A Simple Method for Fabrication of Microstructures Using a PDMS Stamp
Abstract
:1. Introduction
2. Methods/Experiment
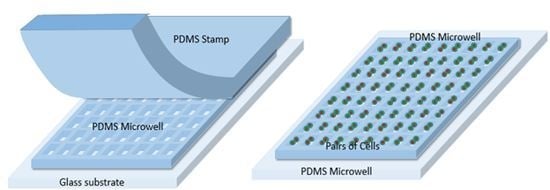
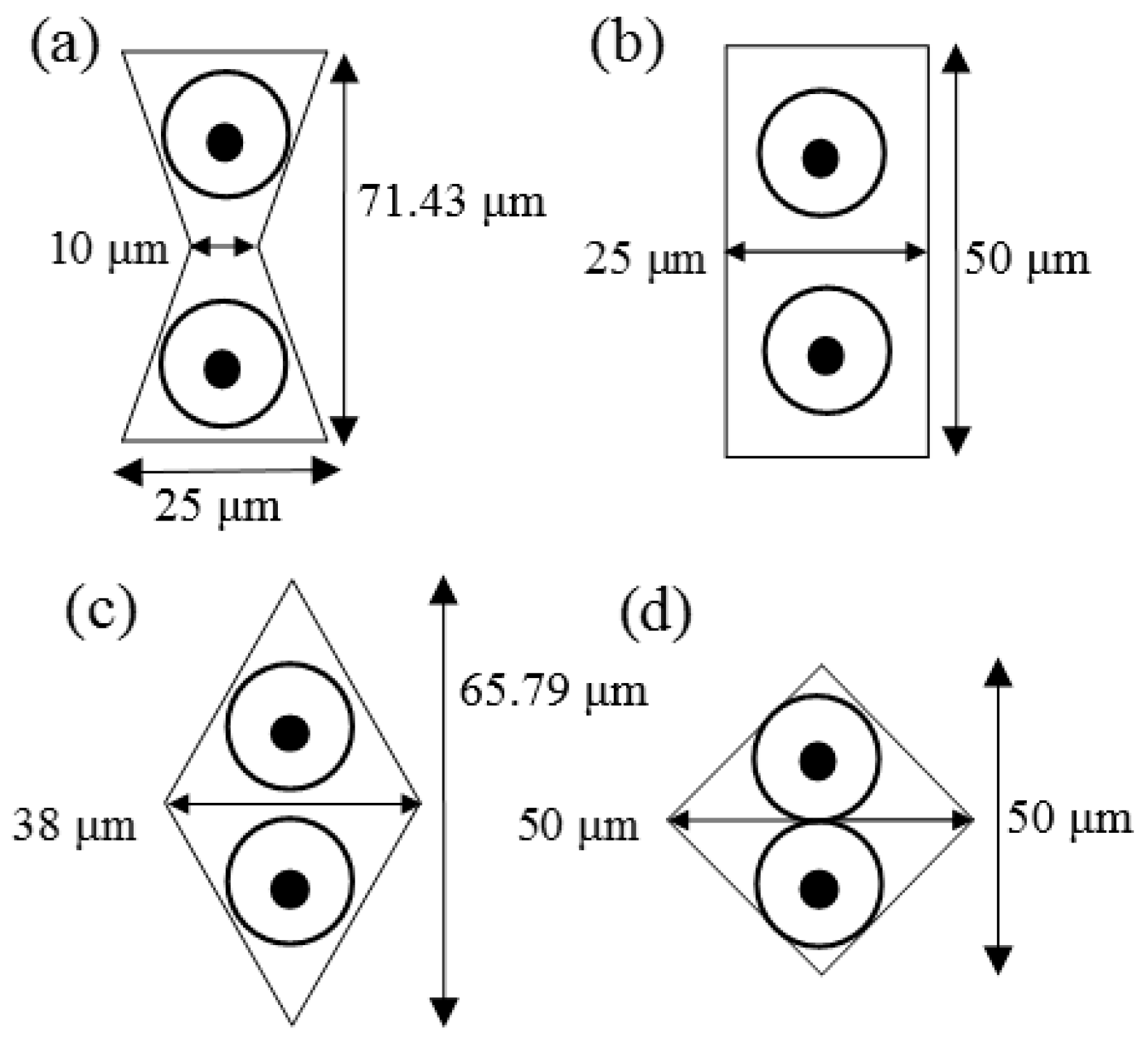
2.1. Microarrays and Microfluidic Channel Design
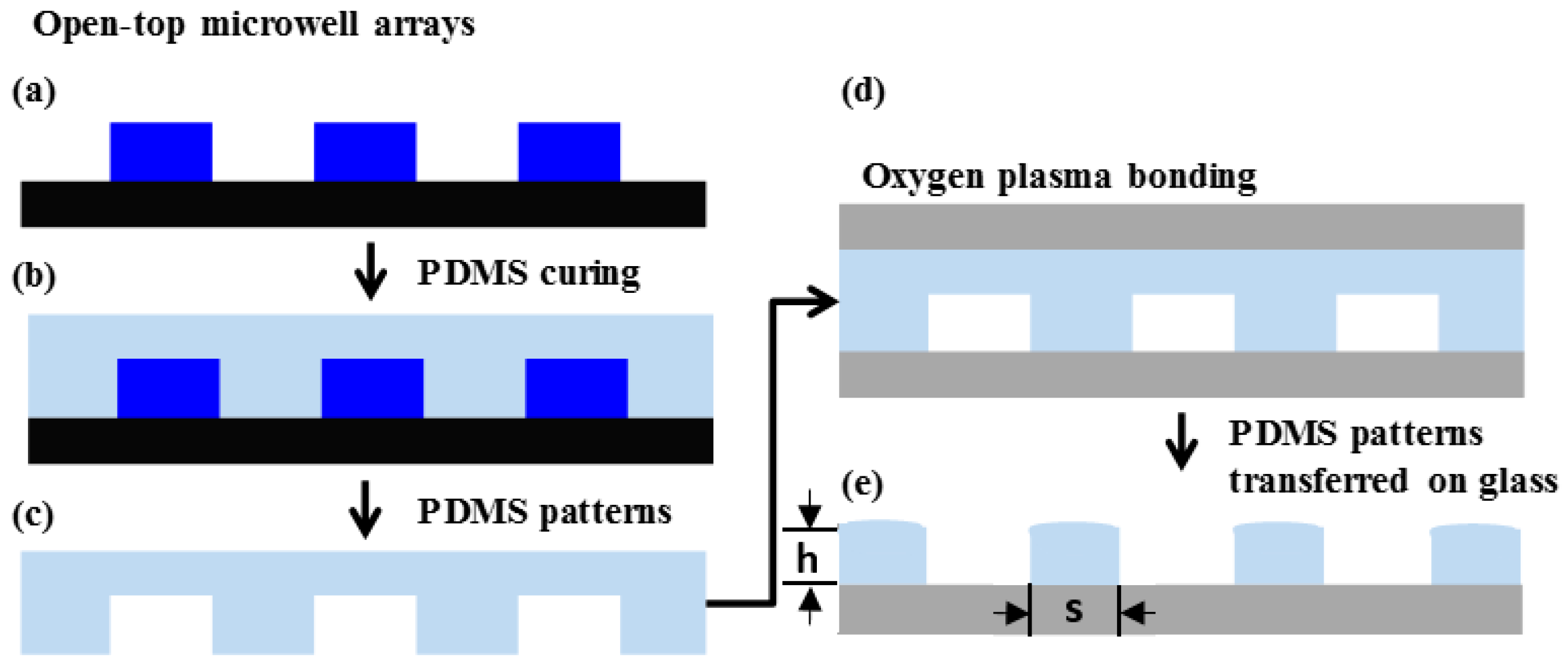
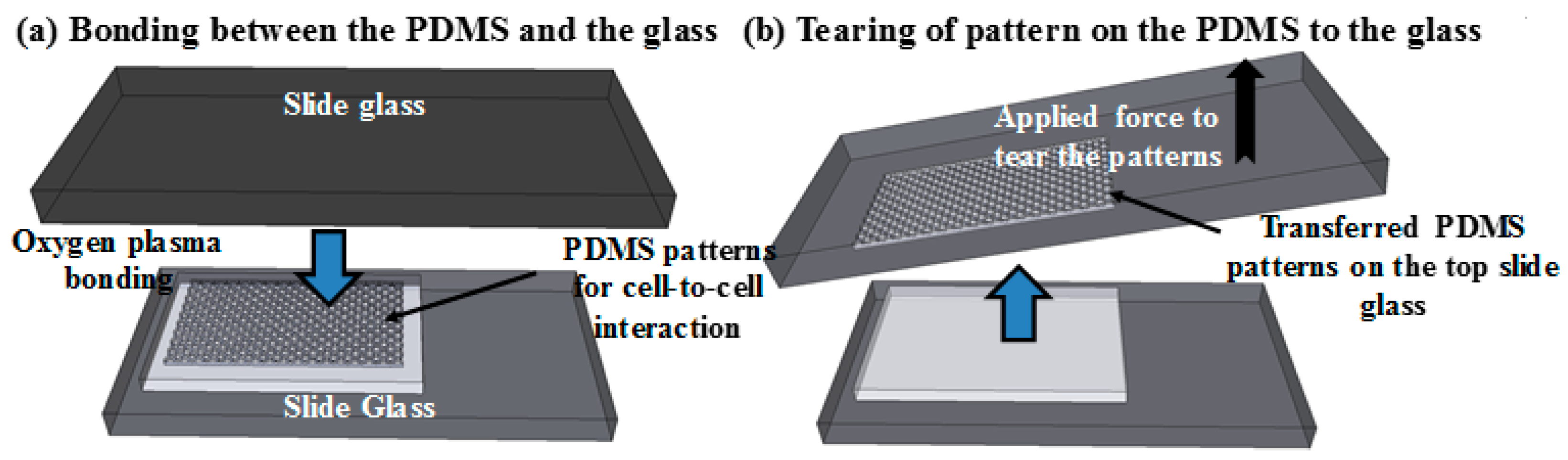
2.2. Fabrication
2.3. Cell-to-Cell Study
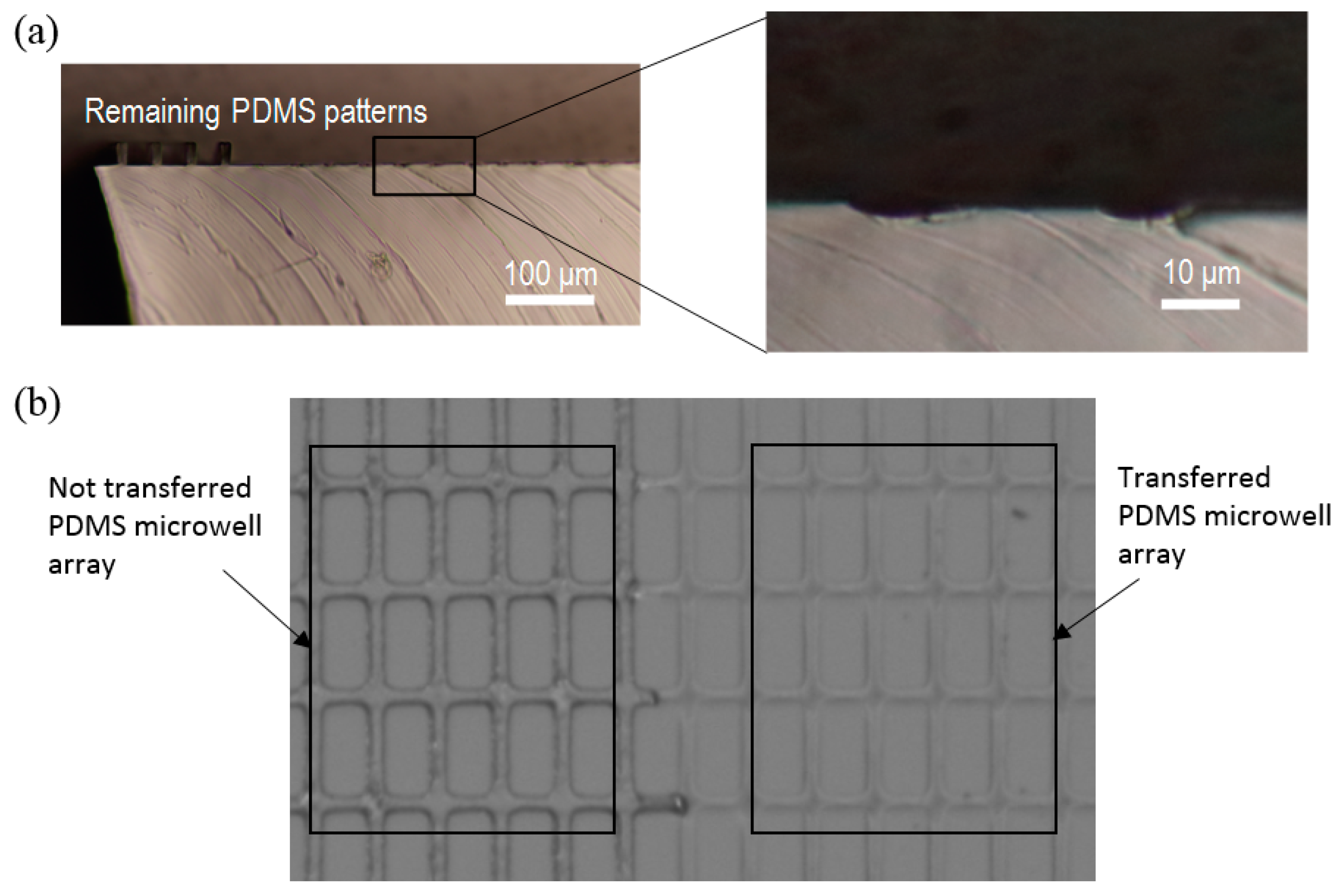
3. Result/Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goral, V.N.; Zhou, C.F.; Lai, F.; Yuen, P.K. A continuous perfusion microplate for cell culture dagger. Lab Chip 2013, 13, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Charnley, M.; Textor, M.; Khademhosseini, A.; Lutolf, M.P. Integration column: Microwell arrays for mammalian cell culture. Integr. Biol. 2009, 1, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Jain, T.; McBride, R.; Head, S.; Saez, E. Highly parallel introduction of nucleic acids into mammalian cells grown in microwell arrays. Lab Chip 2009, 9, 3557–3566. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Lee, H.; Jin, H.K.; Bae, J.S.; Kim, G.M. Micropatterning of neural stem cells and Purkinje neurons using a polydimethylsiloxane (PDMS) stencil. Lab Chip 2012, 12, 5045–5050. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Liu, W.F.; Chen, C.S. Manipulation of cell-cell adhesion using bowtie-shaped microwells. Methods Mol. Biol. 2007, 370, 1–9. [Google Scholar] [PubMed]
- Johnson, L.A.; Rodansky, E.S.; Haak, A.J.; Larsen, S.D.; Neubig, R.R.; Higgins, P.D. Novel Rho/MRTF/SRF inhibitors block matrix-stiffness and TGF-β-induced fibrogenesis in human colonic myofibroblasts. Inflamm. Bowel Dis. 2014, 20, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Charest, J.L.; Jennings, J.M.; King, W.P.; Kowalczyk, A.P.; Garcia, A.J. Cadherin-mediated cell-cell contact regulates keratinocyte differentiation. J. Investig. Dermatol. 2009, 129, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Sakai, Y.; Nakazawa, K. Two-dimensional microarray of HepG2 spheroids using collagen/polyethylene glycol micropatterned chip. J. Mater. Sci. Mater. Med. 2008, 19, 2071–2077. [Google Scholar] [CrossRef] [PubMed]
- Schmid, H.; Wolf, H.; Allenspach, R.; Riel, H.; Karg, S.; Michel, B.; Delamarche, E. Preparation of metallic films on elastomeric stamps and their application for contact processing and contact printing. Adv. Funct. Mater. 2003, 13, 145–153. [Google Scholar] [CrossRef]
- Gray, D.S.; Liu, W.F.; Shen, C.J.; Bhadriraju, K.; Nelson, C.M.; Chen, C.S. Engineering amount of cell-cell contact demonstrates biphasic proliferative regulation through RhoA and the actin cytoskeleton. Exp. Cell Res. 2008, 314, 2846–2854. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Chen, C.S. VE-cadherin simultaneously stimulates and inhibits cell proliferation by altering cytoskeletal structure and tension. J. Cell Sci. 2003, 116, 3571–3581. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, K.; Ahn, B.; Oh, K.W. A new fabrication process for SU-8 thick photoresist structures by simultaneously removing edge-bead and air bubbles. J. Micromech. Microeng. 2011, 21, 125006. [Google Scholar] [CrossRef]
- Kim, M.; Huang, Y.; Choi, K.; Hidrovo, C.H. The improved resistance of PDMS to pressure-induced deformation and chemical solvent swelling for microfluidic devices. Microelectron. Eng. 2014, 124, 66–75. [Google Scholar] [CrossRef]
- Johnston, I.D.; McCluskey, D.K.; Tan, C.K.L.; Tracey, M.C. Mechanical characterization of bulk Sylgard 184 for microfluidics and microengineering. J. Micromech. Microeng. 2014, 24, 035017. [Google Scholar] [CrossRef]
- Mojica, W.D.; Oh, K.W.; Lee, H.; Furlani, E.P.; Sykes, D.; Sands, A.M. Microfluidics enables multiplex evaluation of the same cells for further studies. Cytopathology 2015, 27, 277–283. [Google Scholar] [CrossRef] [PubMed]
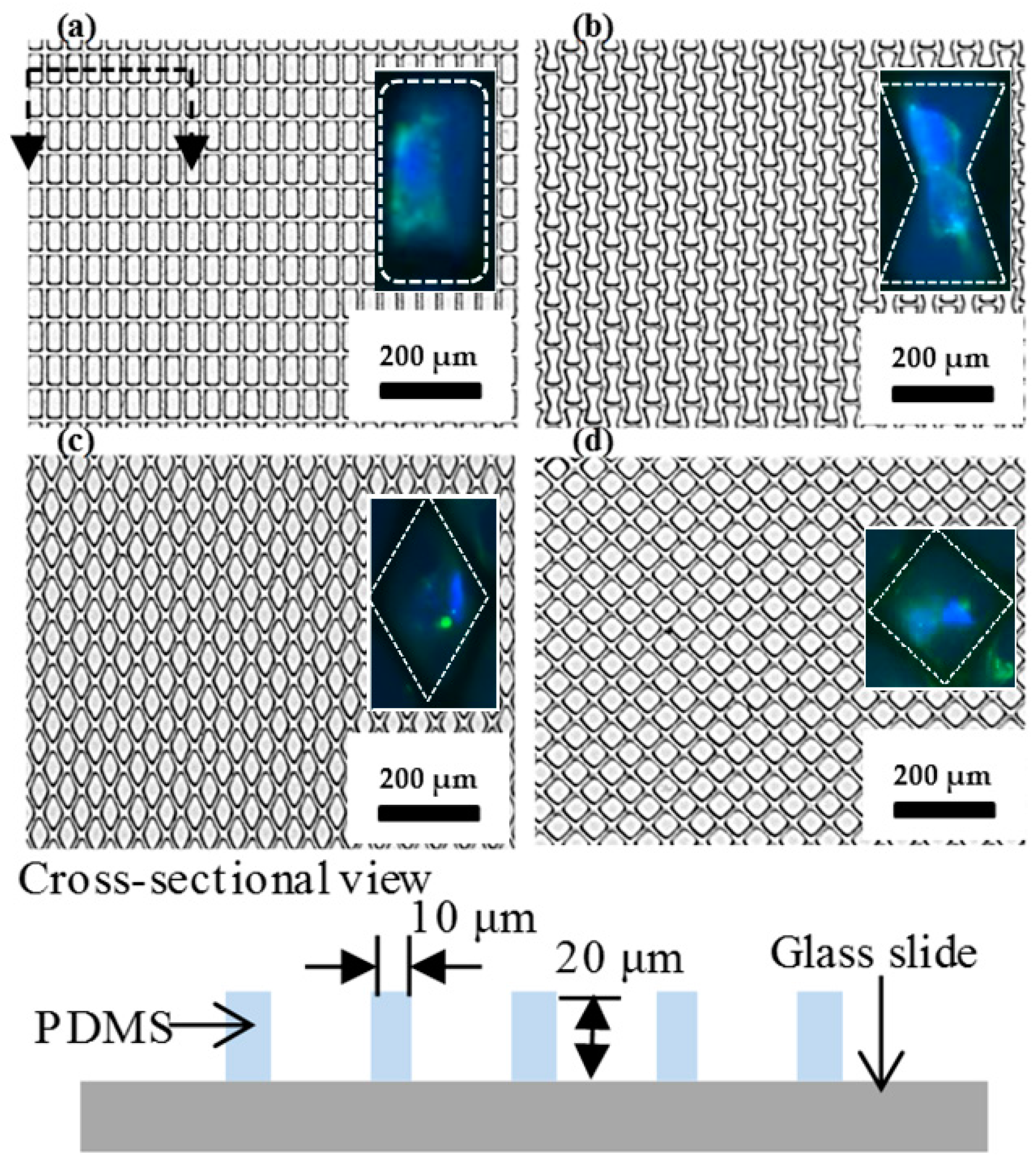



| Shape | Rectangle | Bowtie | Wide Rhombus | Rhombus |
|---|---|---|---|---|
| Density of pattern (#/mm2) | 937.11 | 797.20 | 641.46 | 946.84 |
| Contact area of PDMS (polydimethylsiloxane) (%) | 41.43 | 50.18 | 59.91 | 40.82 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Koh, D.; Xu, L.; Row, S.; Andreadis, S.T.; Oh, K.W. A Simple Method for Fabrication of Microstructures Using a PDMS Stamp. Micromachines 2016, 7, 173. https://doi.org/10.3390/mi7100173
Lee H, Koh D, Xu L, Row S, Andreadis ST, Oh KW. A Simple Method for Fabrication of Microstructures Using a PDMS Stamp. Micromachines. 2016; 7(10):173. https://doi.org/10.3390/mi7100173
Chicago/Turabian StyleLee, Hun, Domin Koh, Linfeng Xu, Sindhu Row, Stelios T. Andreadis, and Kwang W. Oh. 2016. "A Simple Method for Fabrication of Microstructures Using a PDMS Stamp" Micromachines 7, no. 10: 173. https://doi.org/10.3390/mi7100173
APA StyleLee, H., Koh, D., Xu, L., Row, S., Andreadis, S. T., & Oh, K. W. (2016). A Simple Method for Fabrication of Microstructures Using a PDMS Stamp. Micromachines, 7(10), 173. https://doi.org/10.3390/mi7100173