A Novel 3D-Printed and Miniaturized Periodic Counter Current Chromatography System for Continuous Purification of Monoclonal Antibodies
Abstract
1. Introduction
2. Materials and Methods
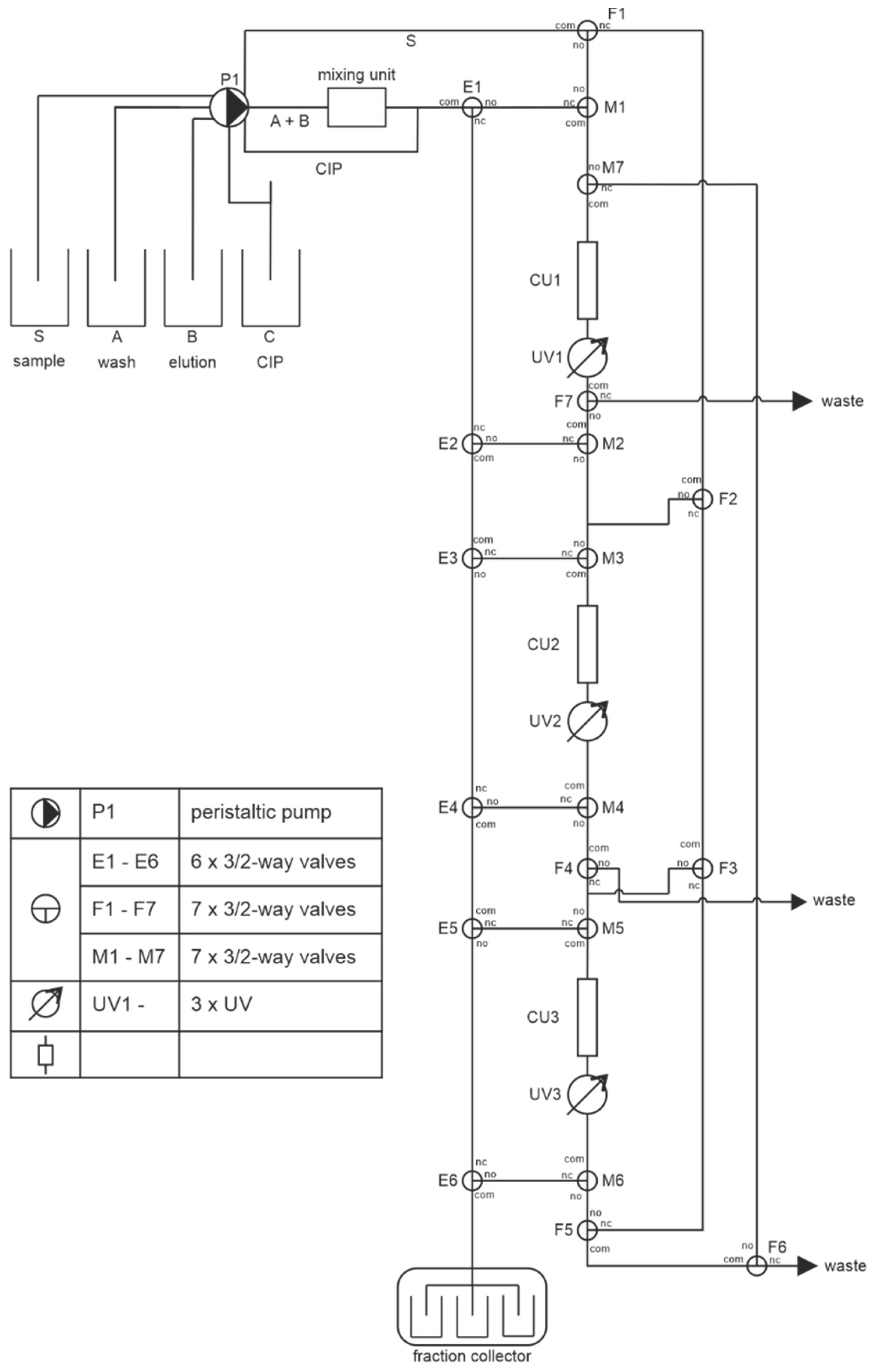
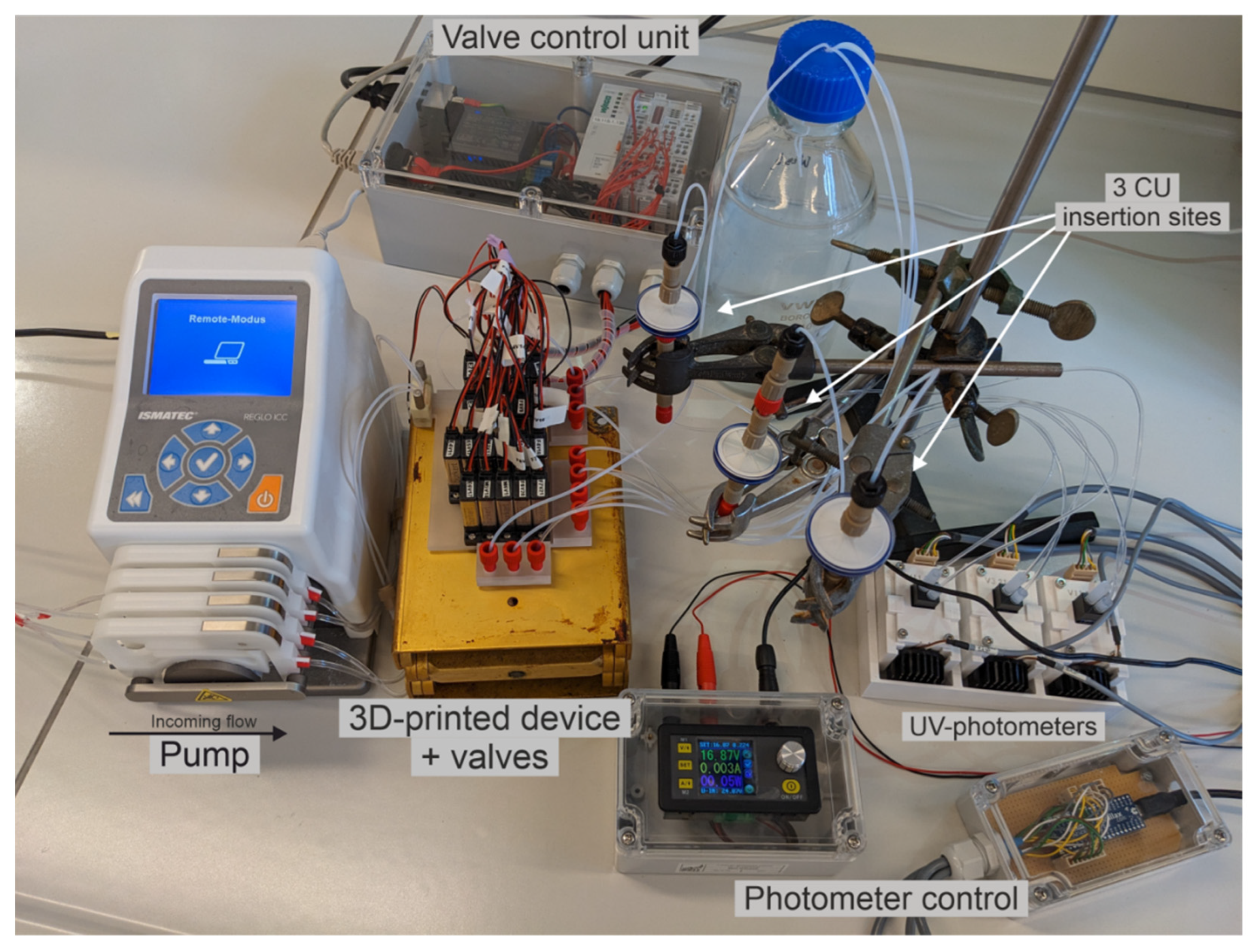
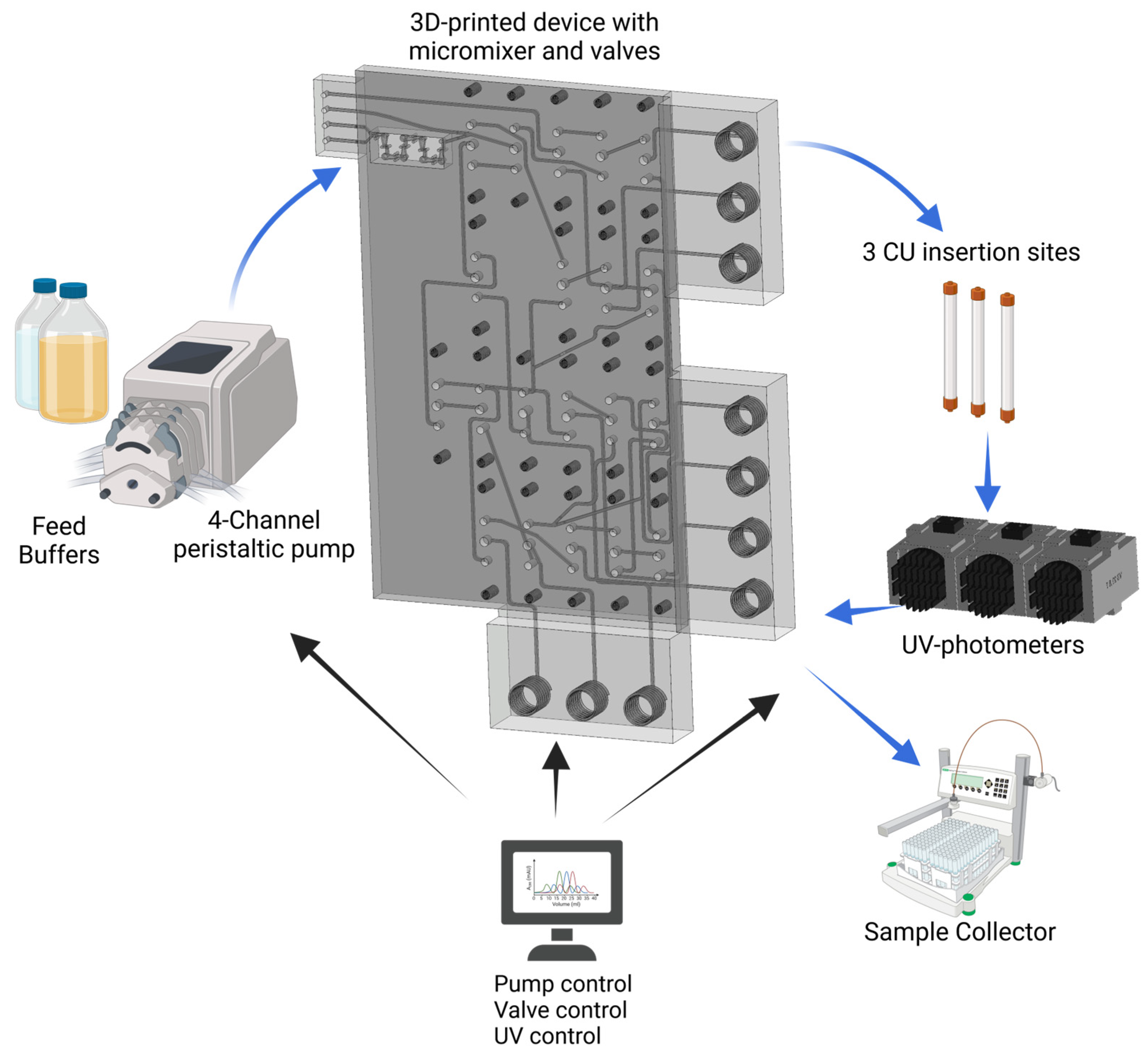
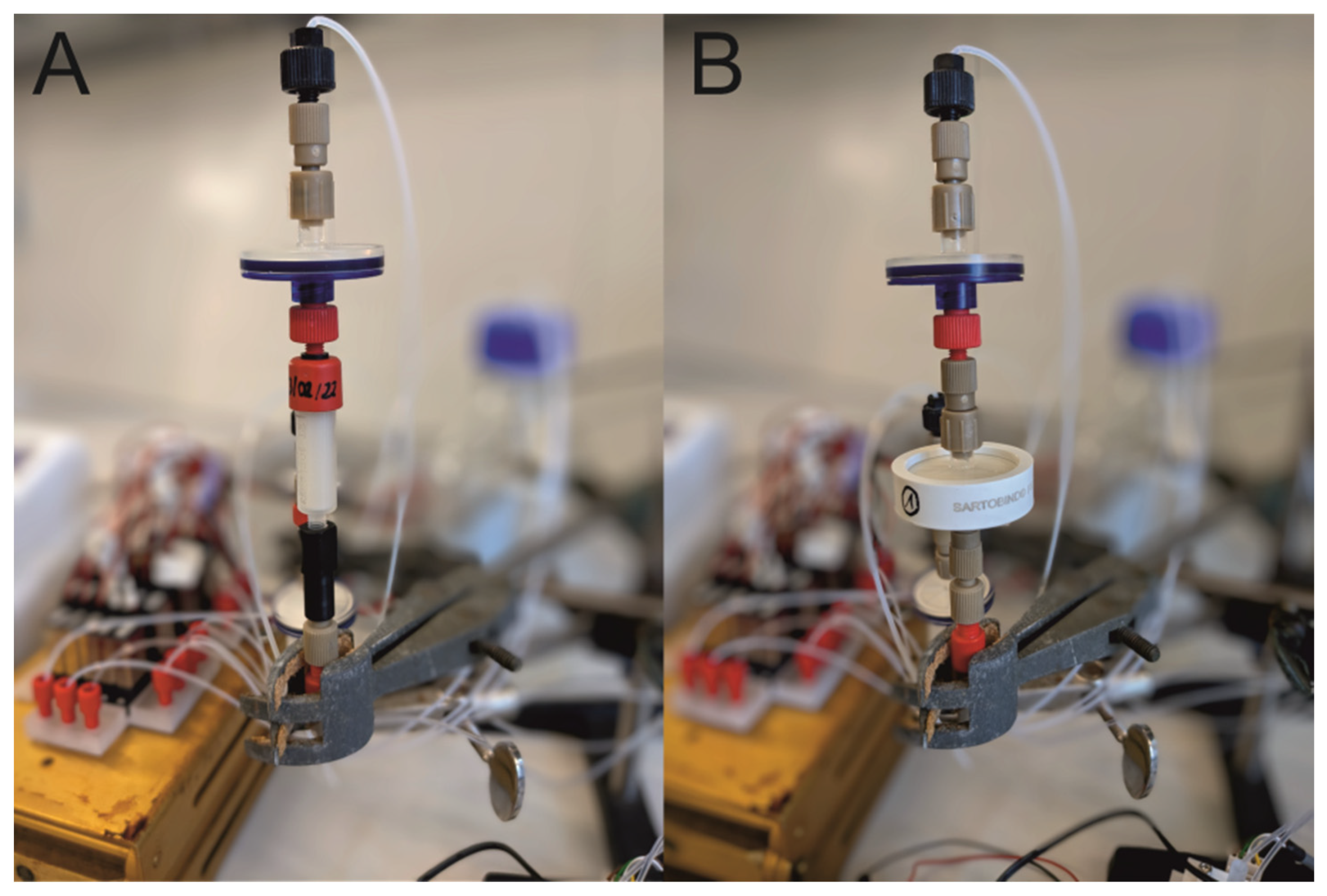
2.1. Fabrication of the Miniaturized 3D-Printed PCCC System
2.2. Experimental Setup of the Continuous Miniaturized Chromatography System
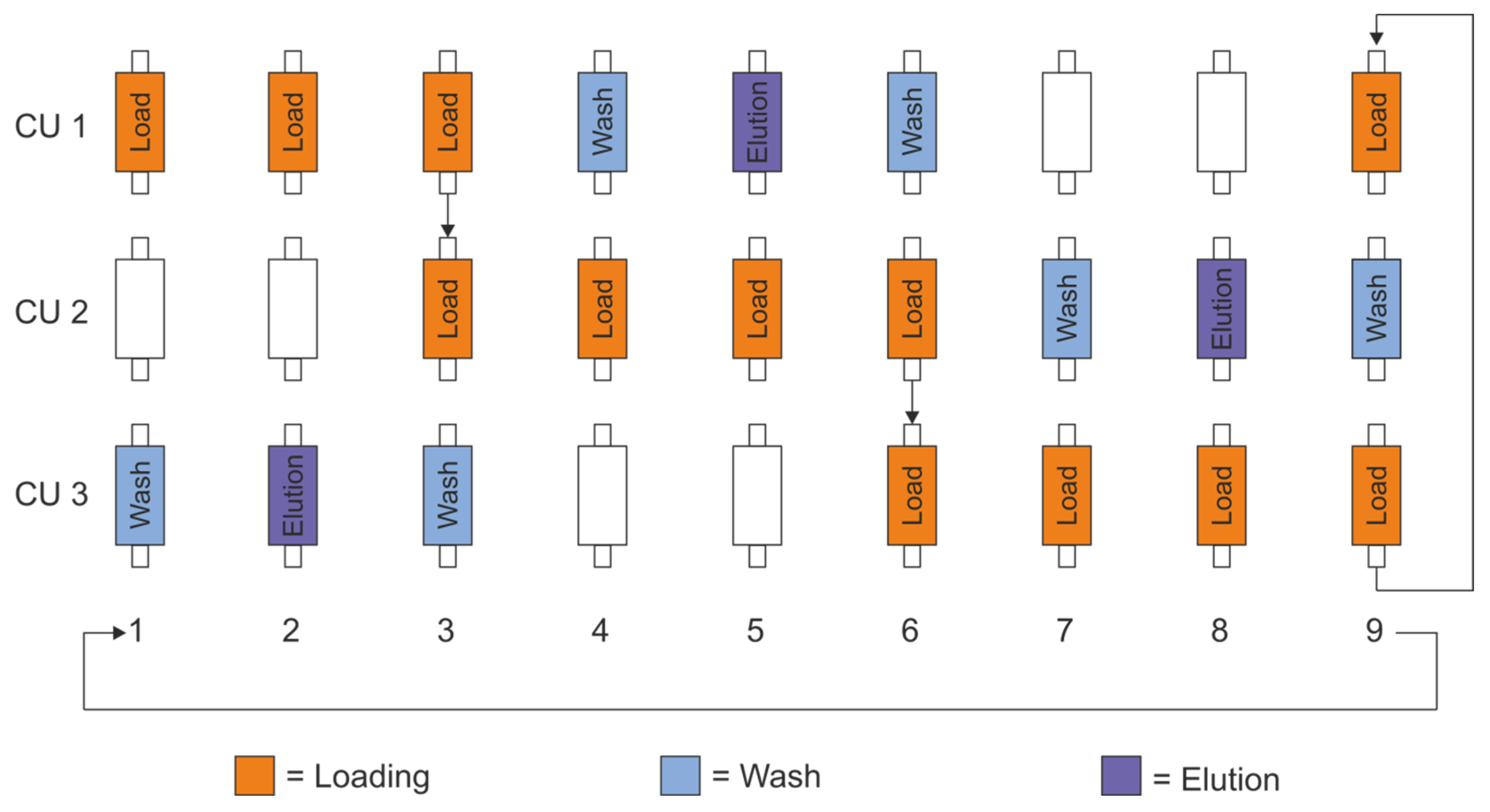
2.3. PCCC Process and System Control
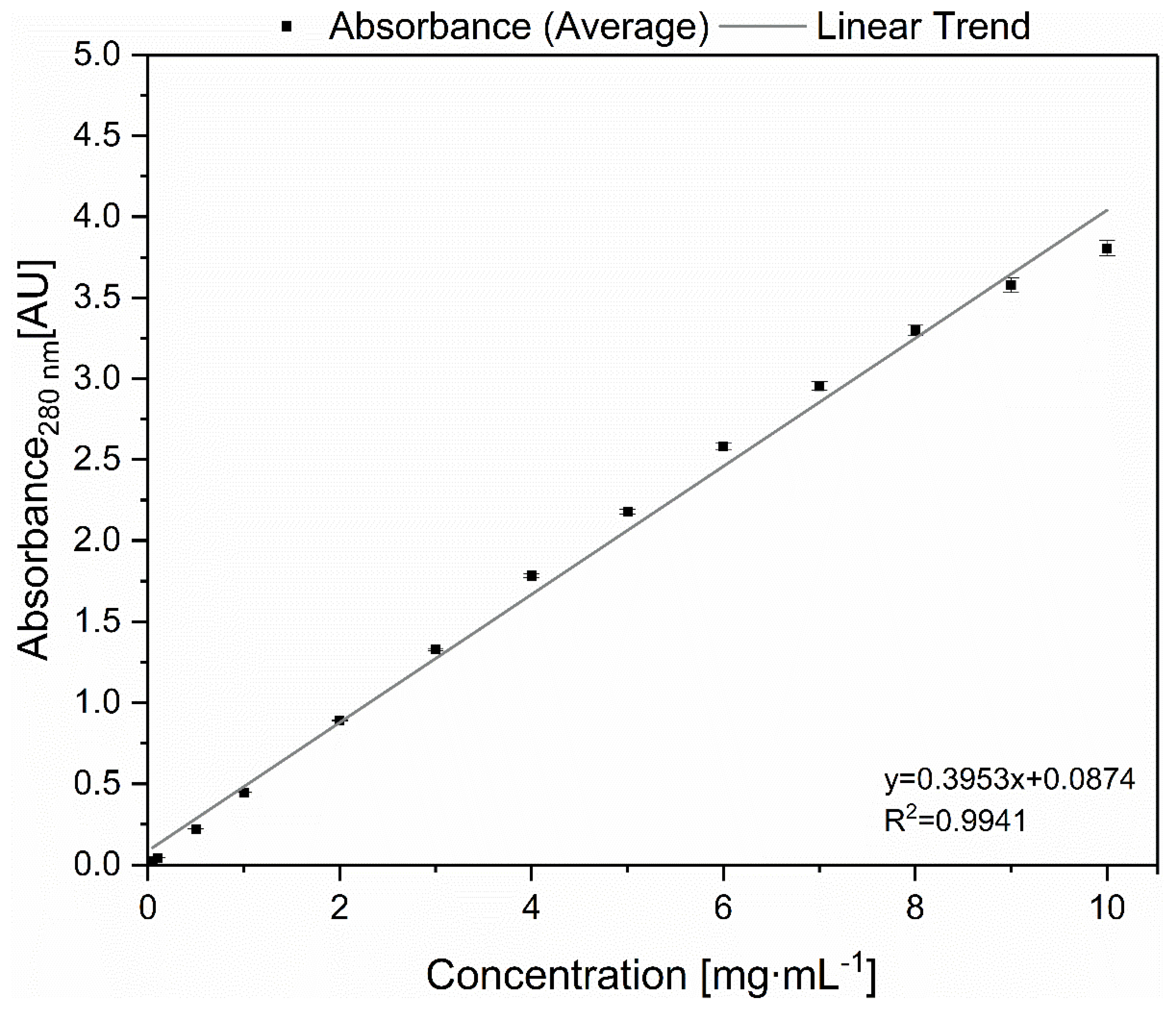
2.4. Determination of Dynamic Binding Capacity
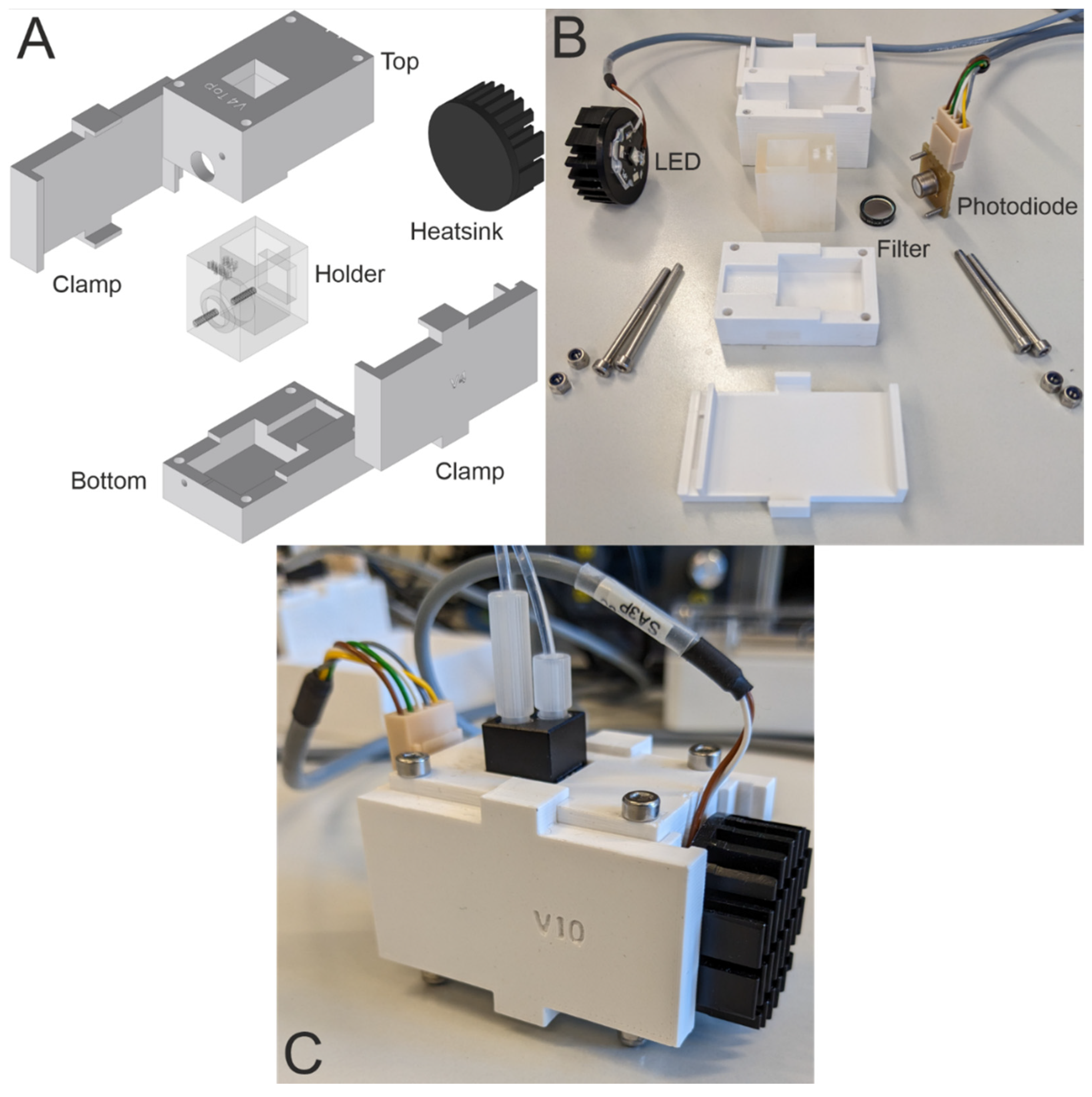
2.5. Fabrication of UV-Photometers
2.6. Purification of mAb
2.7. Analytics via HPLC
3. Results and Discussion
3.1. PCCC Design and Experimental Setup
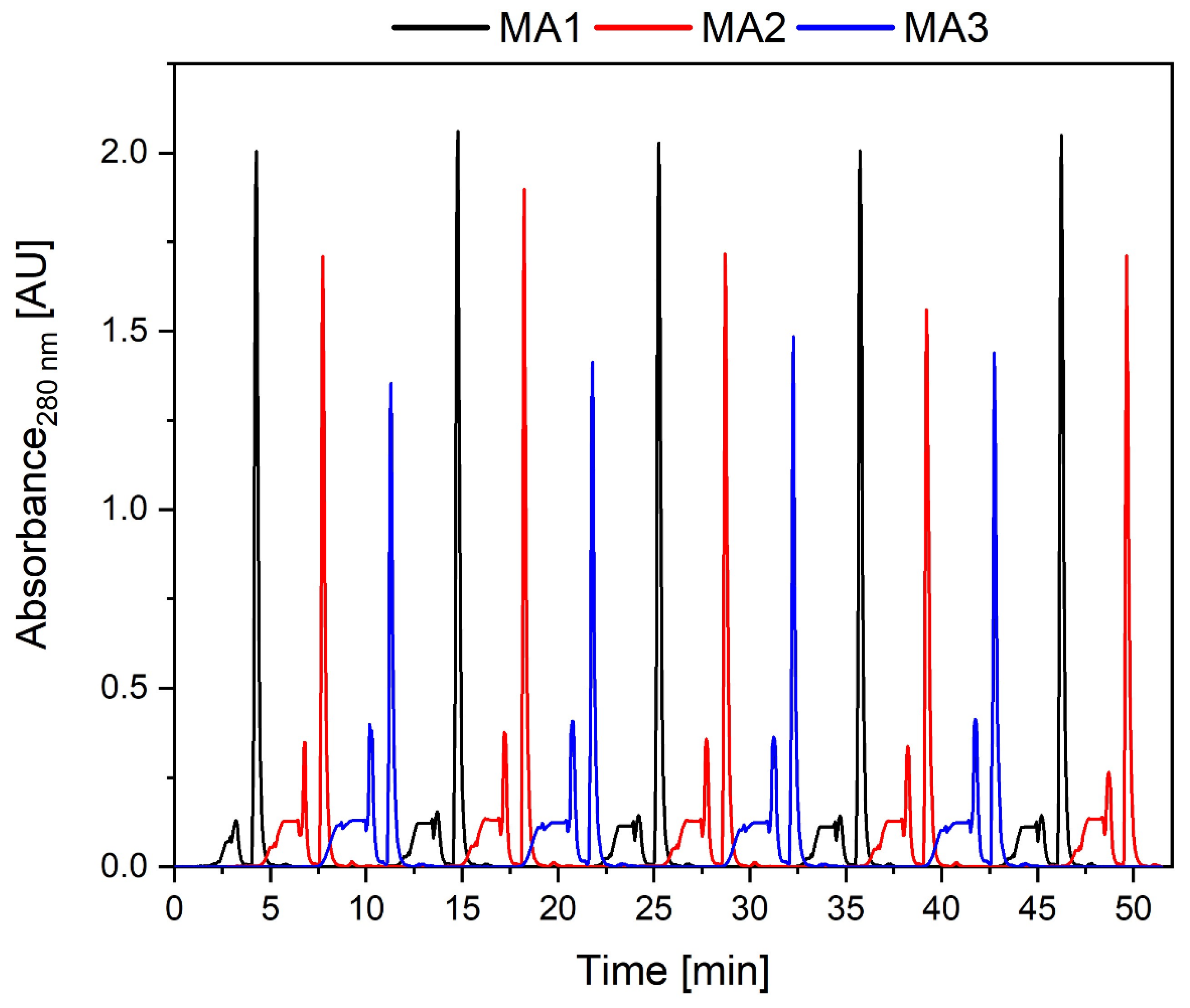
3.2. Protein A Affinity Chromatography
3.3. CEX for Product Polishing
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Tsumoto, K.; Isozaki, Y.; Yagami, H.; Tomita, M. Future perspectives of therapeutic monoclonal antibodies. Immunotherapy 2019, 11, 119–127. [Google Scholar] [CrossRef]
- Nelson, A.L.; Dhimolea, E.; Reichert, J.M. Development trends for human monoclonal antibody therapeutics. Nat. Rev. Drug Discov. 2010, 9, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Rudick, R.A.; Sandrock, A. Natalizumab: Alpha 4-integrin antagonist selective adhesion molecule inhibitors for MS. Expert Rev. Neurother. 2004, 4, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Shirley, M. Daclizumab: A Review in Relapsing Multiple Sclerosis. Drugs 2017, 77, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Lhota, G.; Sissolak, B.; Striedner, G.; Sommeregger, W.; Vorauer-Uhl, K. Quantification of glycated IgG in CHO supernatants: A practical approach. Biotechnol. Prog. 2021, 37, e3124. [Google Scholar] [CrossRef] [PubMed]
- Rau, R. Adalimumab (a fully human anti-tumour necrosis factor alpha monoclonal antibody) in the treatment of active rheumatoid arthritis: The initial results of five trials. Ann. Rheum. Dis. 2002, 61 (Suppl. S2), ii70–ii73. [Google Scholar] [CrossRef]
- Oldfield, V.; Plosker, G.L. Golimumab: In the treatment of rheumatoid arthritis, psoriatic arthritis, and ankylosing spondylitis. BioDrugs 2009, 23, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Grillo-López, A.J. Rituximab (Rituxan/MabThera): The first decade (1993–2003). Expert Rev. Anticancer Ther. 2003, 3, 767–779. [Google Scholar] [CrossRef]
- Kaplon, H.; Chenoweth, A.; Crescioli, S.; Reichert, J.M. Antibodies to watch in 2022. MAbs 2022, 14, 2014296. [Google Scholar] [CrossRef]
- Gupta, S.K.; Shukla, P. Glycosylation control technologies for recombinant therapeutic proteins. Appl. Microbiol. Biotechnol. 2018, 102, 10457–10468. [Google Scholar] [CrossRef]
- Böhl, O.J.; Schellenberg, J.; Bahnemann, J.; Hitzmann, B.; Scheper, T.; Solle, D. Implementation of QbD strategies in the inoculum expansion of a mAb production process. Eng. Life Sci. 2021, 21, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Rehmann, M.S.; Xu, X.; Huang, C.; Tian, J.; Qian, N.-X.; Li, Z.J. Improving titer while maintaining quality of final formulated drug substance via optimization of CHO cell culture conditions in low-iron chemically defined media. MAbs 2018, 10, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Rader, R.A.; Langer, E.S. 30 Years of Upstream Productivity Improvements. BioProcess International, 6 February 2015. Available online: https://www.bioprocessintl.com/expression-platforms/30-years-of-upstream-productivity-improvements (accessed on 3 March 2024).
- Jensen, K. A normally occurring Staphylococcus antibody in human serum. APMIS 2007, 115, 533–539; discussion 540–541. [Google Scholar] [CrossRef]
- Oeding, P.; Grov, A.; Myklestad, B. Immunochemical Studies on Antigen Preparations from Staphylococcus Aureus. 2. Precipitating and Erythrocyte-sensitizing Properties of Protein A (Antigen A) and related Substances. Acta Pathol. Microbiol. Scand. 1964, 62, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, O.; Prabhu, S.; Ierapetritou, M. Comparison between Batch and Continuous Monoclonal Antibody Production and Economic Analysis. Ind. Eng. Chem. Res. 2019, 58, 5851–5863. [Google Scholar] [CrossRef]
- Levy, N.E.; Valente, K.N.; Lee, K.H.; Lenhoff, A.M. Host cell protein impurities in chromatographic polishing steps for monoclonal antibody purification. Biotechnol. Bioeng. 2016, 113, 1260–1272. [Google Scholar] [CrossRef]
- Shukla, A.A.; Hubbard, B.; Tressel, T.; Guhan, S.; Low, D. Downstream processing of monoclonal antibodies—Application of platform approaches. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 848, 28–39. [Google Scholar] [CrossRef]
- Ng, C.K.; Rousset, F.; Valery, E.; Bracewell, D.G.; Sorensen, E. Design of high productivity sequential multi-column chromatography for antibody capture. Food Bioprod. Process. 2014, 92, 233–241. [Google Scholar] [CrossRef]
- Baur, D.; Angarita, M.; Müller-Späth, T.; Morbidelli, M. Optimal model-based design of the twin-column CaptureSMB process improves capacity utilization and productivity in protein A affinity capture. Biotechnol. J. 2016, 11, 135–145. [Google Scholar] [CrossRef]
- Brämer, C.; Lammers, F.; Scheper, T.; Beutel, S. Development and Testing of a 4-Columns Periodic Counter-Current Chromatography System Based on Membrane Adsorbers. Separations 2019, 6, 55. [Google Scholar] [CrossRef]
- Brämer, C.; Schreiber, S.; Scheper, T.; Beutel, S. Continuous purification of Candida antarctica lipase B using 3-membrane adsorber periodic counter-current chromatography. Eng. Life Sci. 2018, 18, 414–424. [Google Scholar] [CrossRef]
- Warikoo, V.; Godawat, R.; Brower, K.; Jain, S.; Cummings, D.; Simons, E.; Johnson, T.; Walther, J.; Yu, M.; Wright, B. Integrated continuous production of recombinant therapeutic proteins. Biotechnol. Bioeng. 2012, 109, 3018–3029. [Google Scholar] [CrossRef]
- Jungbauer, A. Continuous downstream processing of biopharmaceuticals. Trends Biotechnol. 2013, 31, 479–492. [Google Scholar] [CrossRef]
- Nicoud, R.-M. The Amazing Ability of Continuous Chromatography To Adapt to a Moving Environment. Ind. Eng. Chem. Res. 2014, 53, 3755–3765. [Google Scholar] [CrossRef]
- Habib, T.; Brämer, C.; Heuer, C.; Ebbecke, J.; Beutel, S.; Bahnemann, J. 3D-Printed microfluidic device for protein purification in batch chromatography. Lab Chip 2022, 22, 986–993. [Google Scholar] [CrossRef]
- Gross, B.C.; Erkal, J.L.; Lockwood, S.Y.; Chen, C.; Spence, D.M. Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences. Anal. Chem. 2014, 86, 3240–3253. [Google Scholar] [CrossRef]
- Heuer, C.; Preuß, J.-A.; Habib, T.; Enders, A.; Bahnemann, J. 3D printing in biotechnology—An insight into miniaturized and microfluidic systems for applications from cell culture to bioanalytics. Eng. Life Sci. 2021, 22, 744–759. [Google Scholar] [CrossRef] [PubMed]
- 3D Systems. MultiJet Plastic Printers. Available online: https://www.3dsystems.com/sites/default/files/2020-08/3d-systems-mjp-brochure-usen-2020-07-17-web.pdf (accessed on 13 December 2023).
- 3D Systems. Safety Data Sheet VisiJet® M2S-HT90. Available online: https://support.3dsystems.com/s/article/materials-visijet-m2s-ht90?language=en_US (accessed on 13 December 2023).
- 3D Systems. Safety Data Sheet VisiJet® M2 Sup. Available online: https://support.3dsystems.com/s/article/materials-visijet-m2-sup?language=en_US (accessed on 13 December 2023).
- Enders, A.; Siller, I.G.; Urmann, K.; Hoffmann, M.R.; Bahnemann, J. 3D Printed Microfluidic Mixers—A Comparative Study on Mixing Unit Performances. Small 2019, 15, e1804326. [Google Scholar] [CrossRef] [PubMed]
- Brämer, C.; Tünnermann, L.; Gonzalez Salcedo, A.; Reif, O.-W.; Solle, D.; Scheper, T.; Beutel, S. Membrane Adsorber for the Fast Purification of a Monoclonal Antibody Using Protein A Chromatography. Membranes 2019, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Ötes, O.; Flato, H.; Winderl, J.; Hubbuch, J.; Capito, F. Feasibility of using continuous chromatography in downstream processing: Comparison of costs and product quality for a hybrid process vs. a conventional batch process. J. Biotechnol. 2017, 259, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, J.; Zhou, J.X. Process Development for robust removal of aggregates during cation exchange chromatography in monoclonal antibody purification with implementation of quality by design. Prep. Biochem. Biotechnol. 2012, 42, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Steinebach, F.; Müller-Späth, T.; Morbidelli, M. Continuous counter-current chromatography for capture and polishing steps in biopharmaceutical production. Biotechnol. J. 2016, 11, 1126–1141. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.; David, L.; Holzer, M.; Nicoud, R.-M. Designing affinity chromatographic processes for the capture of antibodies. Part I: A simplified approach. J. Chromatogr. A 2017, 1494, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Cytiva. ÄKTA pcc and BioProcess pcc Continuous Chromatography Systems. Available online: https://www.cytivalifesciences.com/en/us/shop/chromatography/chromatography-systems/akta-pcc-and-bioprocess-pcc-continuous-chromatography-systems-p-06265#tech-spec-table (accessed on 13 December 2023).
- Sartorius AG. Resolute® BioSMB PD. Available online: https://shop.sartorius.com/shop/ww/de/eur/punchout-root-process-chromatography/resolute-biosmb-pd/p/BIOSMB-PD-LD100B (accessed on 13 December 2023).
- Zobel, S.; Helling, C.; Ditz, R.; Strube, J. Design and Operation of Continuous Countercurrent Chromatography in Biotechnological Production. Ind. Eng. Chem. Res. 2014, 53, 9169–9185. [Google Scholar] [CrossRef]
- Cytiva. ÄKTA Start Chromatography System. Available online: https://www.cytivalifesciences.com/en/us/shop/chromatography/chromatography-systems/akta-start-p-05773 (accessed on 25 February 2024).

| Method | Direct Load | Indirect Load | Flow Rate | 10% DBC | 50% DBC |
|---|---|---|---|---|---|
| Protein A column | 17 min | 5 min | 1 mL∙min−1 | 17 mg | 21.8 mg |
| CEX MA | 2 min | 0.5 min | 5 mL∙min−1 | 8.78 mg | 10.89 mg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kortmann, C.; Habib, T.; Heuer, C.; Solle, D.; Bahnemann, J. A Novel 3D-Printed and Miniaturized Periodic Counter Current Chromatography System for Continuous Purification of Monoclonal Antibodies. Micromachines 2024, 15, 382. https://doi.org/10.3390/mi15030382
Kortmann C, Habib T, Heuer C, Solle D, Bahnemann J. A Novel 3D-Printed and Miniaturized Periodic Counter Current Chromatography System for Continuous Purification of Monoclonal Antibodies. Micromachines. 2024; 15(3):382. https://doi.org/10.3390/mi15030382
Chicago/Turabian StyleKortmann, Carlotta, Taieb Habib, Christopher Heuer, Dörte Solle, and Janina Bahnemann. 2024. "A Novel 3D-Printed and Miniaturized Periodic Counter Current Chromatography System for Continuous Purification of Monoclonal Antibodies" Micromachines 15, no. 3: 382. https://doi.org/10.3390/mi15030382
APA StyleKortmann, C., Habib, T., Heuer, C., Solle, D., & Bahnemann, J. (2024). A Novel 3D-Printed and Miniaturized Periodic Counter Current Chromatography System for Continuous Purification of Monoclonal Antibodies. Micromachines, 15(3), 382. https://doi.org/10.3390/mi15030382







