Integration of Point-of-Care Technology in the Decoding Process of Single Nucleotide Polymorphism for Healthcare Application † †
Abstract
1. Introduction
2. Common Methods for Genetic Material Preparation
3. Allele-Specific Polymerase Chain Reaction Amplification (AS-PCR)
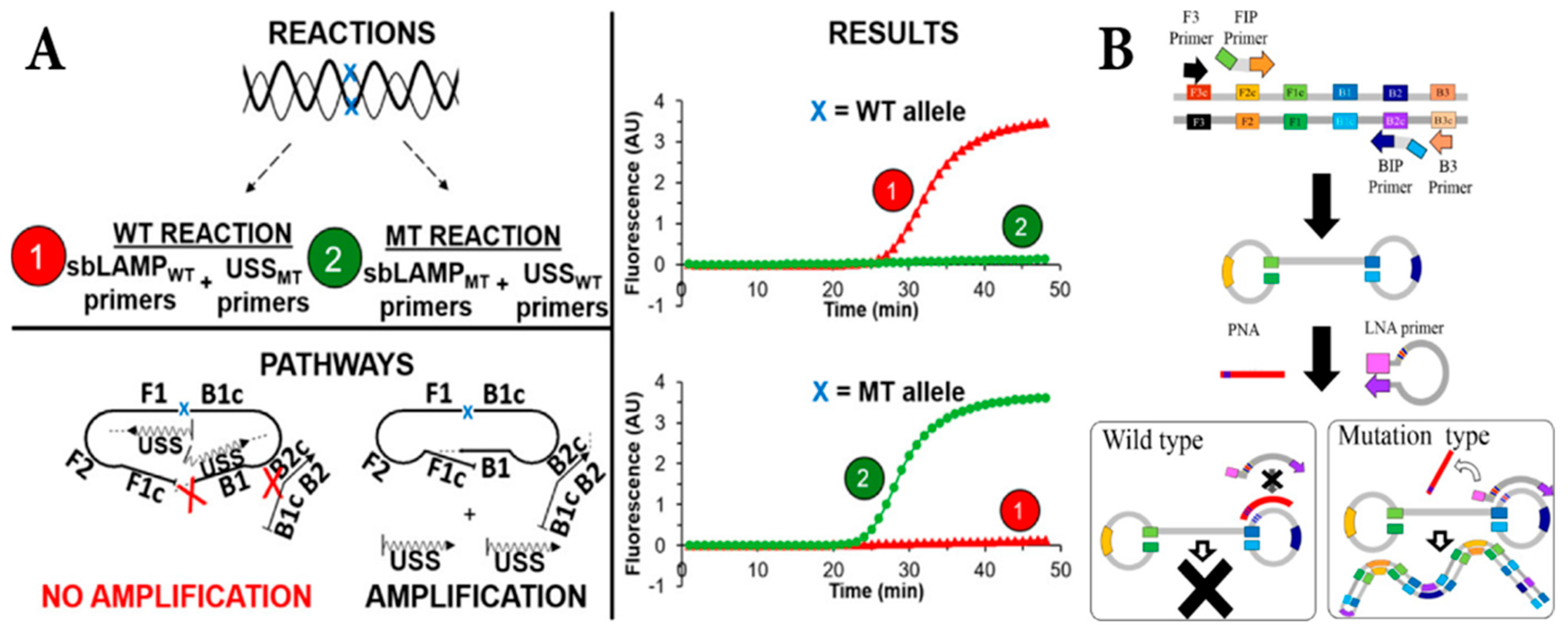
4. Allele-Specific Loop-Mediated Isothermal Amplification (AS-LAMP)
5. Allele-Specific Recombinase Polymerase Amplification (AS-RPA)
6. Point-of-Care Technology Applications for Allele-Specific Amplification
6.1. The Colorimetric Detection Methods Integrating to POCT System
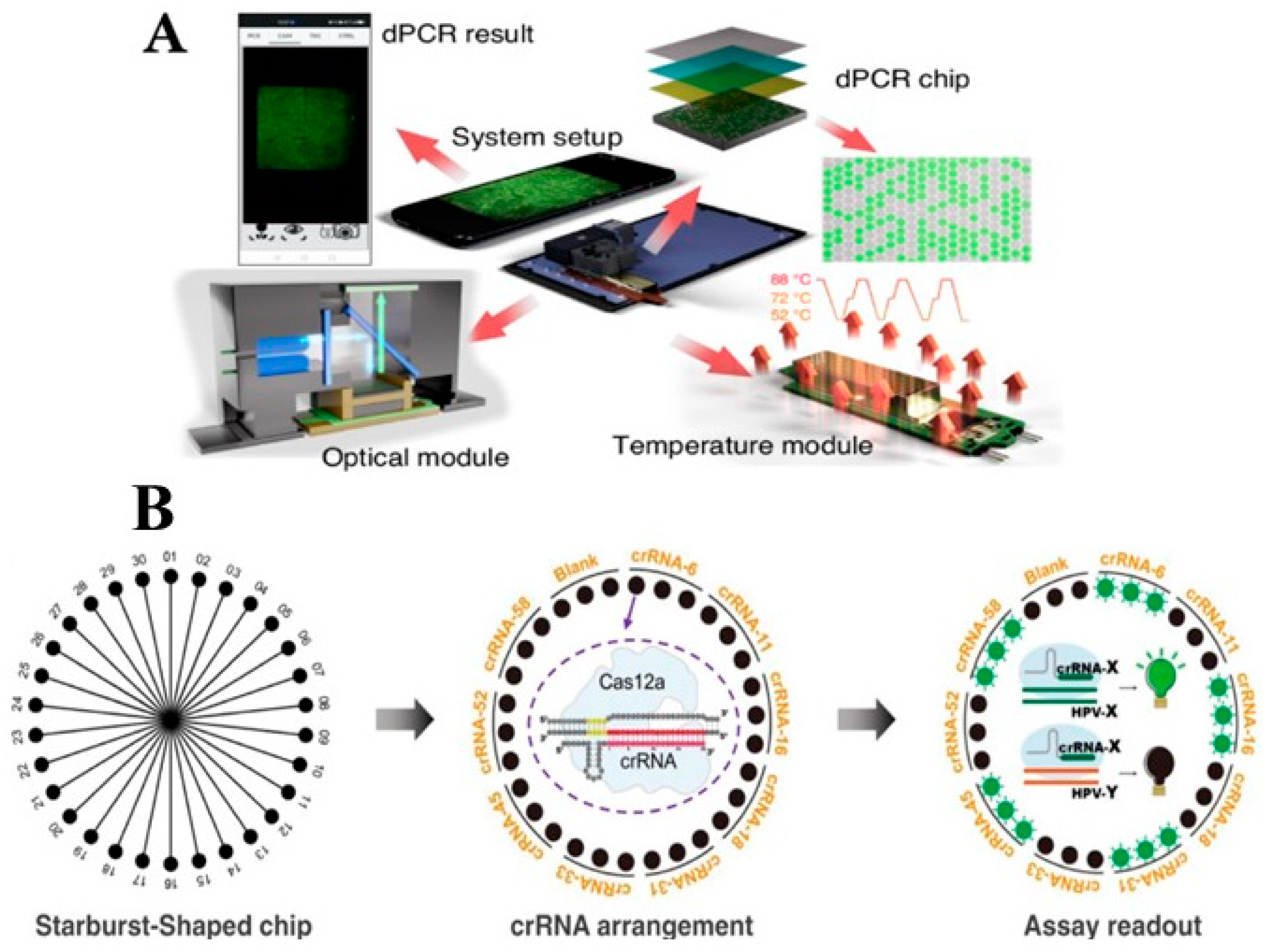
6.2. Typical Point-of-Care Systems
| Amplification Methods | Advantages | Disadvantages | References |
|---|---|---|---|
| PCR |
|
| [52,53] |
| LAMP (Isothermal) (60–72 °C) |
|
| [54,56,57] |
| RPA (Isothermal) (37–42 °C) |
|
| [58,59,60] |
6.3. Point-of-Care Systems Integrated with Allele-Specific Amplification for SNP Detection
7. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shastry, B.S. SNPs in disease gene mapping, medicinal drug development and evolution. J. Hum. Genet. 2007, 52, 871–880. [Google Scholar] [CrossRef]
- Ousmael, K.; Whetten, R.W.; Xu, J.; Nielsen, U.B.; Lamour, K.; Hansen, O.K. Identification and high-throughput genotyping of single nucleotide polymorphism markers in a non-model conifer (Abies nordmanniana (Steven) Spach). Sci. Rep. 2023, 13, 22488. [Google Scholar] [CrossRef]
- He, Y.; Shao, S.; Chen, J. High-fidelity identification of single nucleotide polymorphism by type V CRISPR systems. ACS Sens. 2023, 8, 4478–4483. [Google Scholar] [CrossRef]
- Hyman, L.B.; Christopher, C.R.; Romero, P.A. Competitive SNP-LAMP probes for rapid and robust single-nucleotide polymorphism detection. Cell Rep. Methods 2022, 2, 100242. [Google Scholar] [CrossRef] [PubMed]
- Prakinee, K.; Phaisan, S.; Kongjaroon, S.; Chaiyen, P. Ancestral Sequence Reconstruction for Designing Biocatalysts and Investigating their Functional Mechanisms. JACS Au 2024, 4, 4571–4591. [Google Scholar] [CrossRef]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Arteche-López, A.; Ávila-Fernández, A.; Romero, R.; Riveiro-Álvarez, R.; López-Martínez, M.; Giménez-Pardo, A.; Vélez-Monsalve, C.; Gallego-Merlo, J.; García-Vara, I.; Almoguera, B. Sanger sequencing is no longer always necessary based on a single-center validation of 1109 NGS variants in 825 clinical exomes. Sci. Rep. 2021, 11, 5697. [Google Scholar] [CrossRef]
- Nakae, S.; Hijikata, A.; Tsuji, T.; Yonezawa, K.; Kouyama, K.-I.; Mayanagi, K.; Ishino, S.; Ishino, Y.; Shirai, T. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure 2016, 24, 1960–1971. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Feng, W.; Lin, M.; Chen, S.; Liu, X.; Wang, X.; Chen, Q. Comparative evaluation of PCR-based, LAMP and RPA-CRISPR/Cas12a assays for the rapid detection of Diaporthe aspalathi. Int. J. Mol. Sci. 2024, 25, 5773. [Google Scholar] [CrossRef]
- Yan, S.; Li, C.; Lan, H.; Pan, D.; Wu, Y. Comparison of four isothermal amplification techniques: LAMP, SEA, CPA, and RPA for the identification of chicken adulteration. Food Control 2024, 159, 110302. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Huang, S.; Yin, C.; Wan, L.; Li, J.; Ling, J.; Wu, C.; Dai, L. An open source, PCR based, point-of-care testing platform. Sci. Rep. 2025, 15, 12025. [Google Scholar] [CrossRef]
- Song, Q.; Sun, X.; Dai, Z.; Gao, Y.; Gong, X.; Zhou, B.; Wu, J.; Wen, W. Point-of-care testing detection methods for COVID-19. LOC 2021, 21, 1634–1660. [Google Scholar] [CrossRef]
- Fistera, D.; Kikull, K.; Risse, J.; Herrmann, A.; Brachmann, M.; Kill, C. Point-of-care PCR testing of SARS-CoV-2 in the emergency department: Influence on workflow and efficiency. PLoS ONE 2023, 18, e0288906. [Google Scholar] [CrossRef]
- Xu, D.; Jiang, X.; Zou, T.; Miao, G.; Fu, Q.; Xiang, F.; Feng, L.; Ye, X.; Zhang, L.; Qiu, X. A microfluidic system for rapid nucleic acid analysis based on real-time convective PCR at point-of-care testing. Microfluid. Nanofluid. 2022, 26, 69. [Google Scholar] [CrossRef]
- McCloskey, D.; Erickson, D. Rapid nucleic acid extraction from skin biopsies using a point-of-care device. LOC 2022, 22, 3229–3235. [Google Scholar] [CrossRef]
- Han, K.; Yoon, Y.-J.; Shin, Y.; Park, M.K. Self-powered switch-controlled nucleic acid extraction system. LOC 2016, 16, 132–141. [Google Scholar] [CrossRef]
- Petralia, S.; Sciuto, E.L.; Conoci, S. A novel miniaturized biofilter based on silicon micropillars for nucleic acid extraction. Analyst 2017, 142, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hu, J.; Sun, J.; Wang, B.; Mu, Y. A fast nucleic acid extraction system for point-of-care and integration of digital PCR. Analyst 2019, 144, 7032–7040. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, A.V.; Ramachandran, S.; Vigil, G.; Yager, P.; Böhringer, K. A low cost point-of-care viscous sample preparation device for molecular diagnosis in the developing world; an example of microfluidic origami. LOC 2012, 12, 174–181. [Google Scholar] [CrossRef]
- Alafeef, M.; Moitra, P.; Dighe, K.; Pan, D. RNA-extraction-free nano-amplified colorimetric test for point-of-care clinical diagnosis of COVID-19. Nat. Protoc. 2021, 16, 3141–3162. [Google Scholar] [CrossRef] [PubMed]
- Okayama, H.; Curiel, D.T.; Brantly, M.L.; Holmes, M.D.; Crystal, R.G. Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J. Lab. Clin. Med. 1989, 114, 105–113. [Google Scholar] [PubMed]
- Kwok, S.; Lipka, J.; McKinney, N.; Kellogg, D.; Poiesz, B.; Foung, S.; Sninsky, J. Low incidence of HTLV infections in random blood donors with indeterminate western blot patterns. Transfusion 1990, 30, 491–494. [Google Scholar] [CrossRef]
- Li, H.; Cui, X.; Arnheim, N. Direct electrophoretic detection of the allelic state of single DNA molecules in human sperm by using the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1990, 87, 4580–4584. [Google Scholar] [CrossRef]
- Bundock, P.C.; Cross, M.J.; Shapter, F.M.; Henry, R.J. Robust allele-specific polymerase chain reaction markers developed for single nucleotide polymorphisms in expressed barley sequences. Theor. Appl. Genet. 2006, 112, 358–365. [Google Scholar] [CrossRef]
- Darawi, M.N.; Ai-Vyrn, C.; Ramasamy, K.; Hua, P.P.J.; Pin, T.M.; Kamaruzzaman, S.B.; Majeed, A.B.A. Allele-specific polymerase chain reaction for the detection of Alzheimer’s disease-related single nucleotide polymorphisms. BMC Med. Genet. 2013, 14, 27. [Google Scholar] [CrossRef]
- He, Q.; Chen, M.; Lin, X.; Chen, Z. Allele-specific PCR with a novel data processing method based on difference value for single nucleotide polymorphism genotyping of ALDH2 gene. Talanta 2020, 220, 121432. [Google Scholar] [CrossRef] [PubMed]
- Yongkiettrakul, S.; Kampeera, J.; Chareanchim, W.; Rattanajak, R.; Pornthanakasem, W.; Kiatpathomchai, W.; Kongkasuriyachai, D. Simple detection of single nucleotide polymorphism in Plasmodium falciparum by SNP-LAMP assay combined with lateral flow dipstick. Parasitol. Int. 2017, 66, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Malpartida-Cardenas, K.; Rodriguez-Manzano, J.; Yu, L.-S.; Delves, M.J.; Nguon, C.; Chotivanich, K.; Baum, J.; Georgiou, P. Allele-specific isothermal amplification method using unmodified self-stabilizing competitive primers. Anal. Chem. 2018, 90, 11972–11980. [Google Scholar] [CrossRef]
- Itonaga, M.; Matsuzaki, I.; Warigaya, K.; Tamura, T.; Shimizu, Y.; Fujimoto, M.; Kojima, F.; Ichinose, M.; Murata, S.-I. Novel methodology for rapid detection of KRAS mutation using PNA-LNA mediated loop-mediated isothermal amplification. PLoS ONE 2016, 11, e0151654. [Google Scholar] [CrossRef]
- Natoli, M.E.; Chang, M.M.; Kundrod, K.A.; Coole, J.B.; Airewele, G.E.; Tubman, V.N.; Richards-Kortum, R.R. Allele-specific recombinase polymerase amplification to detect sickle cell disease in low-resource settings. Anal. Chem. 2021, 93, 4832–4840. [Google Scholar] [CrossRef]
- Fujita, T.; Nagata, S.; Fujii, H. Protein or ribonucleoprotein-mediated blocking of recombinase polymerase amplification enables the discrimination of nucleotide and epigenetic differences between cell populations. Commun. Biol. 2021, 4, 988. [Google Scholar] [CrossRef]
- Li, H.; Rothberg, L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 14036–14039. [Google Scholar] [CrossRef]
- Li, Y.; Deng, Z. Silver nanoparticle–DNA bionanoconjugates bearing a discrete number of DNA ligands. Chem. Commun. 2012, 48, 6160–6162. [Google Scholar] [CrossRef]
- Rahnama, S.; Shariati, S.; Divsar, F. Selective aptamer conjugation to silver-coated magnetite nanoparticles for magnetic solid-phase extraction of trace amounts of Pb 2+ ions. RSC Adv. 2021, 11, 4971–4982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, J.; Xiao, M.; Zhang, S.; Ren, S.; Luo, D.; Xi, F.; Liu, H.; Li, Y.; Li, Q. Aptamer-functionalized gold nanoparticles for fast and selective electrochemical sensing of lead in tobacco. Int. J. Electrochem. Sci. 2024, 19, 100858. [Google Scholar] [CrossRef]
- Jazayeri, M.H.; Aghaie, T.; Avan, A.; Vatankhah, A.; Ghaffari, M.R.S. Colorimetric detection based on gold nano particles (GNPs): An easy, fast, inexpensive, low-cost and short time method in detection of analytes (protein, DNA, and ion). Sens. Bio-Sens. Res. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Li, H.; Qiang, W.; Vuki, M.; Xu, D.; Chen, H.-Y. Fluorescence enhancement of silver nanoparticle hybrid probes and ultrasensitive detection of IgE. Anal. Chem. 2011, 83, 8945–8952. [Google Scholar] [CrossRef]
- Díaz, C.R.; Lafuente-Gómez, N.; Coutinho, C.; Pardo, D.; Alarcón-Iniesta, H.; López-Valls, M.; Coloma, R.; Milán-Rois, P.; Domenech, M.; Abreu, M. Development of colorimetric sensors based on gold nanoparticles for SARS-CoV-2 RdRp, E and S genes detection. Talanta 2022, 243, 123393. [Google Scholar] [CrossRef]
- Tatulli, G.; Pompa, P.P. An amplification-free colorimetric test for sensitive DNA detection based on the capturing of gold nanoparticle clusters. Nanoscale 2020, 12, 15604–15610. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano 2018, 13, 1244–1252. [Google Scholar] [CrossRef]
- Gao, X.; Liu, Q.; Zhao, Y.; Li, Z.; Wang, Y.; Zhou, D.; Jiang, K.; Luo, C. Influences of gold and silver nanoparticles in loop-mediated isothermal amplification reactions. J. Exp. Nanosci. 2014, 9, 922–930. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Z.; Chen, J.; Wang, S.; Huang, X. Sensitive and selective detection of glutathione based on resonance light scattering using sensitive gold nanoparticles as colorimetric probes. Analyst 2012, 137, 3132–3137. [Google Scholar] [CrossRef]
- Manajit, O.; Longyant, S.; Sithigorngul, P.; Chaivisuthangkura, P. Development of uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification coupled with nanogold probe (UDG-LAMP-AuNP) for specific detection of Pseudomonas aeruginosa. Mol. Med. Rep. 2018, 17, 5734–5743. [Google Scholar] [CrossRef]
- Jiang, X.; Yang, M.; Liu, J. Capping gold nanoparticles to achieve a protein-like surface for loop-mediated isothermal amplification acceleration and ultrasensitive DNA detection. ACS Appl. Mater. Interfaces 2022, 14, 27666–27674. [Google Scholar] [CrossRef]
- Trinh, T.N.D.; Trinh, K.T.L.; Lee, N.Y. Colorimetric polymerase chain reaction mediated by pH indicator: Rapid detection of drug-resistant nosocomial bacteria. Talanta 2025, 284, 127193. [Google Scholar] [CrossRef]
- Tanner, N.A.; Zhang, Y.; Evans Jr, T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 2015, 58, 59–68. [Google Scholar] [CrossRef]
- González-López, A.; Cima-Cabal, M.D.; Rioboó-Legaspi, P.; Costa-Rama, E.; García-Suárez, M.a.d.M.; Fernández-Abedul, M.T. Electrochemical detection for isothermal loop-mediated amplification of pneumolysin gene of Streptococcus pneumoniae based on the oxidation of phenol red indicator. Anal. Chem. 2022, 94, 13061–13067. [Google Scholar] [CrossRef]
- Tomar, S.; Lavickova, B.; Guiducci, C. Recombinase polymerase amplification in minimally buffered conditions. Biosens. Bioelectron. 2022, 198, 113802. [Google Scholar] [CrossRef] [PubMed]
- Hassanain, W.A.; Johnson, C.L.; Faulds, K.; Keegan, N.; Graham, D. Ultrasensitive Dual ELONA/SERS–RPA Multiplex Diagnosis of Antimicrobial Resistance. Anal. Chem. 2024, 96, 12093–12101. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Kim, Y.T.; Byun, J.-Y.; Ahn, J.; Chung, S.; Gweon, D.-G.; Kim, M.-G.; Seo, T.S. An integrated allele-specific polymerase chain reaction-microarray chip for multiplex single nucleotide polymorphism typing. LOC 2012, 12, 5146–5154. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Kim, Y.T.; Ahn, J.; Kim, K.S.; Gweon, D.-G.; Seo, T.S. Integrated allele-specific polymerase chain reaction–capillary electrophoresis microdevice for single nucleotide polymorphism genotyping. Biosens. Bioelectron. 2012, 35, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, C.J.; Caputo, M.; Hawkins, C.; Balmert, L.C.; Qi, C.; Odorisio, J.; Dembele, E.; Jackson, A.; Abbas, H.; Frediani, J.K. Clinical evaluation of the Diagnostic Analyzer for Selective Hybridization (DASH): A point-of-care PCR test for rapid detection of SARS-CoV-2 infection. PLoS ONE 2022, 17, e0270060. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Wang, X.; Yan, Z.; Xu, Y.; Gaňová, M.; Řezníček, T.; Korabečná, M.; Neuzil, P. SPEED: An integrated, smartphone-operated, handheld digital PCR Device for point-of-care testing. Microsyst. Nanoeng. 2024, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.N.D.; Lee, N.Y. A foldable isothermal amplification microdevice for fuchsin-based colorimetric detection of multiple foodborne pathogens. LOC 2019, 19, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.T.L.; Trinh, T.N.D.; Lee, N.Y. Fully integrated and slidable paper-embedded plastic microdevice for point-of-care testing of multiple foodborne pathogens. Biosens. Bioelectron. 2019, 135, 120–128. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Wang, C.; Feng, Q.; Fan, F.; Zhang, G.; Kang, X.; Qin, X.; Sun, J.; Li, Y. Integrated microcapillary for sample-to-answer nucleic acid pretreatment, amplification, and detection. Anal. Chem. 2014, 86, 10461–10466. [Google Scholar] [CrossRef]
- Lu, W.; Wang, J.; Wu, Q.; Sun, J.; Chen, Y.; Zhang, L.; Zheng, C.; Gao, W.; Liu, Y.; Jiang, X. High-throughput sample-to-answer detection of DNA/RNA in crude samples within functionalized micro-pipette tips. Biosens. Bioelectron. 2016, 75, 28–33. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, D.; Li, T.; Yan, J.; Zhu, J.; He, T.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Microfluidic space coding for multiplexed nucleic acid detection via CRISPR-Cas12a and recombinase polymerase amplification. Nat. Commun. 2022, 13, 6480. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.S.; Woo, H.; Yoo, Y.K.; Lee, D.; Chung, S.; Yoon, D.S.; Lee, K.-B.; Lee, J.H. Rapid deep learning-assisted predictive diagnostics for point-of-care testing. Nat. Commun. 2024, 15, 1695. [Google Scholar] [CrossRef]
- Chae, W.R.; Song, Y.-J.; Lee, N.Y. Polydopamine-mediated gold nanoparticle coating strategy and its application in photothermal polymerase chain reaction. Lab. Chip. 2025, 25, 1429–1438. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, C.; Zhao, M.; Liu, K.; Li, H.; Li, N.; Gao, L.; Yang, X.; Ma, T.; Zhu, J.; et al. A direct isothermal amplification system adapted for rapid SNP genotyping of multifarious sample types. Biosens Bioelectron. 2018, 115, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Li, X.; Mai, L.; Wang, R.; Tang, S.; Hu, L. Visual single nucleotide polymorphism (SNP) detection for ALDH2 genotyping based on multiplex ligation probe amplification (MLPA) and lateral flow assay. Microchem. J. 2023, 194, 109329. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, Y.; Wang, S.; Wang, X.; Li, K.; Wang, Y.; Lu, Z.; Liu, Q.; Wang, W.; Dao, Y.; et al. Development of a CRISPR/Cas12a one-tube POCT method with RCA for multiplex detection of SNPs. Microchem. J. 2025, 210, 113037. [Google Scholar] [CrossRef]
- Thai, D.A.; Lee, N.Y. A point-of-care platform for hair loss-related single nucleotide polymorphism genotyping. Anal. Chim. Acta 2023, 1283, 341973. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trinh, T.N.D.; Nguyen, H.A.; Thi, N.P.A.; Ho, T.X.T.; Trinh, K.T.L.; Tran, N.K.S. Integration of Point-of-Care Technology in the Decoding Process of Single Nucleotide Polymorphism for Healthcare Application †. Micromachines 2025, 16, 1159. https://doi.org/10.3390/mi16101159
Trinh TND, Nguyen HA, Thi NPA, Ho TXT, Trinh KTL, Tran NKS. Integration of Point-of-Care Technology in the Decoding Process of Single Nucleotide Polymorphism for Healthcare Application †. Micromachines. 2025; 16(10):1159. https://doi.org/10.3390/mi16101159
Chicago/Turabian StyleTrinh, Thi Ngoc Diep, Hanh An Nguyen, Nguyen Pham Anh Thi, Thi Xuan Tuy Ho, Kieu The Loan Trinh, and Nguyen Khoi Song Tran. 2025. "Integration of Point-of-Care Technology in the Decoding Process of Single Nucleotide Polymorphism for Healthcare Application †" Micromachines 16, no. 10: 1159. https://doi.org/10.3390/mi16101159
APA StyleTrinh, T. N. D., Nguyen, H. A., Thi, N. P. A., Ho, T. X. T., Trinh, K. T. L., & Tran, N. K. S. (2025). Integration of Point-of-Care Technology in the Decoding Process of Single Nucleotide Polymorphism for Healthcare Application †. Micromachines, 16(10), 1159. https://doi.org/10.3390/mi16101159