Highly Sensitive and Specific Lateral Flow Detection for DNA Methylation Based on GIaI-Mediated Specific-Terminal-Mediated Polymerase Chain Reaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. GlaI Digestion
2.3. STEM-PCR Amplification
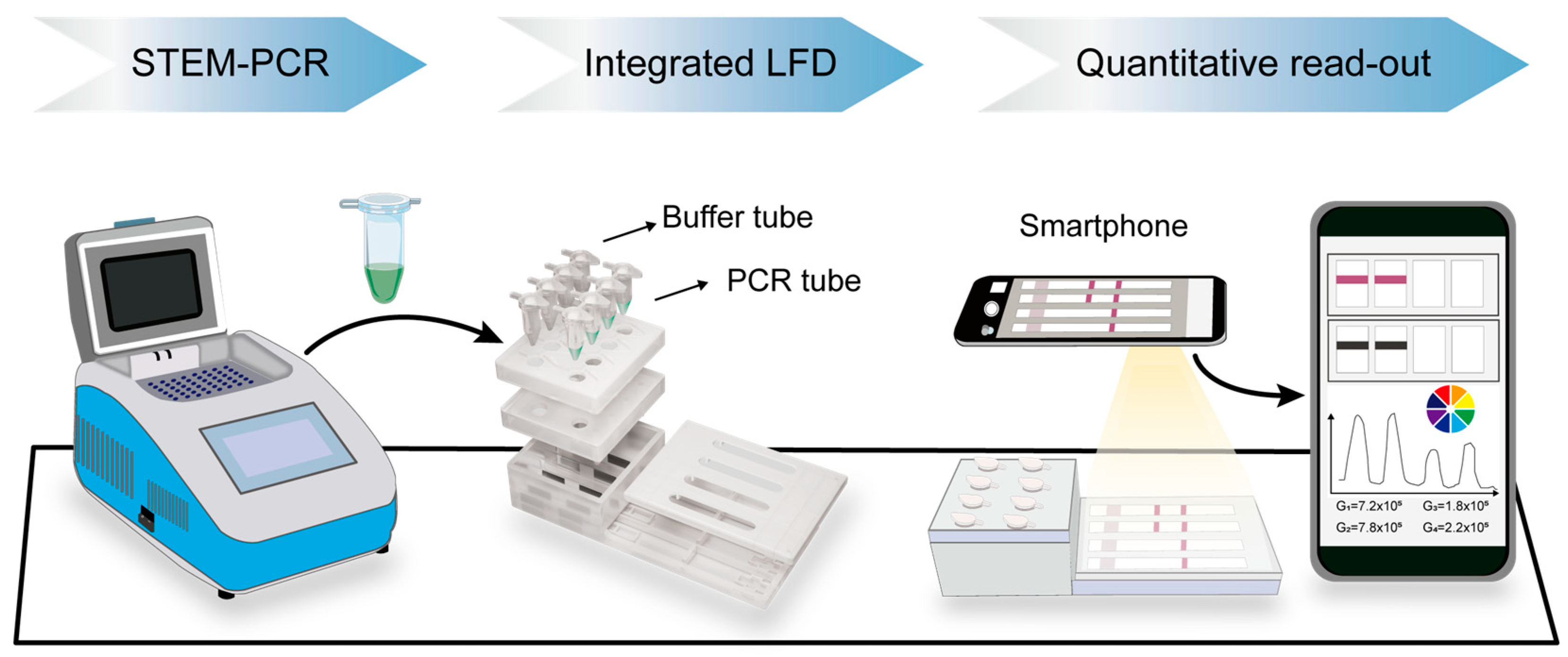
2.4. Lateral Flow Detection (LFD)
3. Results
3.1. Principle of GIaI-Mediated STEM-PCR and Illustration of the Detection Process
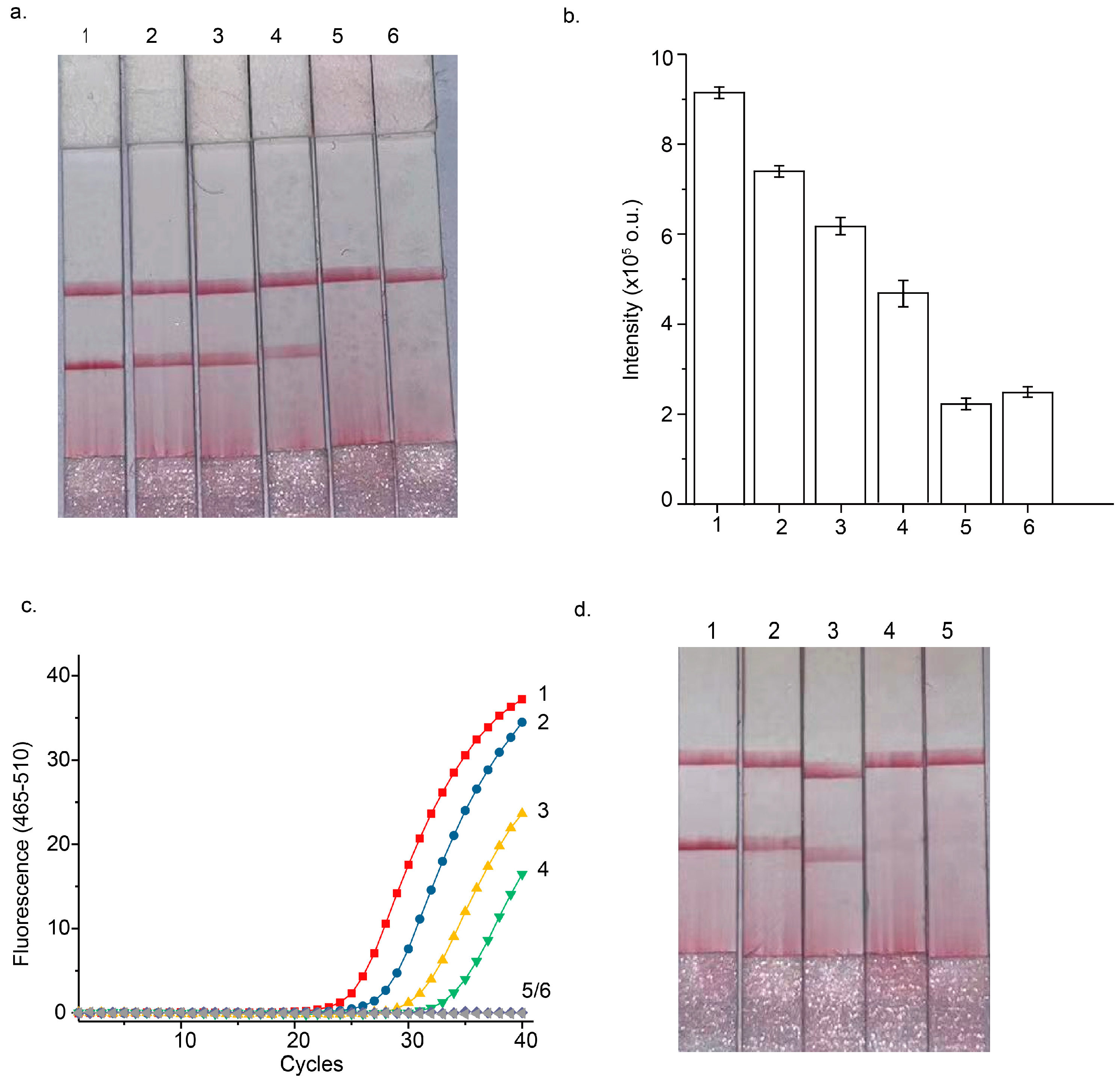
3.2. Optimization of STEM-PCR Assay
3.3. Performance of Septin 9 Methylation Analysis Using the STEM-PCR-LFD Assay
3.4. STEM-PCR-LFD Assay of the Hypermethylated Septin 9 Gene Using CRC Samples
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegfried, Z.; Simon, I. DNA methylation and gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 362–371. [Google Scholar] [PubMed]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA methylation: A historical perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [PubMed]
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA methylation: In the right place at the right time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef]
- Gadwal, A.; Purohit, P.; Khokhar, M.; Vishnoi, J.R.; Pareek, P.; Choudhary, R.; Elhence, P.; Banerjee, M.; Sharma, P. In silico analysis of differentially expressed-aberrantly methylated genes in breast cancer for prognostic and therapeutic targets. Clin. Exp. Med. 2023, 23, 3847–3866. [Google Scholar]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M.; et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [PubMed]
- Koch, A.; Joosten, S.C.; Feng, Z.; de Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar]
- Widschwendter, M.; Jones, A.; Evans, I.; Reisel, D.; Dillner, J.; Sundström, K.; Steyerberg, E.W.; Vergouwe, Y.; Wegwarth, O.; Rebitschek, F.G.; et al. Epigenome-based cancer risk prediction: Rationale, opportunities and challenges. Nat. Rev. Clin. Oncol. 2018, 15, 292–309. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, W.; Mu, Y.; Hu, H.; Dong, K.; Wen, X.; Ye, Z.; Sun, Q.; Yan, B.; Mao, Z.; et al. Ultrasensitive and Quantitative DNA Methylation Detection Method Based on the MutS Protein. Anal. Chem. 2023, 95, 18828–18835. [Google Scholar]
- Zhang, Q.; Wu, Y.; Xu, Q.; Ma, F.; Zhang, C.-Y. Recent advances in biosensors for in vitro detection and in vivo imaging of DNA methylation. Biosens. Bioelectron. 2021, 171, 112712. [Google Scholar]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fang, L.; Yu, G.; Wang, D.; Xiao, C.-L.; Wang, K. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019, 10, 2449. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010, 7, 461–465. [Google Scholar] [CrossRef]
- Cottrell, S.E.; Distler, J.; Goodman, N.S.; Mooney, S.H.; Kluth, A.; Olek, A.; Schwope, I.; Tetzner, R.; Ziebarth, H.; Berlin, K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004, 32, e10. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Cai, Y.; Zhang, S.; Ma, K.; Hua, K.; Cui, Y. A novel DNA methylation biosensor by combination of isothermal amplification and lateral flow device. Sens. Actuators B Chem. 2021, 333, 129624. [Google Scholar] [CrossRef]
- Jamshidi, A.; Liu, M.C.; Klein, E.A.; Venn, O.; Hubbell, E.; Beausang, J.F.; Gross, S.; Melton, C.; Fields, A.P.; Liu, Q.; et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 2022, 40, 1537–1549.e12. [Google Scholar] [CrossRef]
- Booth, M.J.; Ost, T.W.; Beraldi, D.; Bell, N.M.; Branco, M.R.; Reik, W.; Balasubramanian, S. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 2013, 8, 1841–1851. [Google Scholar] [CrossRef]
- Felix, K.; Benjamin, K.; Andre, F.; Simon, R.A. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods 2012, 9, 145–151. [Google Scholar]
- Genereux, D.P.; Johnson, W.C.; Burden, A.F.; Stöger, R.; Laird, C.D. Errors in the bisulfite conversion of DNA: Modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res. 2008, 36, e150. [Google Scholar] [CrossRef]
- Liu, C.; Cui, X.; Zhao, B.S.; Narkhede, P.; Gao, Y.; Liu, J.; Dou, X.; Dai, Q.; Zhang, L.-S.; He, C. DNA 5-Methylcytosine-Specific Amplification and Sequencing. J. Am. Chem. Soc. 2020, 142, 4539–4543. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genetics 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Ye, C.; Irkliyenko, I.; Wang, Y.; Sun, H.-L.; Gao, Y.; Liu, Y.; Beadell, A.; Perea, J.; Goel, A.; et al. Ultrafast bisulfite sequencing detection of 5-methylcytosine in DNA and RNA. Nat. Biotechnol. 2024, 42, 1559–1570. [Google Scholar] [CrossRef]
- Ball, M.P.; Li, J.B.; Gao, Y.; Lee, J.H.; LeProust, E.M.; Park, I.H.; Xie, B.; Daley, G.Q.; Church, G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009, 27, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef]
- Yang, H.; Qiu, J.; Zhen, L.; Huang, Y.; Ren, W.; Gu, H.; Xu, H.; Xu, G. Sensitive GlaI digestion and terminal transferase PCR for DNA methylation detection. Talanta 2022, 247, 123616. [Google Scholar] [CrossRef]
- Zhou, S.; Sun, H.; Huo, D.; Wang, X.; Qi, N.; Peng, L.; Yang, M.; Lu, P.; Hou, C. A novel methyl-dependent DNA endonuclease GlaI coupling with double cascaded strand displacement amplification and CRISPR/Cas12a for ultra-sensitive detection of DNA methylation. Anal. Chim. Acta 2022, 1212, 339914. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, Y.; Tian, W.; Liu, C.; Gao, K.; Li, Z. A novel restriction endonuclease GlaI for rapid and highly sensitive detection of DNA methylation coupled with isothermal exponential amplification reaction. Chem. Sci. 2018, 9, 1344–1351. [Google Scholar] [CrossRef]
- Rand, K.N.; Young, G.P.; Ho, T.; Molloy, P.L. Sensitive and selective amplification of methylated DNA sequences using helper-dependent chain reaction in combination with a methylation-dependent restriction enzymes. Nucleic Acids Res. 2013, 41, e15. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Yue, S.; Bi, S.; Ding, C.; Song, W. Isothermal exponential amplification techniques: From basic principles to applications in electrochemical biosensors. Biosens. Bioelectron. 2018, 110, 207–217. [Google Scholar] [CrossRef]
- Reid, M.S.; Le, X.C.; Zhang, H. Exponential Isothermal Amplification of Nucleic Acids and Assays for Proteins, Cells, Small Molecules, and Enzyme Activities: An EXPAR Example. Angew. Chem. Int. Ed. Engl. 2018, 57, 11856–11866. [Google Scholar] [CrossRef]
- Xu, G.; Yang, H.; Qiu, J.; Reboud, J.; Zhen, L.; Ren, W.; Xu, H.; Cooper, J.M.; Gu, H. Sequence terminus dependent PCR for site-specific mutation and modification detection. Nat. Commun. 2023, 14, 1169. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, B.; Park, J.H.; Lee, K.J.; Kwak, T.J.; Son, T.; Shin, Y.-B.; Im, H.; Kim, M.-G. Rapid PCR kit: Lateral flow paper strip with Joule heater for SARS-CoV-2 detection. Mater. Horiz. 2023, 10, 1697–1704. [Google Scholar] [PubMed]
- Xu, H.; Lan, H.; Pan, D.; Xu, J.; Wang, X. Visual Detection of Chicken Adulteration Based on a Lateral Flow Strip-PCR Strategy. Foods 2022, 11, 2351. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.W.; Le, T.N.; Pham, D.M.; Ko, H.-H.; Chang, H.-C.; Lee, C.-C.; Sharma, N.; Lee, C.-K.; Chiang, W.-H. Recent Advances in Novel Lateral Flow Technologies for Detection of COVID-19. Biosensors 2021, 11, 295. [Google Scholar] [CrossRef]
- Gong, T.; Borgard, H.; Zhang, Z.; Chen, S.; Gao, Z.; Deng, Y. Analysis and Performance Assessment of the Whole Genome Bisulfite Sequencing Data Workflow: Currently Available Tools and a Practical Guide to Advance DNA Methylation Studies. Small Methods 2022, 6, e2101251. [Google Scholar] [CrossRef]



| Name | Sequence (5′-3′) |
|---|---|
| Septin 9-TFP | TGTCAGCCAACGGTATTCATCTTTGCGCAGCTGGATGGG/iSp18/GTCCGCGGCCGCAGCA |
| Septin 9-TSP | Biotin-TGCCAGCCCAGCACCCA |
| Septin 9-probe | CCTTCGAAGTCCGAAATGA-FITC |
| Septin 9-UP | GCCTGTCAGCCAACGGTATTCATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ke, L.; Zhao, H.; Shan, H.; Chen, Y.; Cai, Y.; Wang, Y.; Wei, B.; Du, M. Highly Sensitive and Specific Lateral Flow Detection for DNA Methylation Based on GIaI-Mediated Specific-Terminal-Mediated Polymerase Chain Reaction. Micromachines 2025, 16, 387. https://doi.org/10.3390/mi16040387
Ke L, Zhao H, Shan H, Chen Y, Cai Y, Wang Y, Wei B, Du M. Highly Sensitive and Specific Lateral Flow Detection for DNA Methylation Based on GIaI-Mediated Specific-Terminal-Mediated Polymerase Chain Reaction. Micromachines. 2025; 16(4):387. https://doi.org/10.3390/mi16040387
Chicago/Turabian StyleKe, Lihui, Hang Zhao, Hongbo Shan, Yicheng Chen, Yongsheng Cai, Yang Wang, Bo Wei, and Minghua Du. 2025. "Highly Sensitive and Specific Lateral Flow Detection for DNA Methylation Based on GIaI-Mediated Specific-Terminal-Mediated Polymerase Chain Reaction" Micromachines 16, no. 4: 387. https://doi.org/10.3390/mi16040387
APA StyleKe, L., Zhao, H., Shan, H., Chen, Y., Cai, Y., Wang, Y., Wei, B., & Du, M. (2025). Highly Sensitive and Specific Lateral Flow Detection for DNA Methylation Based on GIaI-Mediated Specific-Terminal-Mediated Polymerase Chain Reaction. Micromachines, 16(4), 387. https://doi.org/10.3390/mi16040387







