An Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic System for the Isolation of High-Purity CD45neg/EpCAMneg Cells from the Blood Samples of Cancer Patients—Demonstration and Initial Exploration of the Clinical Significance of These Cells
Abstract
1. Introduction
2. Materials and Methods
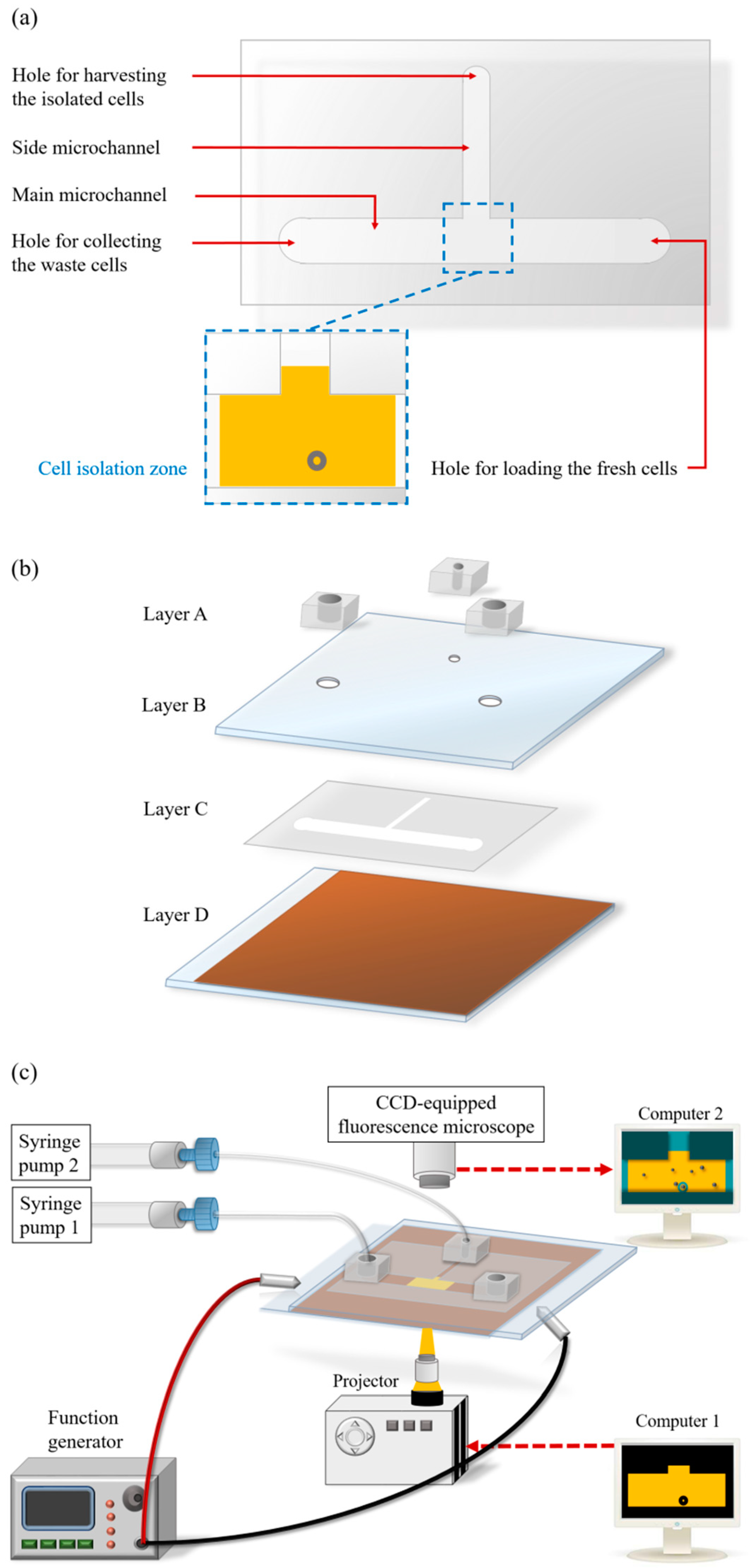
2.1. Design of the ODEP Microfluidic System for High-Purity Isolation of CD45neg/EpCAMneg Cells
2.2. Microfabrication and Experimental Setup
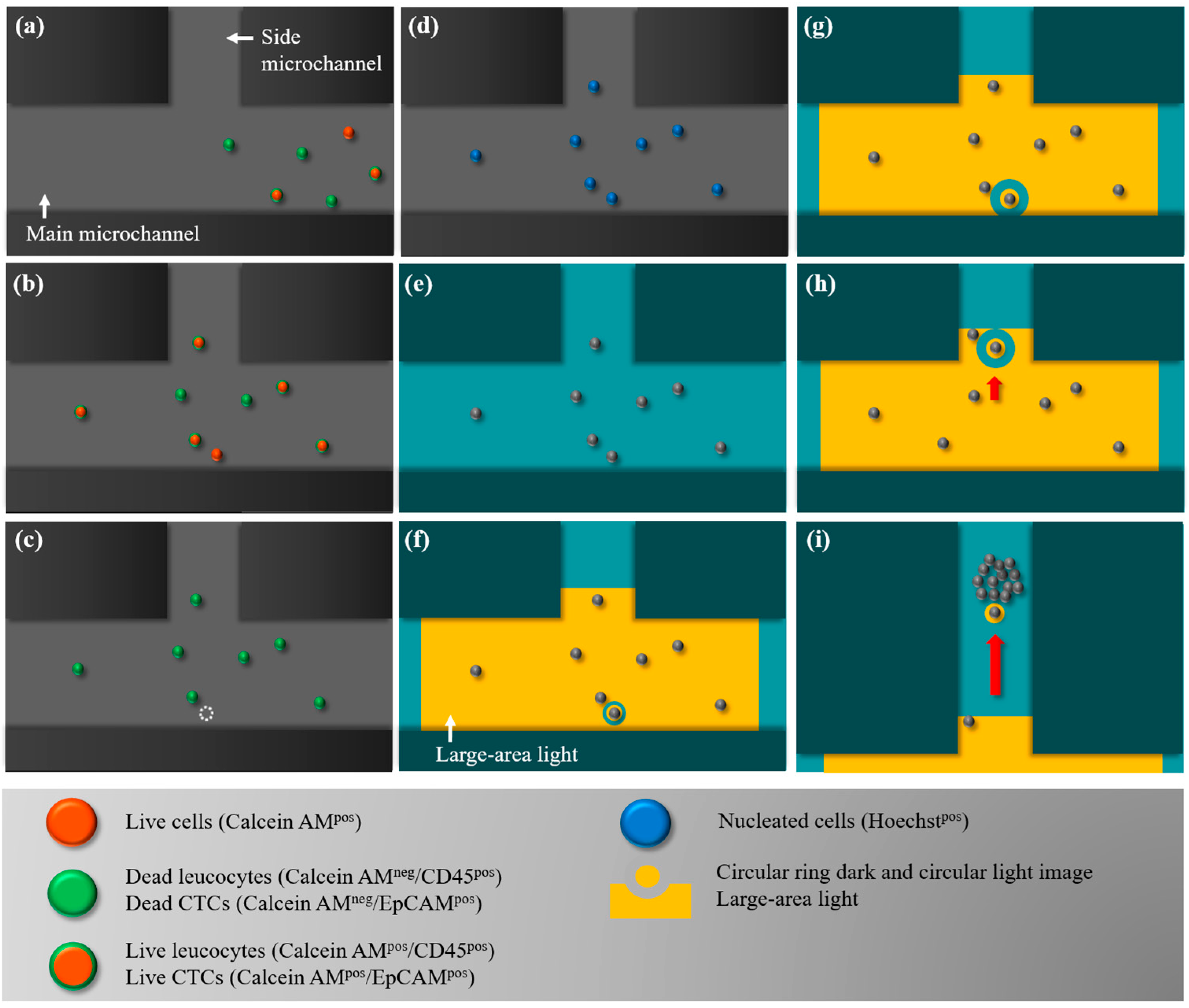
2.3. The Working Scheme for the Isolation and Purification of CD45neg/EpCAMneg Cells
2.4. Working Mechanism and the Optimal Operating Conditions for the ODEP Force-Based Cell Manipulation in This Work
2.5. Evaluation of the CD45neg/EpCAMneg Cell Isolation Performance-Cancer Cell Line Model
2.6. Comparison of the Cancer-Related Gene Expression of the CD45neg/EpCAMneg Cell Populations Isolated from the Blood Samples of Healthy Donors and Head-and-Neck Cancer Patients
2.7. Statistical Analysis
3. Results and Discussions
3.1. Characteristic Features of the Proposed ODEP-Based Microfluidic System for the Isolation and Purification of CD45neg/EpCAMneg Cells
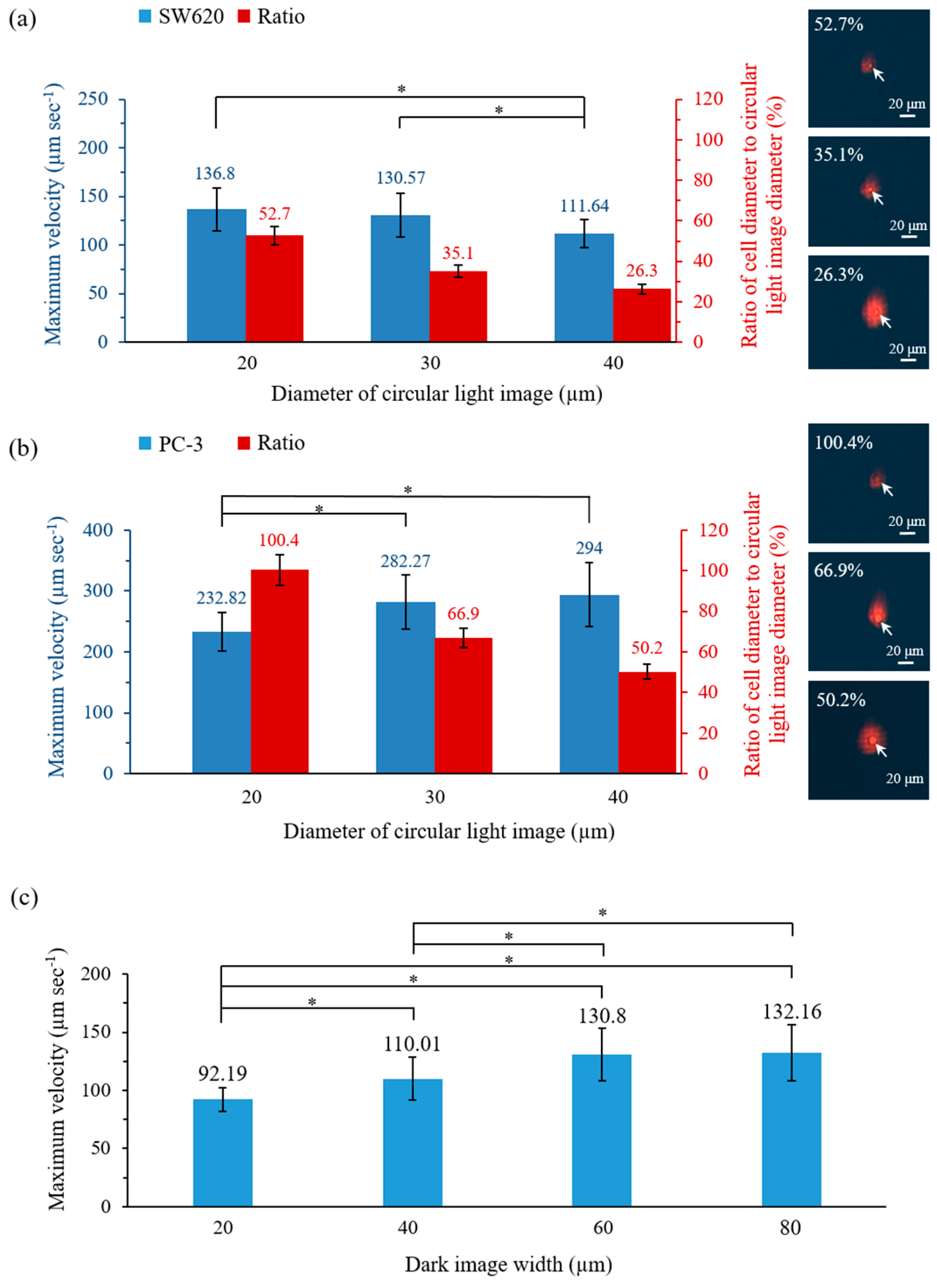
3.2. Optimal Operating Conditions for ODEP Force-Based Cell Manipulation
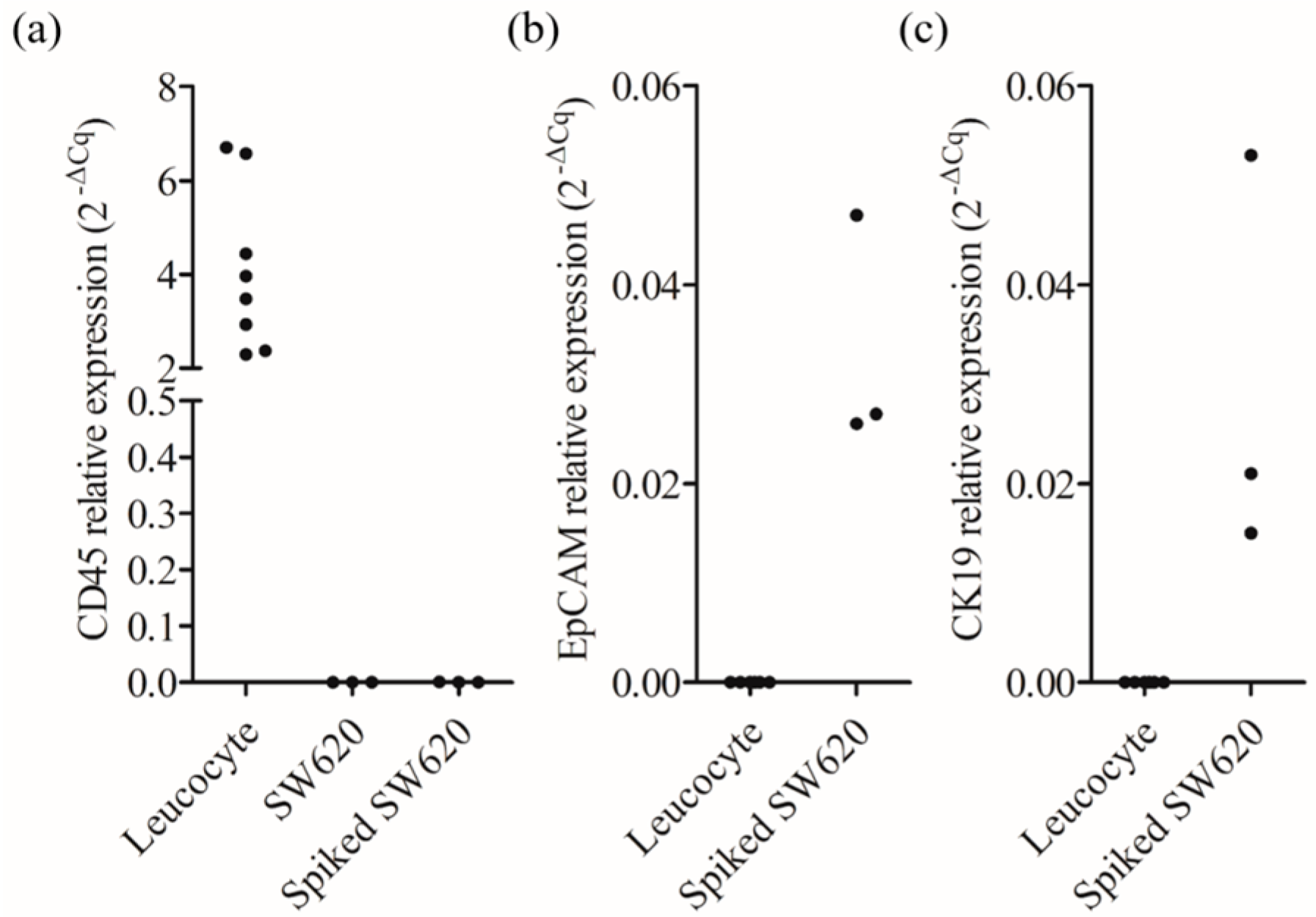
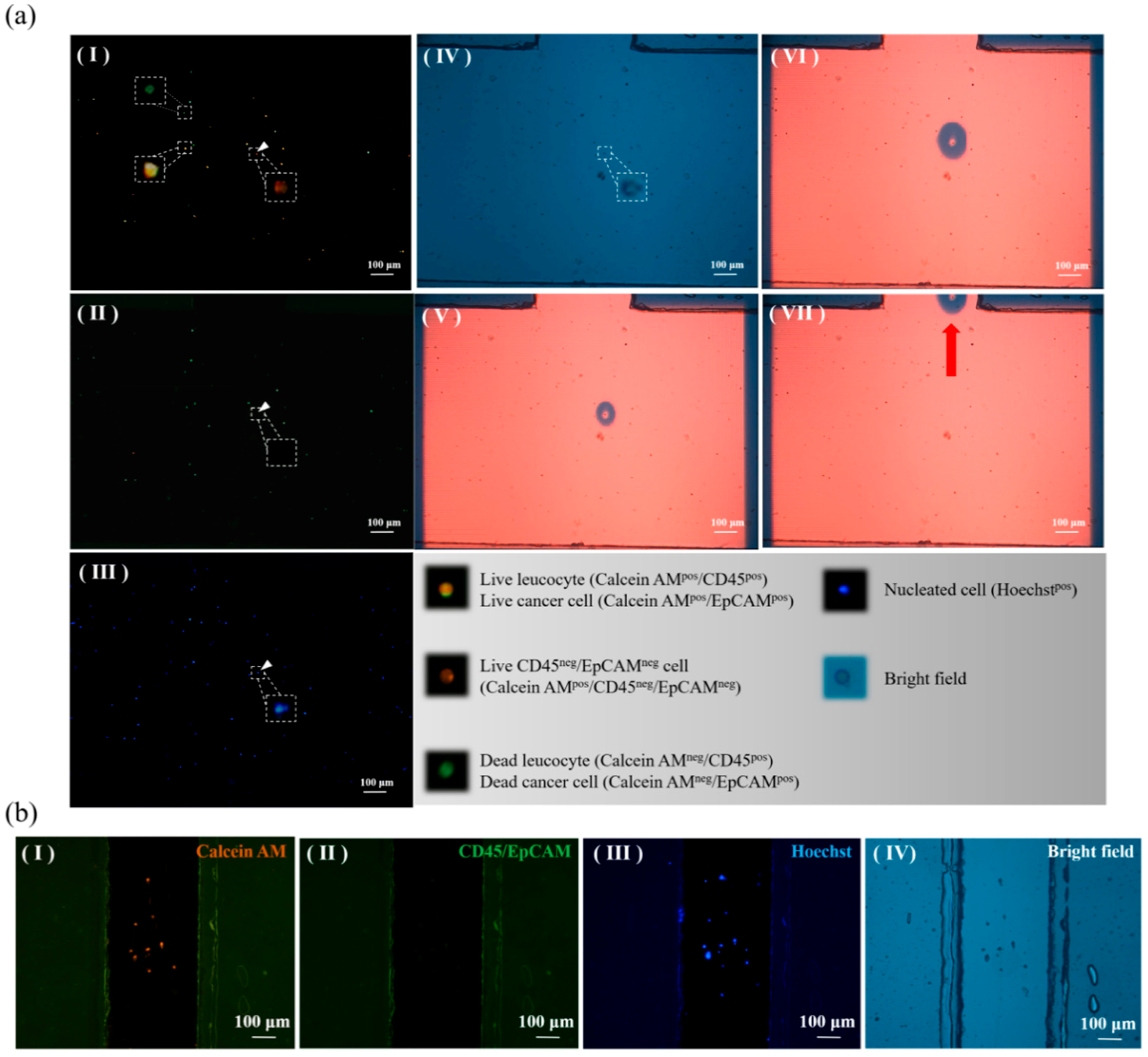
3.3. Performance Evaluation of CD45neg/EpCAMneg Cell Isolation
3.4. Comparison of the Cancer-Related Gene Expression of the CD45neg/EpCAMneg Cells Isolated from the Blood Samples of Healthy Donors and Head-and-Neck Cancer Patients
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146. [Google Scholar]
- Zhang, Y.; Lv, Y.; Niu, Y.; Su, H.; Feng, A. Role of Circulating Tumor Cell (CTC) Monitoring in Evaluating Prognosis of Triple-Negative Breast Cancer Patients in China. Med. Sci. Monit. 2017, 23, 3071–3079. [Google Scholar] [CrossRef] [PubMed]
- Dago, A.E.; Stepansky, A.; Carlsson, A.; Luttgen, M.; Kendall, J.; Baslan, T.; Kolatkar, A.; Wigler, M.; Bethel, K.; Gross, M.E.; et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE 2014, 9, e101777. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Mahalingam, D.; Osmulski, P.; Jadhav, R.R.; Wang, C.M.; Leach, R.J.; Chang, T.C.; Weitman, S.D.; Kumar, A.P.; Sun, L.; et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate 2013, 73, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Van Keymeulen, A.; Lee, M.Y.; Ousset, M.; Brohee, S.; Rorive, S.; Giraddi, R.R.; Wuidart, A.; Bouvencourt, G.; Dubois, C.; Salmon, I.; et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 2015, 525, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Chiu, T.K.; Chou, W.P.; Huang, S.B.; Wang, H.M.; Lin, Y.C.; Hsieh, C.H.; Wu, M.H. Application of optically-induced-dielectrophoresis in microfluidic system for purification of circulating tumour cells for gene expression analysis- Cancer cell line model. Sci. Rep. 2016, 6, 32851. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A.; Gorges, T.M.; Pantel, K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol. Med. 2015, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Haglund, T.A.; Rogers, A.J.; Ghanim, A.F.; Sethu, P. Review: Microfluidics technologies for blood-based cancer liquid biopsies. Anal. Chim. Acta 2018, 1012, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Yousuff, C.; Ho, E.; Hussain, K.I.; Hamid, N. Microfluidic Platform for Cell Isolation and Manipulation Based on Cell Properties. Micromachines 2017, 8, 15. [Google Scholar] [CrossRef]
- Hvichia, G.E.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Muller, V.; Loges, S.; Neves, R.P.; Stoecklein, N.H.; et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer 2016, 138, 2894–2904. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Jia, C.; Yang, J.; Li, G.; Mao, H.; Jin, Q.; Zhao, J. A microfluidic chip integrated with a high-density PDMS-based microfiltration membrane for rapid isolation and detection of circulating tumor cells. Biosens. Bioelectron. 2015, 71, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.; Schutze, K.; Capron, F.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.I.; Morgan, B.; Trautwein, J.; et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra147. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Herrler, M.; Burgess, D.; Manna, E.; Krag, D.; Burke, J.F. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Res. 2008, 10, R69. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Fritsche, H.; Muller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Janicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Mostert, B.; Sleijfer, S.; Foekens, J.A.; Gratama, J.W. Circulating tumor cells (CTCs): Detection methods and their clinical relevance in breast cancer. Cancer Treat. Rev. 2009, 35, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.M.; Siewert, C.; Scharl, A.; Harms, T.; Heinze, R.; Ohl, S.; Radbruch, A.; Miltenyi, S.; Schmitz, J. Immunomagnetic enrichment of disseminated epithelial tumor cells from peripheral blood by MACS. Exp. Hematol. 1998, 26, 252–264. [Google Scholar] [PubMed]
- Zhu, D.M.; Wu, L.; Suo, M.; Gao, S.; Xie, W.; Zan, M.H.; Liu, A.; Chen, B.; Wu, W.T.; Ji, L.W.; et al. Engineered red blood cells for capturing circulating tumor cells with high performance. Nanoscale 2018, 10, 6014–6023. [Google Scholar] [CrossRef] [PubMed]
- Pezzi, H.M.; Niles, D.J.; Schehr, J.L.; Beebe, D.J.; Lang, J.M. Integration of Magnetic Bead-Based Cell Selection into Complex Isolations. ACS Omega 2018, 3, 3908–3917. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial–mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 2003, 15, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Mikolajczyk, S.D.; Millar, L.S.; Tsinberg, P.; Coutts, S.M.; Zomorrodi, M.; Pham, T.; Bischoff, F.Z.; Pircher, T.J. Detection of EpCAM-Negative and Cytokeratin-Negative Circulating Tumor Cells in Peripheral Blood. J. Oncol. 2011, 2011, 252361. [Google Scholar] [CrossRef] [PubMed]
- Danila, D.C.; Heller, G.; Gignac, G.A.; Gonzalez-Espinoza, R.; Anand, A.; Tanaka, E.; Lilja, H.; Schwartz, L.; Larson, S.; Fleisher, M.; et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin. Cancer Res. 2007, 13, 7053–7058. [Google Scholar] [CrossRef] [PubMed]
- Lustberg, M.B.; Balasubramanian, P.; Miller, B.; Garcia-Villa, A.; Deighan, C.; Wu, Y.; Carothers, S.; Berger, M.; Ramaswamy, B.; Macrae, E.R.; et al. Heterogeneous atypical cell populations are present in blood of metastatic breast cancer patients. Breast Cancer Res. 2014, 16, R23. [Google Scholar] [CrossRef] [PubMed]
- Antfolk, M.; Laurell, T. Continuous flow microfluidic separation and processing of rare cells and bioparticles found in blood—A review. Anal. Chim. Acta 2017, 965, 9–35. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.A.; Lee, T.Y.; Lee, S.H.; Jung, H.I. Two-stage microfluidic chip for selective isolation of circulating tumor cells (CTCs). Biosens. Bioelectron. 2015, 67, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Chen, J.F.; Lu, Y.T.; Zhang, Y.; Song, J.; Hou, S.; Ke, Z.; Tseng, H.R. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc. Chem. Res. 2014, 47, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Huang, W.; Jalal, S.I.; Chan, B.D.; Mahmood, A.; Shahda, S.; O’Neil, B.H.; Matei, D.E.; Savran, C.A. Circulating tumor cell detection using a parallel flow micro-aperture chip system. Lab Chip 2015, 15, 1677–1688. [Google Scholar] [CrossRef] [PubMed]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Wu, M.H.; Lin, Y.H.; Hsieh, C.H.; Yang, C.L.; Lin, H.C.; Tseng, C.P.; Lee, G.B. High-purity and label-free isolation of circulating tumor cells (CTCs) in a microfluidic platform by using optically-induced-dielectrophoretic (ODEP) force. Lab Chip 2013, 13, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Chiou, P.Y.; Ohta, A.T.; Wu, M.C. Massively parallel manipulation of single cells and microparticles using optical images. Nature 2005, 436, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.-P.; Wang, H.-M.; Chang, J.-H.; Chiu, T.-K.; Hsieh, C.-H.; Liao, C.-J.; Wu, M.-H. The utilization of optically-induced-dielectrophoresis (ODEP)-based virtual cell filters in a microfluidic system for continuous isolation and purification of circulating tumour cells (CTCs) based on their size characteristics. Sens. Actuators B Chem. 2017, 241, 245–254. [Google Scholar] [CrossRef]
- Svoboda, K.; Block, S.M. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 247–285. [Google Scholar] [CrossRef] [PubMed]
- Ohta, A.T.; Chiou, P.Y.; Phan, H.L.; Sherwood, S.W.; Yang, J.M.; Lau, A.N.K.; Hsu, H.Y.; Jamshidi, A.; Wu, M.C. Optically controlled cell discrimination and trapping using optoelectronic tweezers. IEEE J. Sel. Top. Quant. 2007, 13, 235–243. [Google Scholar] [CrossRef]
- Ohta, A.T.; Chiou, P.Y.; Han, T.H.; Liao, J.C.; Bhardwaj, U.; McCabe, E.R.B.; Yu, F.Q.; Sun, R.; Wu, M.C. Dynamic cell and microparticle control via optoelectronic tweezers. J. Microelectromech. Syst. 2007, 16, 491–499. [Google Scholar] [CrossRef]
- Pethig, R. Review article-dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4. [Google Scholar] [CrossRef] [PubMed]
- Neale, S.L.; Ohta, A.T.; Hsu, H.Y.; Valley, J.K.; Jamshidi, A.; Wu, M.C. Trap profiles of projector based optoelectronic tweezers (OET) with HeLa cells. Opt. Express 2009, 17, 5232–5239. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.C.; Wu, M.H.; Chang, P.H.; Hsu, H.C.; Chang, G.J.; Huang, W.K.; Wu, C.E.; Hsieh, J.C. A Prognostic Model Based on Circulating Tumour Cells is Useful for Identifying the Poorest Survival Outcome in Patients with Metastatic Colorectal Cancer. Int. J. Biol. Sci. 2018, 14, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.C.; Hsu, H.C.; Hsieh, C.H.; Wang, H.M.; Huang, C.Y.; Wu, M.H.; Tseng, C.P. A negative selection system PowerMag for effective leukocyte depletion and enhanced detection of EpCAM positive and negative circulating tumor cells. Clin. Chim. Acta Int. J. Clin. Chem. 2013, 419, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Chiu, T.-K.; Chao, A.C.; Chou, W.-P.; Liao, C.-J.; Wang, H.-M.; Chang, J.-H.; Chen, P.-H.; Wu, M.-H. Optically-induced-dielectrophoresis (ODEP)-based cell manipulation in a microfluidic system for high-purity isolation of integral circulating tumor cell (CTC) clusters based on their size characteristics. Sens. Actuators B Chem. 2018, 258, 1161–1173. [Google Scholar] [CrossRef]
- Hou, S.; Zhao, H.; Zhao, L.; Shen, Q.; Wei, K.S.; Suh, D.Y.; Nakao, A.; Garcia, M.A.; Song, M.; Lee, T.; et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 2013, 25, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.S.; Kwon, K.; Kim, S.I.; Han, H.; Sohn, J.; Lee, S.; Jung, H.I. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 2011, 11, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, X.-L.; Hu, N.; Yang, J.; Luo, H.-Y.; Jiang, F.; Liao, Y.-J. Research Progress on Microfluidic Chip of Cell Separation Based on Dielectrophoresis. Chin. J. Anal. Chem. 2015, 43, 300–309. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Lee, G.-B. An integrated cell counting and continuous cell lysis device using an optically induced electric field. Sens. Actuators B Chem. 2010, 145, 854–860. [Google Scholar] [CrossRef]
- Huang, S.H.; Hung, L.Y.; Lee, G.B. Continuous nucleus extraction by optically-induced cell lysis on a batch-type microfluidic platform. Lab Chip 2016, 16, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Song-Bin, H.; Shing-Lun, L.; Jian-Ting, L.; Min-Hsien, W. Label-free Live and Dead Cell Separation Method Using a High-Efficiency Optically-Induced Dielectrophoretic (ODEP) Force-based Microfluidic Platform. Int. J. Autom. Smart Technol. 2014, 4, 83–91. [Google Scholar] [CrossRef]
- Xie, S.; Wang, X.; Jiao, N.; Tung, S.; Liu, L. Programmable micrometer-sized motor array based on live cells. Lab Chip 2017, 17, 2046–2053. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.-H.; Lin, Y.-H.; Lee, G.-B. A microfluidic platform for manipulation and separation of oil-in-water emulsion droplets using optically induced dielectrophoresis. J. Micromech. Microeng. 2010, 20, 045026. [Google Scholar] [CrossRef]
- Liao, C.-J.; Hsieh, C.-H.; Wang, H.-M.; Chou, W.-P.; Chiu, T.-K.; Chang, J.-H.; Chao, A.C.; Wu, M.-H. Isolation of label-free and viable circulating tumour cells (CTCs) from blood samples of cancer patients through a two-step process: Negative selection-type immunomagnetic beads and spheroid cell culture-based cell isolation. RSC Adv. 2017, 7, 29339–29349. [Google Scholar] [CrossRef]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Vincent, T.; Neve, E.P.A.; Johnson, J.R.; Kukalev, A.; Rojo, F.; Albanell, J.; Pietras, K.; Virtanen, I.; Philipson, L.; Leopold, P.L.; et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat. Cell Biol. 2009, 11, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Li, S.L. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, O.; Deidesheimer, T.; Muehlenberg, M.; Deicke, P.; Dietel, M. Immunohistochemical differentiation of metastatic breast carcinomas from metastatic adenocarcinomas of other common primary sites. Histopathology 1996, 29, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Teknos, T.N.; Pan, Q.T. Epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 2013, 49, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13. [Google Scholar] [CrossRef] [PubMed]
- Poruk, K.E.; Valero, V.; Saunders, T.; Blackford, A.L.; Griffin, J.F.; Poling, J.; Hruban, R.H.; Anders, R.A.; Herman, J.; Zheng, L.; et al. Circulating Tumor Cell Phenotype Predicts Recurrence and Survival in Pancreatic Adenocarcinoma. Ann. Surg. 2016, 264, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Mitra, A.; Brownlee, Z.; Xia, X.; Bellister, S.; Overman, M.J.; Kopetz, S.; Ellis, L.M.; Meng, Q.H.; Li, S. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res. 2015, 21, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, C.R.; Le Moulec, S.; Billiot, F.; Loriot, Y.; Ngo-Camus, M.; Vielh, P.; Fizazi, K.; Massard, C.; Farace, F. Vimentin and Ki67 expression in circulating tumour cells derived from castrate-resistant prostate cancer. BMC Cancer 2016, 16, 168. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Chen, Y.W.; Hsu, H.S.; Tseng, L.M.; Huang, P.I.; Lu, K.H.; Chen, D.T.; Tai, L.K.; Yung, M.C.; Chang, S.C.; et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem. Biophys. Res. Commun. 2009, 385, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Snyder, V.; Reed-Newman, T.C.; Arnold, L.; Thomas, S.M.; Anant, S. Cancer Stem Cell Metabolism and Potential Therapeutic Targets. Front. Oncol. 2018, 8, 203. [Google Scholar] [CrossRef] [PubMed]
| Biological Function | Gene Name | Healthy Donor | Head-and-Neck Cancer Patient |
|---|---|---|---|
| Positive/Total | |||
| EMT-related | EpCAM | 0/5 | 0/8 |
| CK19 | 0/5 | 1/8 | |
| Vimentin | 1/5 | 6/8 | |
| SNAIL1 | 0/5 | 1/8 | |
| MDR-related | MRP1 | 0/5 | 3/8 |
| MRP2 | 0/5 | 0/8 | |
| MRP4 | 0/5 | 0/8 | |
| MRP5 | 0/5 | 2/8 | |
| MRP7 | 0/5 | 0/8 | |
| CSC-related | NANOG | 1/5 | 2/8 |
| OCT4 | 1/5 | 4/8 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, C.-J.; Hsieh, C.-H.; Chiu, T.-K.; Zhu, Y.-X.; Wang, H.-M.; Hung, F.-C.; Chou, W.-P.; Wu, M.-H. An Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic System for the Isolation of High-Purity CD45neg/EpCAMneg Cells from the Blood Samples of Cancer Patients—Demonstration and Initial Exploration of the Clinical Significance of These Cells. Micromachines 2018, 9, 563. https://doi.org/10.3390/mi9110563
Liao C-J, Hsieh C-H, Chiu T-K, Zhu Y-X, Wang H-M, Hung F-C, Chou W-P, Wu M-H. An Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic System for the Isolation of High-Purity CD45neg/EpCAMneg Cells from the Blood Samples of Cancer Patients—Demonstration and Initial Exploration of the Clinical Significance of These Cells. Micromachines. 2018; 9(11):563. https://doi.org/10.3390/mi9110563
Chicago/Turabian StyleLiao, Chia-Jung, Chia-Hsun Hsieh, Tzu-Keng Chiu, Yu-Xian Zhu, Hung-Ming Wang, Feng-Chun Hung, Wen-Pin Chou, and Min-Hsien Wu. 2018. "An Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic System for the Isolation of High-Purity CD45neg/EpCAMneg Cells from the Blood Samples of Cancer Patients—Demonstration and Initial Exploration of the Clinical Significance of These Cells" Micromachines 9, no. 11: 563. https://doi.org/10.3390/mi9110563
APA StyleLiao, C.-J., Hsieh, C.-H., Chiu, T.-K., Zhu, Y.-X., Wang, H.-M., Hung, F.-C., Chou, W.-P., & Wu, M.-H. (2018). An Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic System for the Isolation of High-Purity CD45neg/EpCAMneg Cells from the Blood Samples of Cancer Patients—Demonstration and Initial Exploration of the Clinical Significance of These Cells. Micromachines, 9(11), 563. https://doi.org/10.3390/mi9110563





