klein, J.B. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin. Appl. 2010, 4, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Grant, R.; Ansa-Addo, E.; Stratton, D.; Antwi-Baffour, S.; Jorfi, S.; Kholia, S.; Krige, L.; Lange, S.; Inal, J. A filtration-based protocol to isolate human Plasma Membrane-derived Vesicles and exosomes from blood plasma. J. Immunol. Methods 2011, 371, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, M.L.; Ilmer, M.; Silva, L.P.; Hawke, D.H.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Colao, I.L.; Corteling, R.; Bracewell, D.; Wall, I. Manufacturing Exosomes: A Promising Therapeutic Platform. Trends Mol. Med. 2018, 24, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Willis, G.R.; Kourembanas, S.; Mitsialis, S.A. Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front. Cardiovasc. Med. 2017, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Yakimchuk, K. Exosomes: Isolation and characterization methods and specific markers. Mater. Methods 2015, 5, 1450. [Google Scholar] [CrossRef]
- Rekker, K.; Saare, M.; Roost, A.M.; Kubo, A.-L.; Zarovni, N.; Chiesi, A.; Salumets, A.; Peters, M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014, 47, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shin, H.; Kim, J.; Kim, J.; Park, J. Isolation of High-Purity Extracellular Vesicles by Extracting Proteins Using Aqueous Two-Phase System. PLoS ONE 2015, 10, e0129760. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Han, C.; Labuz, J.M.; Kim, J.; Kim, J.; Cho, S.; Gho, Y.S.; Takayama, S.; Park, J. High-yield isolation of extracellular vesicles using aqueous two-phase system. Sci. Rep. 2015, 5, 13103. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Inoshima, Y.; Matsuda, T.; Ishiguro, N. Comparison of Methods for Isolating Exosomes from Bovine Milk. J. Vet. Med. Sci. 2012, 74, 1523–1525. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Kim, D.-K.; Nishida, H.; An, S.Y.; Shetty, A.K.; Bartosh, T.J.; Prockop, D.J. Chromatographically isolated CD63+ CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc. Natl. Acad. Sci. USA 2016, 113, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef] [PubMed]
- Farajollahi, M.M.; Cook, D.B.; Hamzehlou, S.; Self, C.H. Reduction of non-specific binding in immunoassays requiring long incubations. Scand. J. Clin. Lab. Investig. 2012, 72, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, H.-J.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.J.; Liu, X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013, 13, 2879–2882. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773–3780. [Google Scholar] [CrossRef] [PubMed]
- Shiddiky, M.J.A.; Vaidyanathan, R.; Rauf, S.; Tay, Z.; Trau, M. Molecular Nanoshearing: An Innovative Approach to Shear off Molecules with AC-Induced Nanoscopic Fluid Flow. Sci. Rep. 2014, 4, 3716. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Han, D.; Park, M.C.; Mun, J.Y.; Choi, J.; Chun, H.; Kim, S.; Hong, J.W. Separation of extracellular nanovesicles and apoptotic bodies from cancer cell culture broth using tunable microfluidic systems. Sci. Rep. 2017, 7, 9907. [Google Scholar] [CrossRef] [PubMed]
- Sitar, S.; Kejžar, A.; Pahovnik, D.; Kogej, K.; Tušek-Žnidarič, M.; Lenassi, M.; Žagar, E. Size Characterization and Quantification of Exosomes by Asymmetrical-Flow Field-Flow Fractionation. Anal. Chem. 2015, 87, 9225–9233. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Urrea, J.A.; Tuoriniemi, J.; Hassellöv, M. Applications of particle-tracking analysis to the determination of size distributions and concentrations of nanoparticles in environmental, biological and food samples. TrAC Trends Anal. Chem. 2011, 30, 473–483. [Google Scholar] [CrossRef]
- Goldburg, W.I. Dynamic light scattering. Am. J. Phys. 1999, 67, 1152–1160. [Google Scholar] [CrossRef]
- Tang, Y.; Zeng, X.; Liang, J. Surface Plasmon Resonance: An Introduction to a Surface Spectroscopy Technique. J. Chem. Educ. 2010, 87, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.E.; Manangon, E.; Hood, J.L.; Wickline, S.A.; Fernandez, D.P.; Johnson, W.P.; Gale, B.K. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal. Bioanal. Chem. 2014, 406, 7855–7866. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, S.K.; Wang, C.S. The self-preserving particle size distribution for coagulation by brownian motion. J. Colloid Interface Sci. 1966, 22, 126–132. [Google Scholar] [CrossRef]
- Soo, C.Y.; Song, Y.; Zheng, Y.; Campbell, E.C.; Riches, A.C.; Gunn-Moore, F.; Powis, S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 2012, 136, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Berne, B.J.; Pecora, R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics; Dover Publications: Mineola, NY, USA, 2000; p. 376. [Google Scholar]
- Lawrie, A.S.; Albanyan, A.; Cardigan, R.A.; Mackie, I.J.; Harrison, P. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009, 96, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Boyd, R.D.; Pichaimuthu, S.K.; Cuenat, A. New approach to inter-technique comparisons for nanoparticle size measurements; using atomic force microscopy, nanoparticle tracking analysis and dynamic light scattering. Colloids Surf. A Physicochem. Eng. Asp. 2011, 387, 35–42. [Google Scholar] [CrossRef]
- Mason, S.; La, S.; Mytych, D.; Swanson, S.J.; Ferbas, J. Validation of the BIACORE 3000 platform for detection of antibodies against erythropoietic agents in human serum samples. Curr. Med. Res. Opin. 2003, 19, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiar, R. Surface Plasmon Resonance Spectroscopy: A Versatile Technique in a Biochemist’s Toolbox. J. Chem. Educ. 2013, 90, 203–209. [Google Scholar] [CrossRef]
- Chung, C.Y.; Khurana, V.; Auluck, P.K.; Tardiff, D.F.; Mazzulli, J.R.; Soldner, F.; Baru, V.; Lou, Y.; Freyzon, Y.; Cho, S.; et al. Identification and Rescue of α-Synuclein Toxicity in Parkinson Patient-Derived Neurons. Science 2013, 342, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, J.V.; Ahmed, A.; Griffiths, H.H.; Pollock, N.M.; Hooper, N.M.; Millner, P.A. A label-free electrical impedimetric biosensor for the specific detection of Alzheimer’s amyloid-beta oligomers. Biosens. Bioelectron. 2014, 56, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Rama, E.C.; González-García, M.B.; Costa-García, A. Competitive electrochemical immunosensor for amyloid-beta 1-42 detection based on gold nanostructurated Screen-Printed Carbon Electrodes. Sens. Actuators B Chem. 2014, 201, 567–571. [Google Scholar] [CrossRef]
- Oh, J.; Yoo, G.; Chang, Y.W.; Kim, H.J.; Jose, J.; Kim, E.; Pyun, J.-C.; Yoo, K.-H. A carbon nanotube metal semiconductor field effect transistor-based biosensor for detection of amyloid-beta in human serum. Biosens. Bioelectron. 2013, 50, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.; Vanderstichele, H.; Andreasen, N.; De Meyer, G.; Wallin, A.; Holmberg, B.; Rosengren, L.; Vanmechelen, E.; Blennow, K. Simultaneous measurement of beta-amyloid(1-42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin. Chem. 2005, 51, 336. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, S.; Zetterberg, H.; Mattsson, N.; Johansson, P.; Minthon, L.; Blennow, K.; Olsson, M.; Hansson, O. Detailed comparison of amyloid PET and CSF biomarkers for identifying early Alzheimer disease. Neurology 2015, 85, 1240. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.; Luo, X.; Forsgren, L.; Morozova-Roche, L.A.; Davis, J.J. The robust electrochemical detection of a Parkinson’s disease marker in whole blood sera. Chem. Sci. 2012, 3, 3468–3473. [Google Scholar] [CrossRef]
- Masařík, M.; Stobiecka, A.; Kizek, R.; Jelen, F.; Pechan, Z.; Hoyer, W.; Jovin, T.M.; Subramaniam, V.; Paleček, E. Sensitive Electrochemical Detection of Native and Aggregated α-Synuclein Protein Involved in Parkinson’s Disease. Electroanalysis 2004, 16, 1172–1181. [Google Scholar] [CrossRef]
- An, Y.; Tang, L.; Jiang, X.; Chen, H.; Yang, M.; Jin, L.; Zhang, S.; Wang, C.; Zhang, W. A Photoelectrochemical Immunosensor Based on Au-Doped TiO2 Nanotube Arrays for the Detection of α-Synuclein. Chem. A Eur. J. 2010, 16, 14439–14446. [Google Scholar] [CrossRef] [PubMed]
- Chae, M.-S.; Park, J.H.; Son, H.W.; Hwang, K.S.; Kim, T.G. IGZO-based electrolyte-gated field-effect transistor for in situ biological sensing platform. Sens. Actuators B Chem. 2018, 262, 876–883. [Google Scholar] [CrossRef]
- Lue, L.-F.; Guerra, A.; Walker, D.G. Amyloid Beta and Tau as Alzheimer’s Disease Blood Biomarkers: Promise from New Technologies. Neurol. Ther. 2017, 6, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Li, Y.; Buller, B.; Katakowski, M.; Zhang, Y.; Wang, X.; Shang, X.; Zhang, Z.G.; Chopp, M. Exosome-Mediated Transfer of miR-133b from Multipotent Mesenchymal Stromal Cells to Neural Cells Contributes to Neurite Outgrowth. Stem Cells 2012, 30, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Yuyama, K.; Sun, H.; Usuki, S.; Sakai, S.; Hanamatsu, H.; Mioka, T.; Kimura, N.; Okada, M.; Tahara, H.; Furukawa, J.-I.; et al. A potential function for neuronal exosomes: Sequestering intracerebral amyloid-β peptide. FEBS Lett. 2014, 589, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Schmechel, D.E.; Saunders, A.M.; Strittmatter, W.J.; Crain, B.J.; Hulette, C.M.; Joo, S.H.; Pericak-Vance, M.A.; Goldgaber, D.; Roses, A.D. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 9649. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.S.; Troncoso, J.C.; Fangmark Tucker, S.M. Maximizing the Potential of Plasma Amyloid-Beta as a Diagnostic Biomarker for Alzheimer’s Disease. Neuromol. Med. 2008, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Blennow, K.; Zetterberg, H. Understanding Biomarkers of Neurodegeneration: Ultrasensitive detection techniques pave the way for mechanistic understanding. Nat. Med. 2015, 21, 217. [Google Scholar] [CrossRef] [PubMed]
- Marques, O.; Outeiro, T.F. Alpha-synuclein: From secretion to dysfunction and death. Cell Death Dis. 2012, 3, e350. [Google Scholar] [CrossRef] [PubMed]
- Terwel, D.; Dewachter, I.; Van Leuven, F. Axonal transport, tau protein, and neurodegeneration in Alzheimer’s disease. Neuromol. Med. 2002, 2, 151–165. [Google Scholar] [CrossRef]
- Dai, Y.; Molazemhosseini, A.; Liu, C. A Single-Use, In Vitro Biosensor for the Detection of T-Tau Protein, A Biomarker of Neuro-Degenerative Disorders, in PBS and Human Serum Using Differential Pulse Voltammetry (DPV). Biosensors 2017, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Simeoli, R.; Montague, K.; Jones, H.R.; Castaldi, L.; Chambers, D.; Kelleher, J.H.; Vacca, V.; Pitcher, T.; Grist, J.; Al-Ahdal, H.; et al. Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat. Commun. 2017, 8, 1778. [Google Scholar] [CrossRef] [PubMed]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef] [PubMed]
- Ngolab, J.; Trinh, I.; Rockenstein, E.; Mante, M.; Florio, J.; Trejo, M.; Masliah, D.; Adame, A.; Masliah, E.; Rissman, R.A. Brain-derived exosomes from dementia with Lewy bodies propagate α-synuclein pathology. Acta Neuropathol. Commun. 2017, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.A.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Chiasserini, D.; van Weering, J.R.T.; Piersma, S.R.; Pham, T.V.; Malekzadeh, A.; Teunissen, C.E.; de Wit, H.; Jiménez, C.R. Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. J. Proteomics 2014, 106, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.Y.; et al. Exosome-associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef] [PubMed]
- Winston, C.N.; Goetzl, E.J.; Akers, J.C.; Carter, B.S.; Rockenstein, E.M.; Galasko, D.; Masliah, E.; Rissman, R.A. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2016, 3, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Stern, R.A.; Tripodis, Y.; Baugh, C.M.; Fritts, N.G.; Martin, B.M.; Chaisson, C.; Cantu, R.C.; Joyce, J.A.; Shah, S.; Ikezu, T.; et al. Preliminary Study of Plasma Exosomal Tau as a Potential Biomarker for Chronic Traumatic Encephalopathy. J. Alzheimer’s Dis. 2016, 51, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Ingato, D.; Lee, J.U.; Sim, S.J.; Kwon, Y.J. Good things come in small packages: Overcoming challenges to harness extracellular vesicles for therapeutic delivery. J. Control. Release 2016, 241, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Heinzelman, P.; Bilousova, T.; Campagna, J.; John, V. Nanoscale Extracellular Vesicle Analysis in Alzheimer’s Disease Diagnosis and Therapy. Int. J. Alzheimer’ Dis. 2016, 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Han, S.I.; Hwang, K.S.; Kwak, R.; Lee, J.H. Microfluidic paper-based biomolecule preconcentrator based on ion concentration polarization. Lab Chip 2016, 16, 2219–2227. [Google Scholar] [CrossRef] [PubMed]
- Han, S.I.; Yoo, Y.K.; Lee, J.; Kim, C.; Lee, K.; Lee, T.H.; Kim, H.; Yoon, D.S.; Hwang, K.S.; Kwak, R.; et al. High-ionic-strength pre-concentration via ion concentration polarization for blood-based biofluids. Sens. Actuators B Chem. 2018, 268, 485–493. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).