Operation of Droplet-Microfluidic Devices with a Lab Centrifuge
Abstract
:1. Introduction
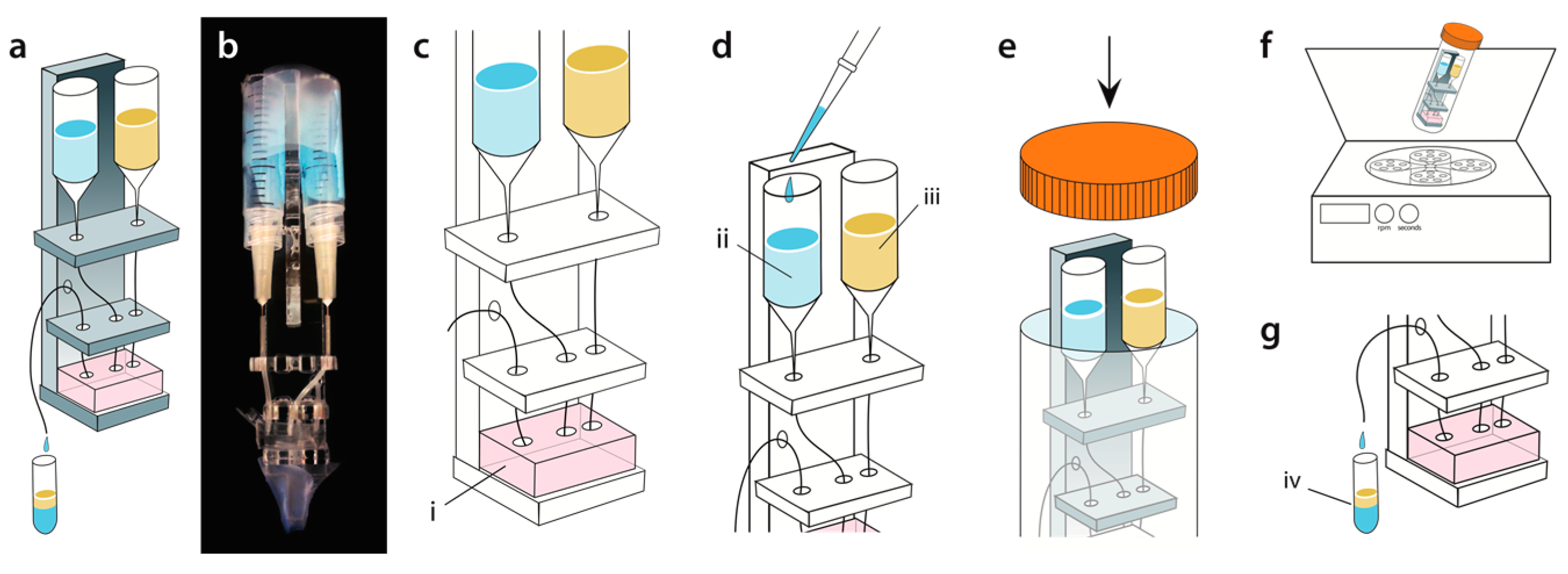
2. Materials and Methods
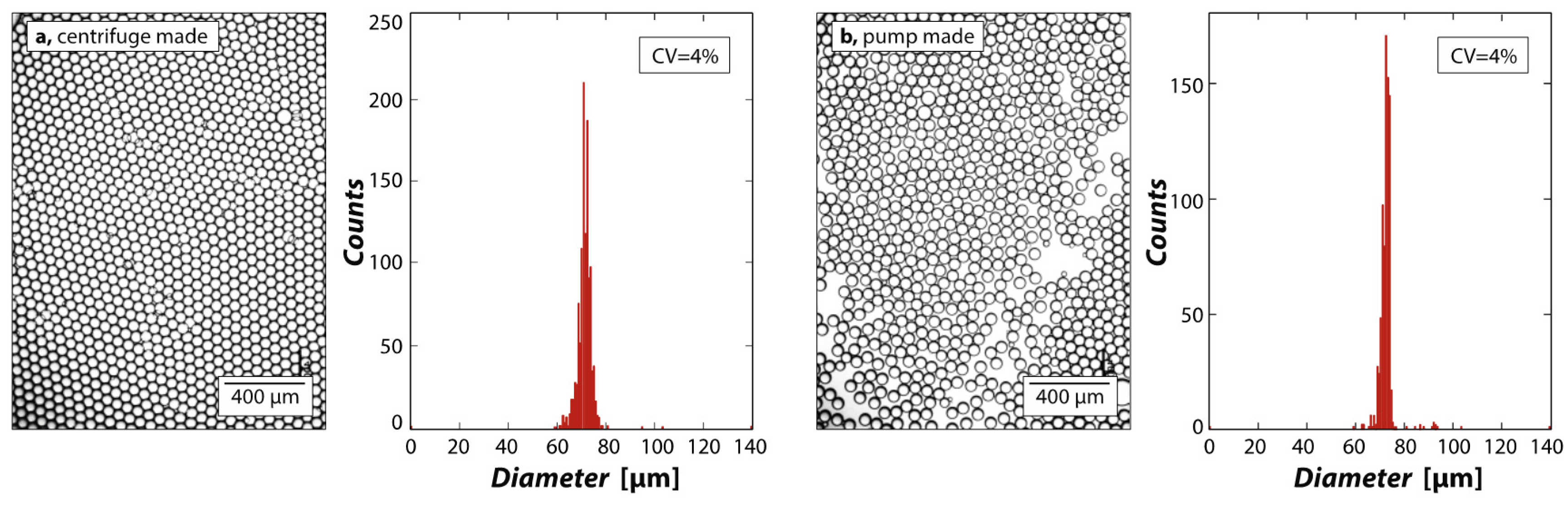
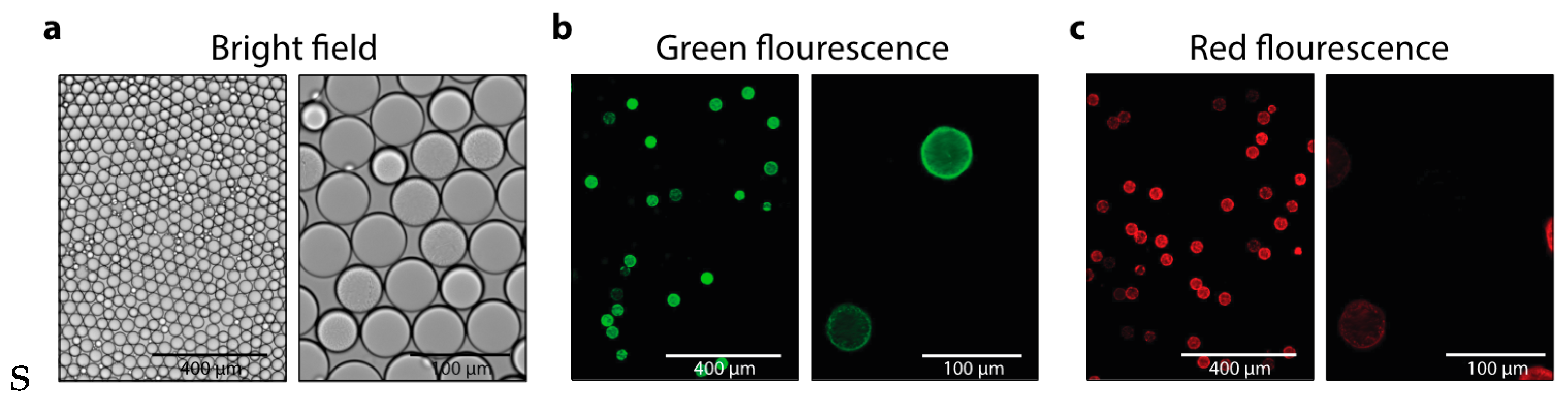
3. Results
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baroud, C.; Gallaire, F.; Dangla, R. Dynamics of microfluidic droplets. Lab Chip 2010, 16, 2032–2045. [Google Scholar] [CrossRef] [PubMed]
- Seemann, R.; Brinkmann, M.; Pfohl, T.; Herminghaus, S. Droplet based microfluidics. Rep. Prog. Phys. 2011, 75, 016601. [Google Scholar] [CrossRef] [PubMed]
- Theberge, A.B.; Courtois, F.; Schaerli, Y.; Fischlenechner, M.; Abell, C.; Hollfelder, F.; Huck, W.T. Microdroplets in microfluidics: An evolving platform for discoveries in chemistry and biology. Angew. Chem. 2010, 49, 5846–5868. [Google Scholar] [CrossRef] [PubMed]
- Kintses, B.; Hein, C.; Mohamed, M.F.; Fischlechner, M.; Courtois, F.; Lainé, C.; Hollfelder, F. Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem. Biol. 2012, 19, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Agresti, J.J.; Antipov, E.; Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, J.-C.; Marquez, M.; Klibanov, A.M.; Griffiths, A.D.; Weitz, D.A. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 4004–4009. [Google Scholar] [CrossRef] [PubMed]
- Huebner, A.; Srisa-Art, M.; Holt, D.; Abell, C.; Hollfelder, F.; deMello, A.J.; Edel, J. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem. Commun. 2007, 1218–1220. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.T.; Rotem, A.; Heyman, J.A.; Weitz, D.A. Droplet microfluidics for high-throughput biological assays. Lab Chip 2012, 12, 2146–2155. [Google Scholar] [CrossRef] [PubMed]
- Brouzes, E.; Medkova, M.; Savenelli, N.; Marran, D.; Twardowski, M.; Hutchison, J.B.; Rothberg, J.M.; Link, D.R.; Perrimon, N.; Samuels, M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in Poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.; Whitesides, G.M. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar]
- Wu, Z.; Jensen, H.; Gamby, J.; Bai, X.; Girault, H.H. A flexible sample introduction method for polymer microfluidic chips using a push/pull pressure pump. Lab Chip 2004, 4, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Stiles, T.; Fallon, R.; Vestad, T.; Oakey, J.; Marr, D.; Squier, J.; Jimenez, R. Hydrodynamic focusing for vacuum-pumped microfluidics. Microfluid. Nanofluid. 2005, 1, 280–283. [Google Scholar] [CrossRef]
- Abate, A.R.; Weitz, D.A. Syringe-vacuum microfluidics: A portable technique to create monodisperse emulsions. Biomicrofluidics 2011, 5, 014107. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lin, J.-M.; Knopp, D. Using a circular groove surrounded inlet to generate monodisperse droplets inside a microfluidic chip in a gravity-driven manner. J. Micromech. Microeng. 2008, 18, 095014. [Google Scholar] [CrossRef]
- Duffy, D.C.; Gillis, H.L.; Lin, J.; Sheppard, N.F.; Kellogg, G.J. Microfabricated Centrifugal Microfluidic Systems: Characterization and Multiple Enzymatic Assays. Anal. Chem. 1999, 71, 4669–4678. [Google Scholar] [CrossRef]
- Ducrée, J.; Haeberle, S.; Lutz, S.; Pausch, S.; von Stetten, F.; Zengerle, R. The centrifugal microfluidic Bio-Disk platform. J. Micromech. Microeng. 2007, 17, S103. [Google Scholar] [CrossRef]
- Hoffmann, J.; Mark, D.; Zengerle, R.; von Stetten, F. Liquid reagent storage and release for centrifuge operated LAB-on-a-chip systems based on a burstable seal. In Proceedings of the 15th International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers’09), Denver, CO, USA, 21–25 June 2009.
- Schwemmer, F.; Blanchet, C.E.; Spilotros, A.; Kosse, D.; Zehnle, S.; Mertens, H.D.; Graewert, M.A.; Rössle, M.; Paust, N.; Svergun, D.I. LabDisk for SAXS: A centrifugal microfluidic sample preparation platform for small-angle X-ray scattering. Lab Chip 2016, 16, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Chen, Y.F.; Lee, G.B. CE chips fabricated by injection molding and polyethylene/thermoplastic elastomer film packaging methods. Electrophoresis 2007, 28, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.; Heim, U. Hot embossing as a method for the fabrication of polymer high aspect ratio structures. Sens. Actuators A Phys. 2000, 83, 130–135. [Google Scholar] [CrossRef]
- Fiorini, G.S.; Chiu, D.T. Disposable microfluidic devices: fabrication, function, and application. BioTechniques 2005, 38, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Tice, J.D.; Ismagilov, R.F. A Microfluidic System for Controlling Reaction Networks in Time. Angew. Chem. 2003, 115, 792–796. [Google Scholar] [CrossRef]
- Zhao, C.-X. Multiphase flow microfluidics for the production of single or multiple emulsions for drug delivery. Adv. Drug Deliv. Rev. 2013, 65, 1420–1446. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Wang, F. Flow rate effect on droplet control in a co-flowing microfluidic device. Microfluid. Nanofluid. 2007, 3, 341–346. [Google Scholar] [CrossRef]
- Song, H.; Chen, D.L.; Ismagilov, R.F. Reactions in droplets in microfluidic channels. Angew. Chem. Int. Ed. 2006, 45, 7336–7356. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.K.; Shum, H.C.; Rowat, A.C.; Lee, D.; Agresti, J.J.; Utada, A.S.; Chu, L.-Y.; Kim, J.-W.; Fernandez-Nieves, A.; Martinez, C.J.; et al. Designer emulsions using microfluidics. Mater. Today 2008, 11, 18–27. [Google Scholar] [CrossRef]
- Chu, L.Y.; Utada, A.S.; Shah, R.K.; Kim, J.W.; Weitz, D.A. Controllable monodisperse multiple emulsions. Angew. Chem. Int. Ed. 2007, 46, 8970–8974. [Google Scholar] [CrossRef] [PubMed]
- Duncanson, W.J.; Lin, T.; Abate, A.R.; Seiffert, S.; Shah, R.K.; Weitz, D.A. Microfluidic synthesis of advanced microparticles for encapsulation and controlled release. Lab Chip 2012, 12, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.; Kwon, K.W.; Park, M.C.; Lee, S.H.; Kim, S.M.; Suh, K.Y. Soft Lithography for Microfluidics: A Review. Biochip J. 2008, 2, 1–11. [Google Scholar]
- Abate, A.R.; Lee, D.; Do, T.; Holtze, C.; Weitz, D.A. Glass coating for PDMS microfluidic channels by sol-gel methods. Lab Chip 2008, 8, 516–518. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Mak, S.Y.; Sauret, A.; Shum, H.C. Syringe-pump-induced fluctuation in all-aqueous microfluidic system implications for flow rate accuracy. Lab Chip 2014, 14, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Garstecki, P.; Fuerstman, M.J.; Stone, H.A.; Whitesides, G.M. Formation of droplets and bubbles in a microfluidic T-junction—scaling and mechanism of break-up. Lab Chip 2006, 6, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.A.; Abate, A.R. Flow focusing geometry generates droplets through a plug and squeeze mechanism. Lab Chip 2012, 12, 5130–5132. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Matharoo, H.S.; Makkar, D.; Kumar, R. Droplet formation via squeezing mechanism in a microfluidic flow-focusing device. Comput. Fluids 2014, 100, 218–226. [Google Scholar] [CrossRef]
- Piccin, E.; Ferraro, D.; Sartori, P.; Chiarello, E.; Pierno, M.; Mistura, G. Generation of water-in-oil and oil-in-water microdroplets in polyester-toner microfluidic devices. Sens. Actuators B Chem. 2014, 196, 525–531. [Google Scholar] [CrossRef]
- Edd, J.F.; Carlo, D.D.; Humphry, K.J.; Köster, S.; Irimia, D.; Weitz, D.; Toner, M. Controlled encapsulation of single cells into monodisperse picoliter drops. Lab Chip 2008, 8, 1262–1264. [Google Scholar] [CrossRef] [PubMed]
- Köster, S.; Angilè, F.E.; Duan, H.; Agresti, J.J.; Wintner, A.; Shmitz, C.; Rowat, A.C.; Merten, C.A.; Pisignano, D.; Griffiths, A.D.; et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip 2008, 8, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.J.; Neild, A.; deMello, A.; Liu, A.; Ai, Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip 2015, 15, 3439–3459. [Google Scholar] [CrossRef] [PubMed]
- Clausell-Tormos, J.; Lieber, D.; Baret, J.; El-Harrak, A.; Miller, O.J.; Frenz, L.; Blouwolff, J.; Humphry, K.J.; Köster, S.; Duan, H.; et al. Droplet-Based Microfluidic Platforms for the Encapsulation and Screening of Mammalian Cells and Multicellular Organisms. Chem. Biol. 2008, 15, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Eun, Y.; Utada, A.S.; Copeland, M.F.; Takeuchi, S.; Weibel, D. Encapsulating Bacteria in Agarose Microparticles Using Microfluidics for High-Throughput Cell Analysis Isolation. ACS Chem. Biol. 2011, 6, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Joensson, H.N.; Svahn, H.A. Droplet microfluidics—A tool for single-cell analysis. Angew. Chem. 2012, 51, 12176–12192. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, N.; Sukovich, D.; Abate, A.R. Operation of Droplet-Microfluidic Devices with a Lab Centrifuge. Micromachines 2016, 7, 161. https://doi.org/10.3390/mi7090161
Ahmed N, Sukovich D, Abate AR. Operation of Droplet-Microfluidic Devices with a Lab Centrifuge. Micromachines. 2016; 7(9):161. https://doi.org/10.3390/mi7090161
Chicago/Turabian StyleAhmed, Noorsher, David Sukovich, and Adam R. Abate. 2016. "Operation of Droplet-Microfluidic Devices with a Lab Centrifuge" Micromachines 7, no. 9: 161. https://doi.org/10.3390/mi7090161
APA StyleAhmed, N., Sukovich, D., & Abate, A. R. (2016). Operation of Droplet-Microfluidic Devices with a Lab Centrifuge. Micromachines, 7(9), 161. https://doi.org/10.3390/mi7090161





