An Integrated Microfluidic System for One-Stop Multiplexed Exosomal PD-L1 and MMP9 Automated Analysis with Deep Learning Model YOLO
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Microfluidic System Design and Fabrication
2.3. Preparation of Anti-CD63 Immuno-Magnetic Beads
2.4. Cell Culture and Supernatant Preparation
2.5. Isolation of Model Exosome Samples
2.6. On-Chip Immunoassay Procedure
2.7. Image Acquisition and Automated Data Analysis
3. Results and Discussion
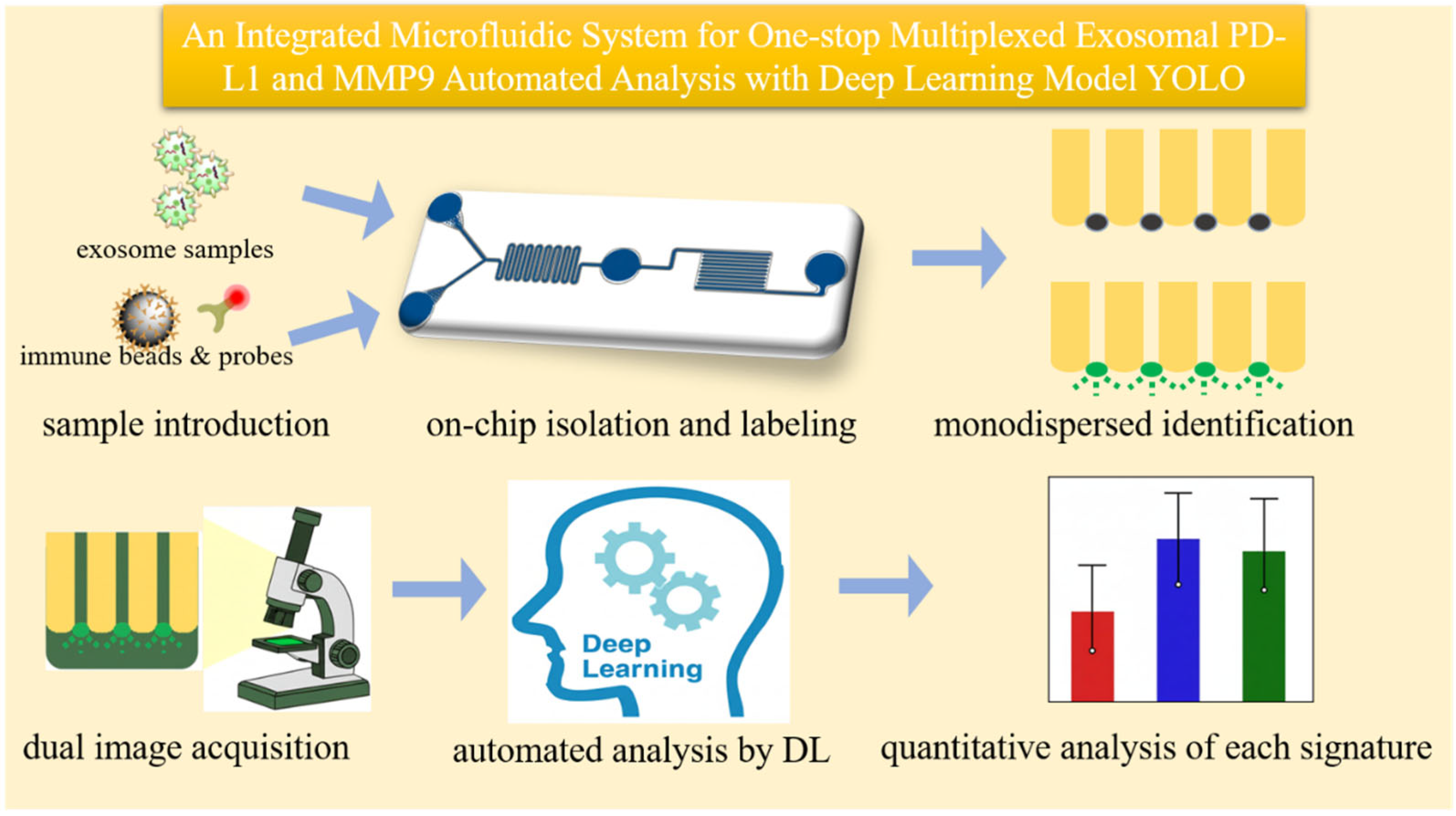
3.1. Principle of the Integrated On-Chip Analysis
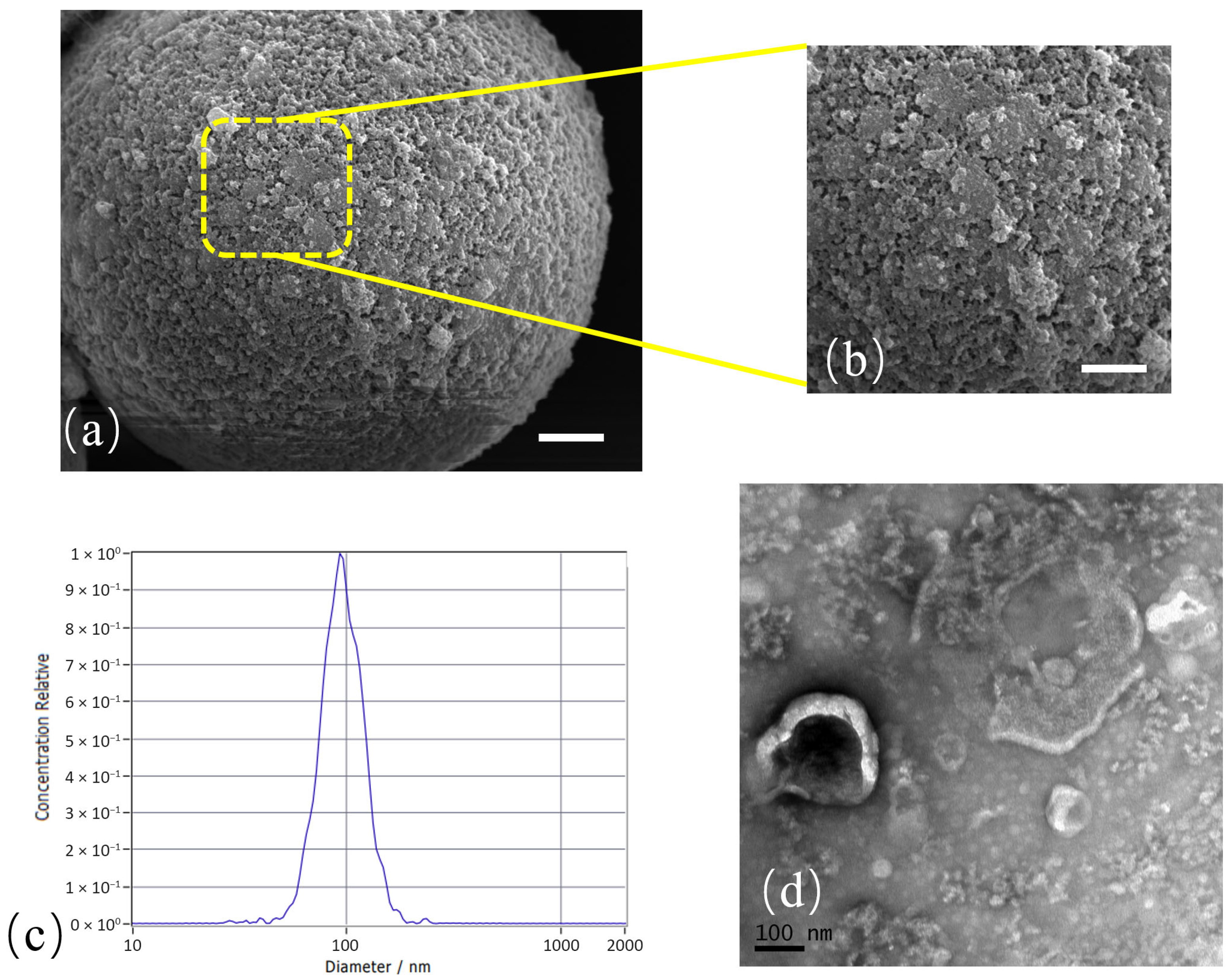
3.2. Characterization of Isolated Exosomes
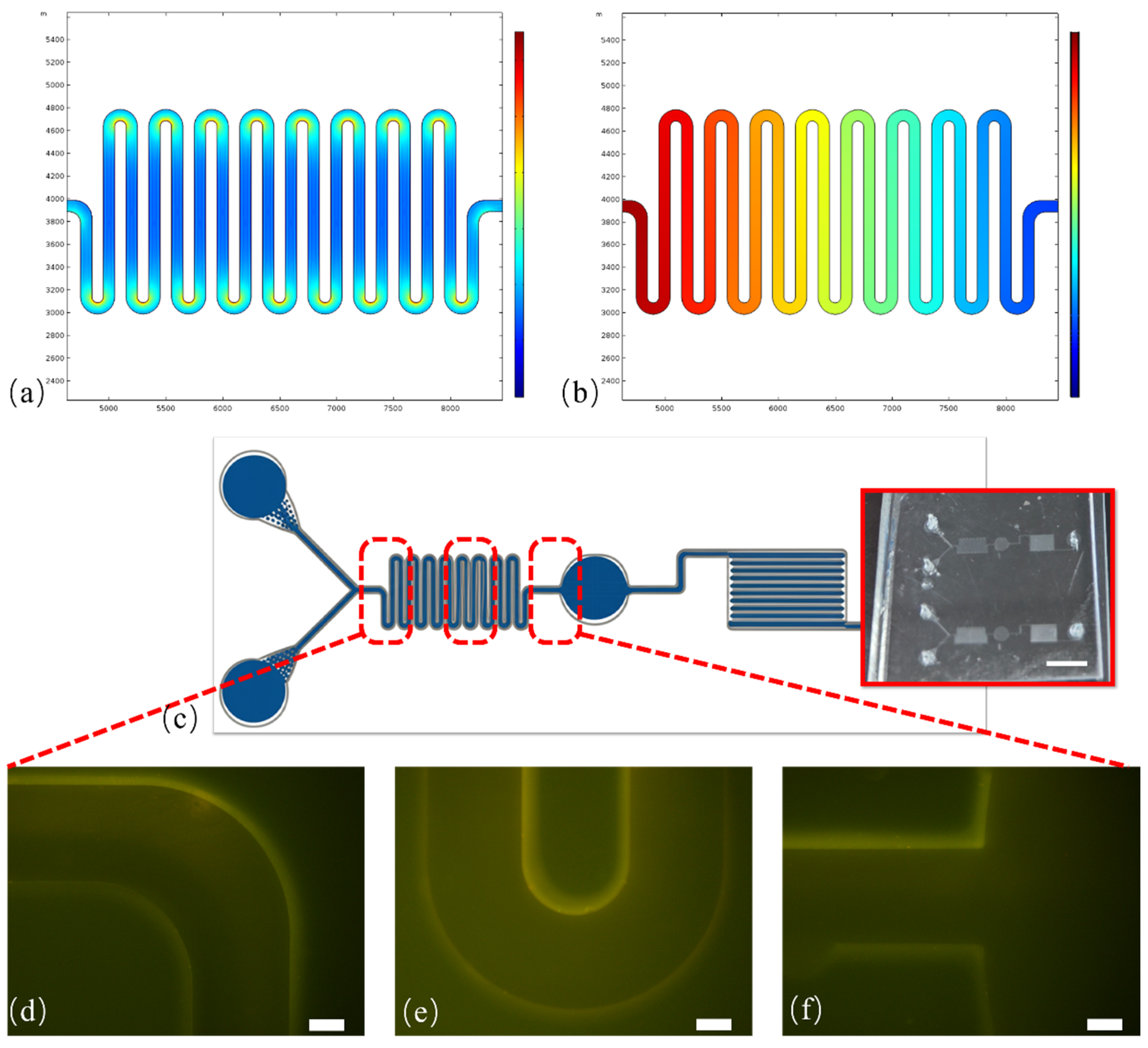
3.3. System Optimization and Analytical Performance
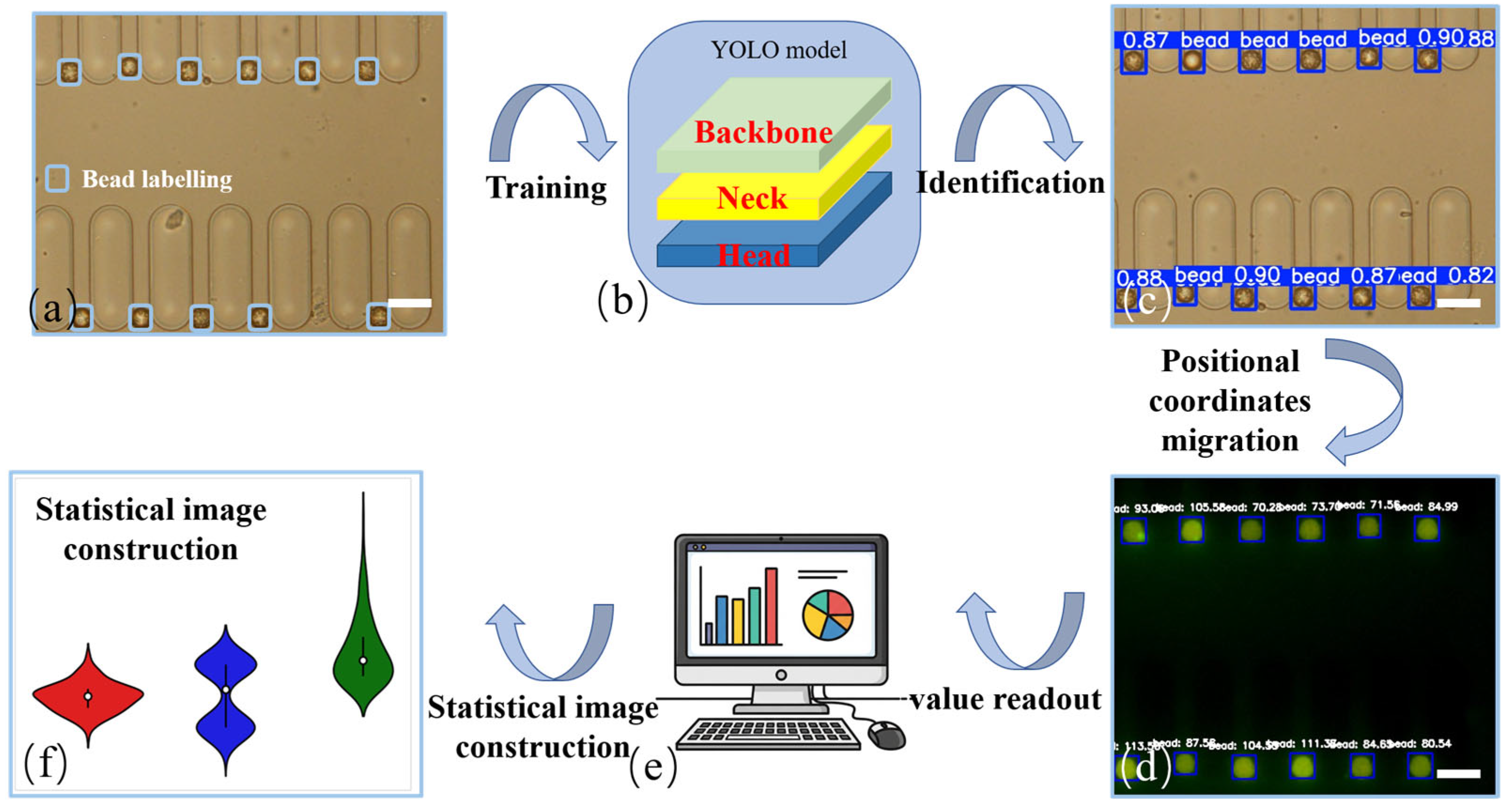
3.4. Automated Data Quantification Workflow
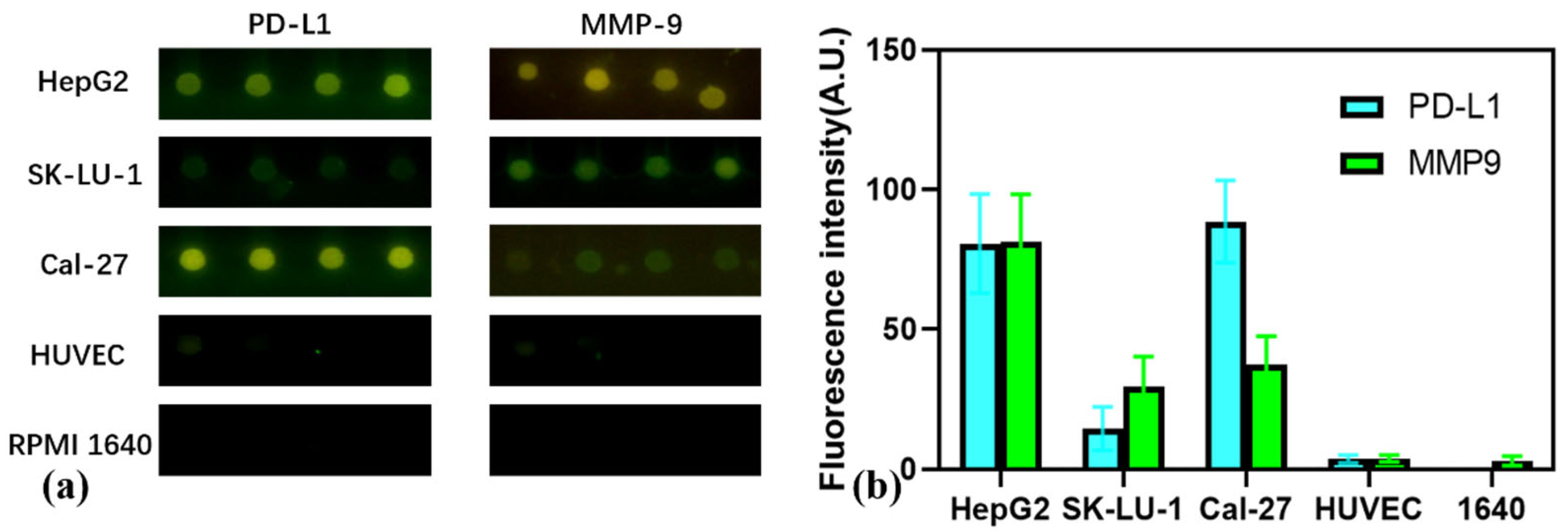
3.5. Multiplexed Profiling of Exosomal Markers Across Cancer Cell Lines
3.6. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.; Huang, J.; Sun, Y.; Xu, W.; Qian, H. Emerging role of extracellular vesicles in diabetic retinopathy. Theranostics 2024, 14, 1631–1646. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2019, 16, e1903916. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- Lu, Y.; Ye, L.; Jian, X.; Yang, D.; Zhang, H.; Tong, Z.; Wu, Z.; Shi, N.; Han, Y.; Mao, H. Integrated microfluidic system for isolating exosome and analyzing protein marker PD-L1. Biosens. Bioelectron. 2022, 204, 113879. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Lu, Y.; Wang, K.; Cheng, Z.; Qu, Y.; Qiu, S.; Zhou, L.; Wu, Z.; Liu, H.; Zhao, J.; et al. Rapid Isolation and Multiplexed Detection of Exosome Tumor Markers Via Queued Beads Combined with Quantum Dots in a Microarray. Nano-Micro Lett. 2019, 11, 59. [Google Scholar] [CrossRef]
- Qiu, S.; Shen, C.; Jian, X.; Lu, Y.; Tong, Z.; Wu, Z.; Mao, H.; Zhao, J. Single-cell level point mutation analysis of circulating tumor cells through droplet microfluidics. Chin. Chem. Lett. 2022, 33, 2701–2704. [Google Scholar] [CrossRef]
- Zhou, T.; Fang, G.; Wang, Z.; Qiao, Z.; Nie, N.; Fu, B.; Tseng, P.-H.; Sun, X.; Chen, Y.-C. Digital Lasing Biochip for Tumor-Derived Exosome Analysis. Anal. Chem. 2025, 97, 5605–5611. [Google Scholar] [CrossRef]
- Yu, Q.; Stamenkovic, I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Wang, X.; Ren, T.; Li, Z.; Wu, B.; Li, M. MMP9 in pan-cancer and computational study to screen for MMP9 inhibitors. Am. J. Transl. Res. 2024, 16, 7071–7086. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Pang, Y.; Shi, J.; Yang, X.; Wang, C.; Sun, Z.; Xiao, R. Personalized detection of circling exosomal PD-L1 based on Fe3O4@TiO2 isolation and SERS immunoassay. Biosens. Bioelectron. 2020, 148, 111800. [Google Scholar] [CrossRef]
- Borchers, A.; Pieler, T. Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs. Genes 2010, 1, 413–426. [Google Scholar] [CrossRef]
- Guo, P.; Li, H.; Zhang, X.; Liu, Y.; Xue, S.; Yong, V.W.; Xue, M. Matrix metalloproteinase-9 in hemorrhagic transformation after acute ischemic stroke (Review). Mol. Med. Rep. 2025, 32, 225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shu, X.; Zhang, Y.; Song, C.; Wu, Y.; Cui, K.; Zhang, X.; Sun, Y.; Shen, H.; Wei, Q.; et al. Astrocyte-derived MMP-9 is a key mediator of pseudorabies virus penetration of the blood-brain barrier and tight junction disruption. Vet. Res. 2025, 56, 72. [Google Scholar] [CrossRef]
- Chen, C.; Skog, J.; Hsu, C.-H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef]
- Dilsiz, N. A comprehensive review on recent advances in exosome isolation and characterization: Toward clinical applications. Transl. Oncol. 2024, 50, 102121. [Google Scholar] [CrossRef] [PubMed]
- Havers, M.; Broman, A.; Lenshof, A.; Laurell, T. Advancement and obstacles in microfluidics-based isolation of extracellular vesicles. Anal. Bioanal. Chem. 2022, 415, 1265–1285. [Google Scholar] [CrossRef]
- Lang, J.B.; Buck, M.C.; Rivière, J.; Stambouli, O.; Sachenbacher, K.; Choudhary, P.; Dietz, H.; Giebel, B.; Bassermann, F.; Oostendorp, R.A.J.; et al. Comparative analysis of extracellular vesicle isolation methods from human AML bone marrow cells and AML cell lines. Front. Oncol. 2022, 12, 949261. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ma, Z.; Xu, J.; Xue, T.; Lv, X.; Zhu, G.; Huang, B. Exosome Isolation and Detection: From Microfluidic Chips to Nanoplasmonic Biosensor. ACS Appl. Mater. Interfaces 2024, 16, 22776–22793. [Google Scholar] [CrossRef]
- Shetty, K.H.; Desai, D.T.; Patel, H.P.; Shah, D.O.; Willcox, M.D.P.; Maulvi, F.A. Contact lens as an emerging platform for non-invasive bio-sensing: A review. Sens. Actuators A Phys. 2024, 376, 115617. [Google Scholar] [CrossRef]
- Wang, C.; Qiu, J.; Liu, M.; Wang, Y.; Yu, Y.; Liu, H.; Zhang, Y.; Han, L. Microfluidic Biochips for Single-Cell Isolation and Single-Cell Analysis of Multiomics and Exosomes. Adv. Sci. 2024, 11, e2401263. [Google Scholar] [CrossRef] [PubMed]
- Zhand, S.; Goss, D.M.; Cheng, Y.Y.; Warkiani, M.E. Recent Advances in Microfluidics for Nucleic Acid Analysis of Small Extracellular Vesicles in Cancer. Adv. Healthc. Mater. 2024, 14, e2401295. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Amevor, F.K.; Zhao, X.; Mou, C.; Pang, J.; Peng, X.; Liu, A.; Lan, X.; Liu, L. Potential therapeutic effects of milk-derived exosomes on intestinal diseases. J. Nanobiotechnol. 2023, 21, 496. [Google Scholar] [CrossRef]
- Xiao, F.-J.; Wang, L.-S.; Lu, Y.; Du, L.; Wen, L.; Cheng, X.; Zhang, W.-Y. Large-scale separation and purification of exosomes using ionexchange chromatography. Biomed. Res. Ther. 2024, 11, 6348–6356. [Google Scholar] [CrossRef]
- Yin, W.; Ma, H.; Qu, Y.; Wang, S.; Zhao, R.; Yang, Y.; Guo, Z.-N. Targeted exosome-based nanoplatform for new-generation therapeutic strategies. Biomed. Mater. 2024, 19, 032002. [Google Scholar] [CrossRef] [PubMed]
- Hettiarachchi, S.; Ouyang, L.; Cha, H.; Hansen, H.H.W.B.; An, H.; Nguyen, N.-T.; Zhang, J. Viscoelastic microfluidics for enhanced separation resolution of submicron particles and extracellular vesicles. Nanoscale 2024, 16, 3560–3570. [Google Scholar] [CrossRef]
- Chen, H.; Liu, H.; Xing, L.; Fan, D.; Chen, N.; Ma, P.; Zhang, X. Deep Learning-driven Microfluidic-SERS to Characterize the Heterogeneity in Exosomes for Classifying Non-Small Cell Lung Cancer Subtypes. ACS Sens. 2025, 10, 2872–2882. [Google Scholar] [CrossRef]
- Kim, J.; Son, H.Y.; Lee, S.; Rho, H.W.; Kim, R.; Jeong, H.; Park, C.; Mun, B.; Moon, Y.; Jeong, E.; et al. Deep learning-assisted monitoring of trastuzumab efficacy in HER2-Overexpressing breast cancer via SERS immunoassays of tumor-derived urinary exosomal biomarkers. Biosens. Bioelectron. 2024, 258, 116347. [Google Scholar] [CrossRef]
- Quan, H.; Wang, S.; Xi, X.; Zhang, Y.; Ding, Y.; Li, Y.; Lin, J.; Liu, Y. Deep learning enhanced multiplex detection of viable foodborne pathogens in digital microfluidic chip. Biosens. Bioelectron. 2024, 245, 115837. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xie, W.; Huang, Y.; Mo, J.; Dong, H.; Chen, X.; Zhang, Z.; Shang, J. Paper microfluidics with deep learning for portable intelligent nucleic acid amplification tests. Talanta 2023, 258, 124470. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Abbasi, S.M.T.; Mehmood, N.; Li, L.; Qu, F.; Cheng, G.; Hu, D.; Ho, Y.P.; Yuan, W.; Ho, H.P. Deep-qGFP: A Generalist Deep Learning Assisted Pipeline for Accurate Quantification of Green Fluorescent Protein Labeled Biological Samples in Microreactors. Small Methods 2023, 8, e2301293. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, H.; Zeng, Z.; Hui, J.; Ji, J.; Mao, H.; Shi, Q.; Yang, X. A deep learning-based integrated analytical system for tumor exosome on-chip isolation and automated image identification. Talanta Open 2025, 11, 100398. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, J.; Wu, X.; Qu, C.; Fang, X.; Yang, Y.; Yuan, Y.; Liu, H.; Han, Z. Microfluidics-based label-free SERS profiling of exosomes with machine learning for osteosarcoma diagnosis. Talanta 2025, 294, 128276. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Zhang, W.; Shi, Q.; Hui, J.; Wang, J.; Song, Y.; Yang, X. An Integrated Microfluidic System for One-Stop Multiplexed Exosomal PD-L1 and MMP9 Automated Analysis with Deep Learning Model YOLO. Micromachines 2025, 16, 1208. https://doi.org/10.3390/mi16111208
Lu Y, Zhang W, Shi Q, Hui J, Wang J, Song Y, Yang X. An Integrated Microfluidic System for One-Stop Multiplexed Exosomal PD-L1 and MMP9 Automated Analysis with Deep Learning Model YOLO. Micromachines. 2025; 16(11):1208. https://doi.org/10.3390/mi16111208
Chicago/Turabian StyleLu, Yunxing, Wenjing Zhang, Qiang Shi, Jianan Hui, Jieyu Wang, Yiman Song, and Xiaoyue Yang. 2025. "An Integrated Microfluidic System for One-Stop Multiplexed Exosomal PD-L1 and MMP9 Automated Analysis with Deep Learning Model YOLO" Micromachines 16, no. 11: 1208. https://doi.org/10.3390/mi16111208
APA StyleLu, Y., Zhang, W., Shi, Q., Hui, J., Wang, J., Song, Y., & Yang, X. (2025). An Integrated Microfluidic System for One-Stop Multiplexed Exosomal PD-L1 and MMP9 Automated Analysis with Deep Learning Model YOLO. Micromachines, 16(11), 1208. https://doi.org/10.3390/mi16111208





