Miniaturization of CRISPR/Cas12-Based DNA Sensor Array by Non-Contact Printing
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
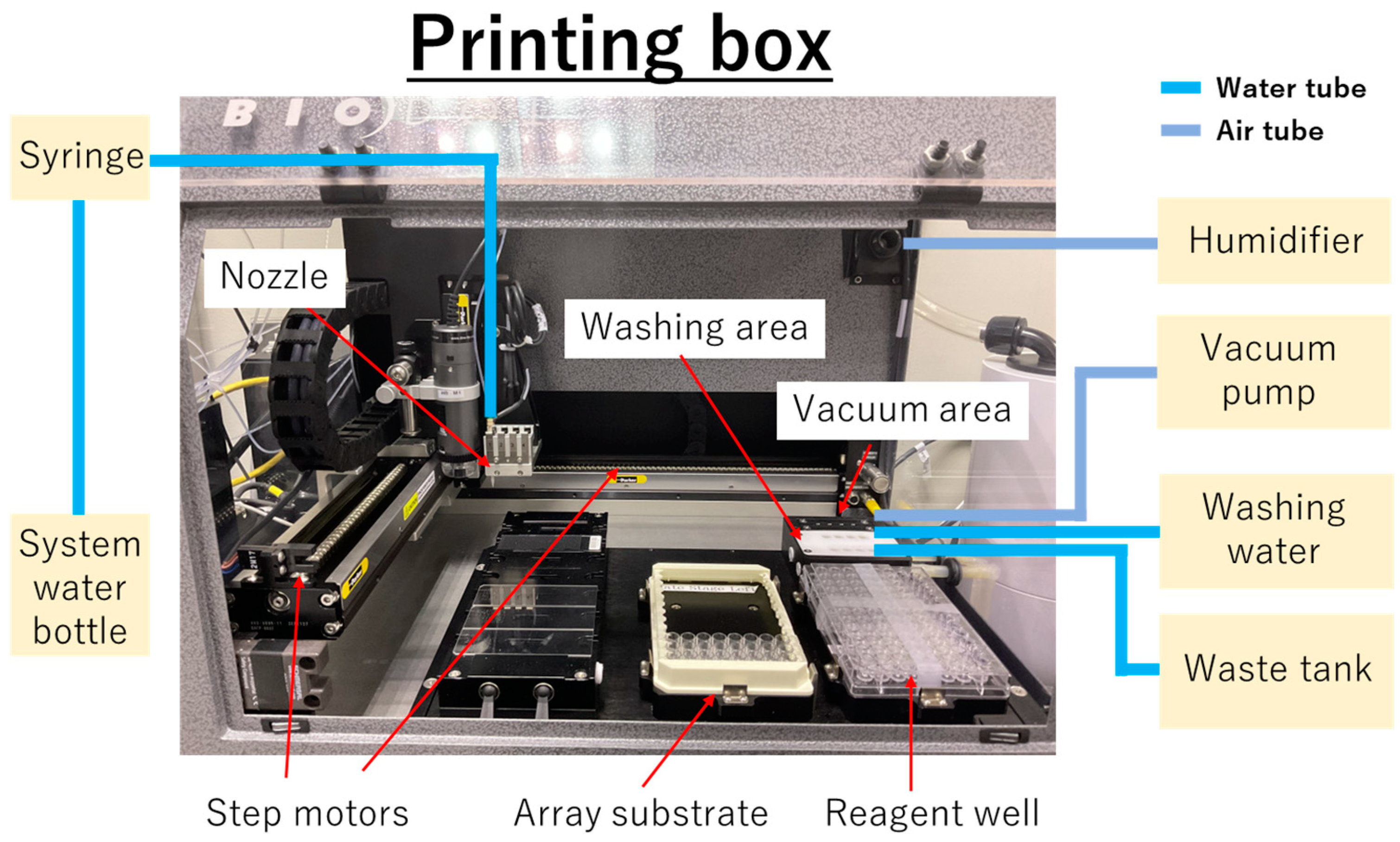
2.2. Instrumentation
2.3. Preparation of Target dsDNA from pEGFP-N1 and pVenus-N1 Plasmids
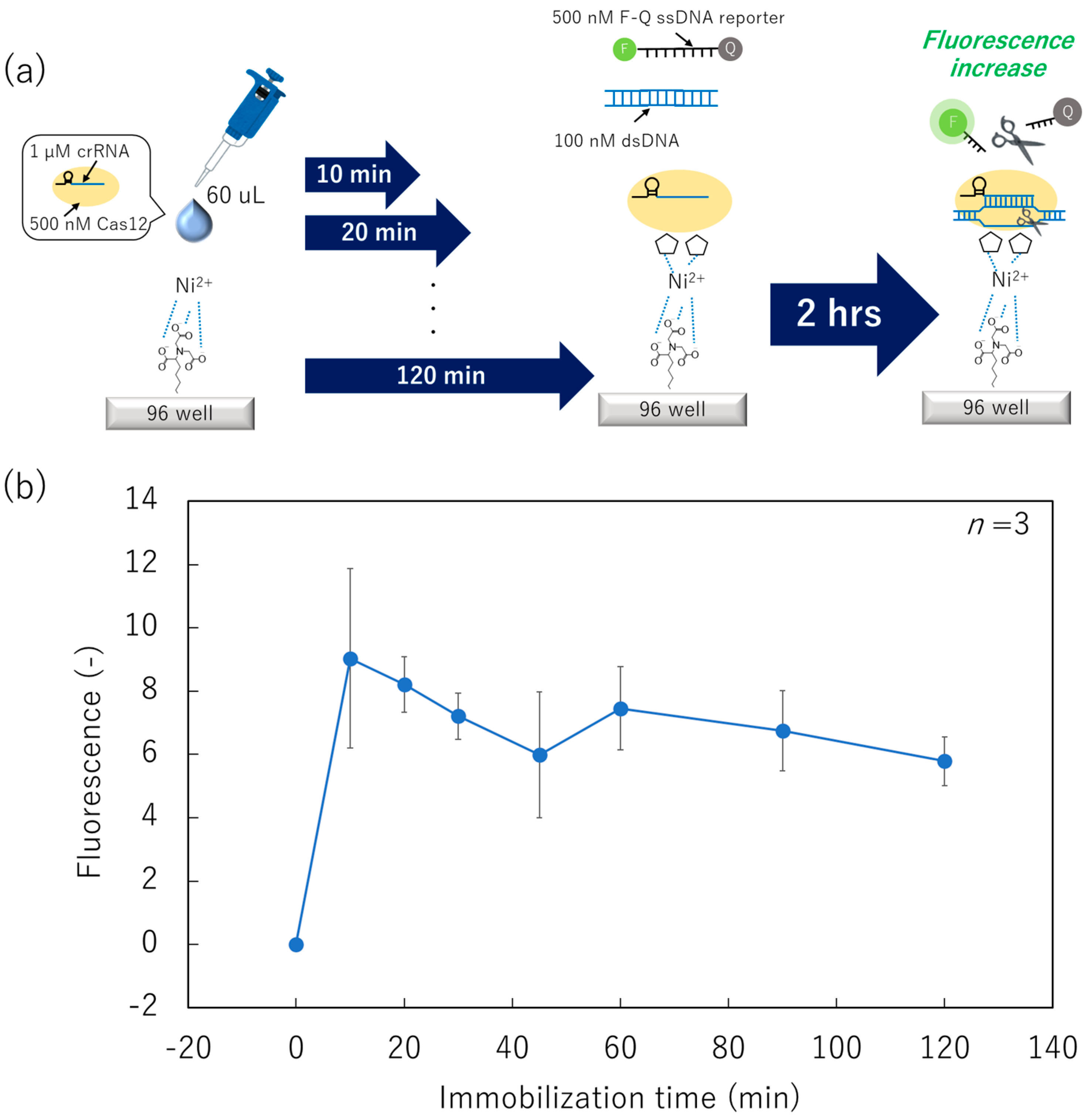
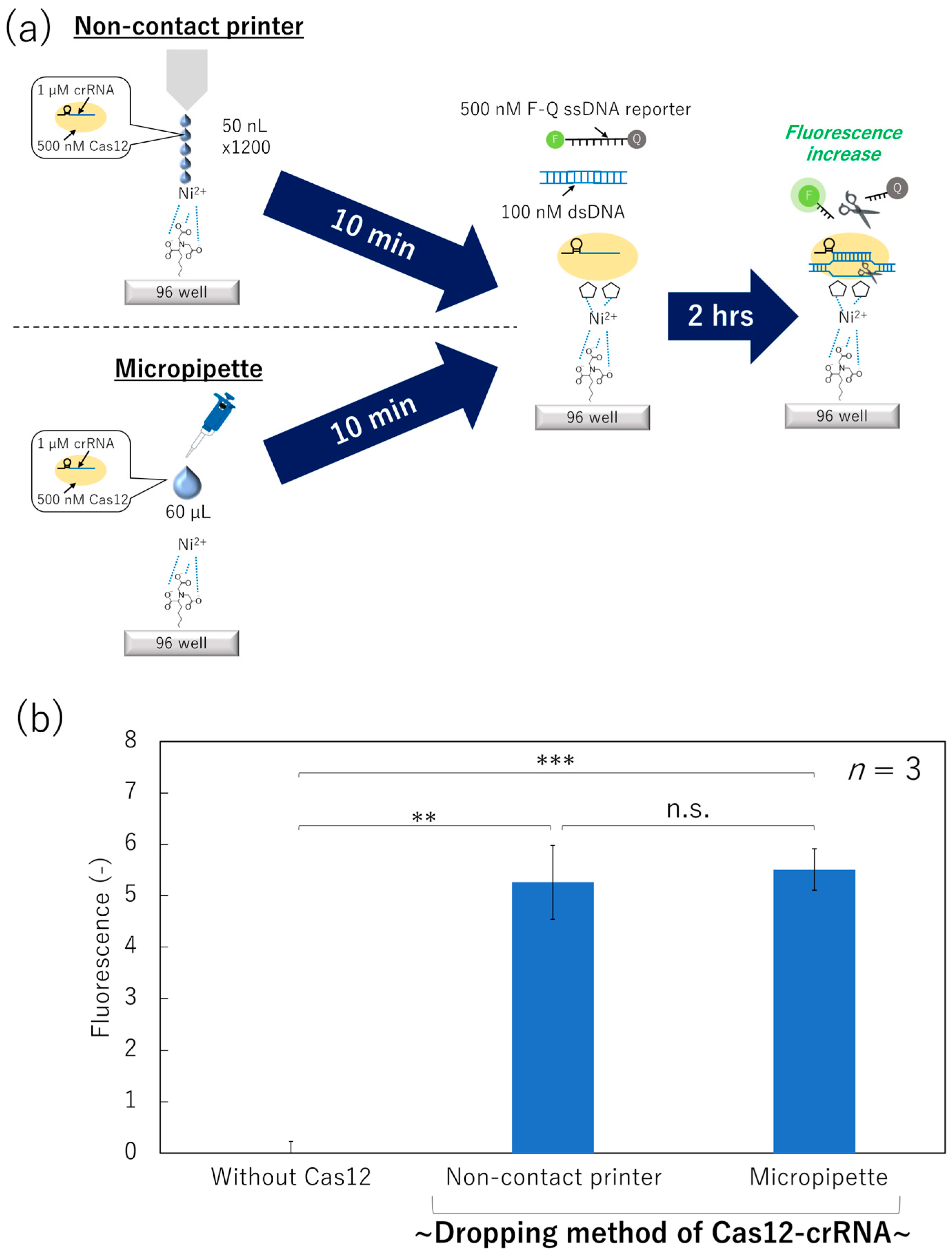
2.4. Immobilization Process of the ssDNA Reporter and the Cas12–crRNA on the Bottom Surface of the 96-Well ELISA Plate
2.5. Non-Contact Printing of the Cas12–crRNA
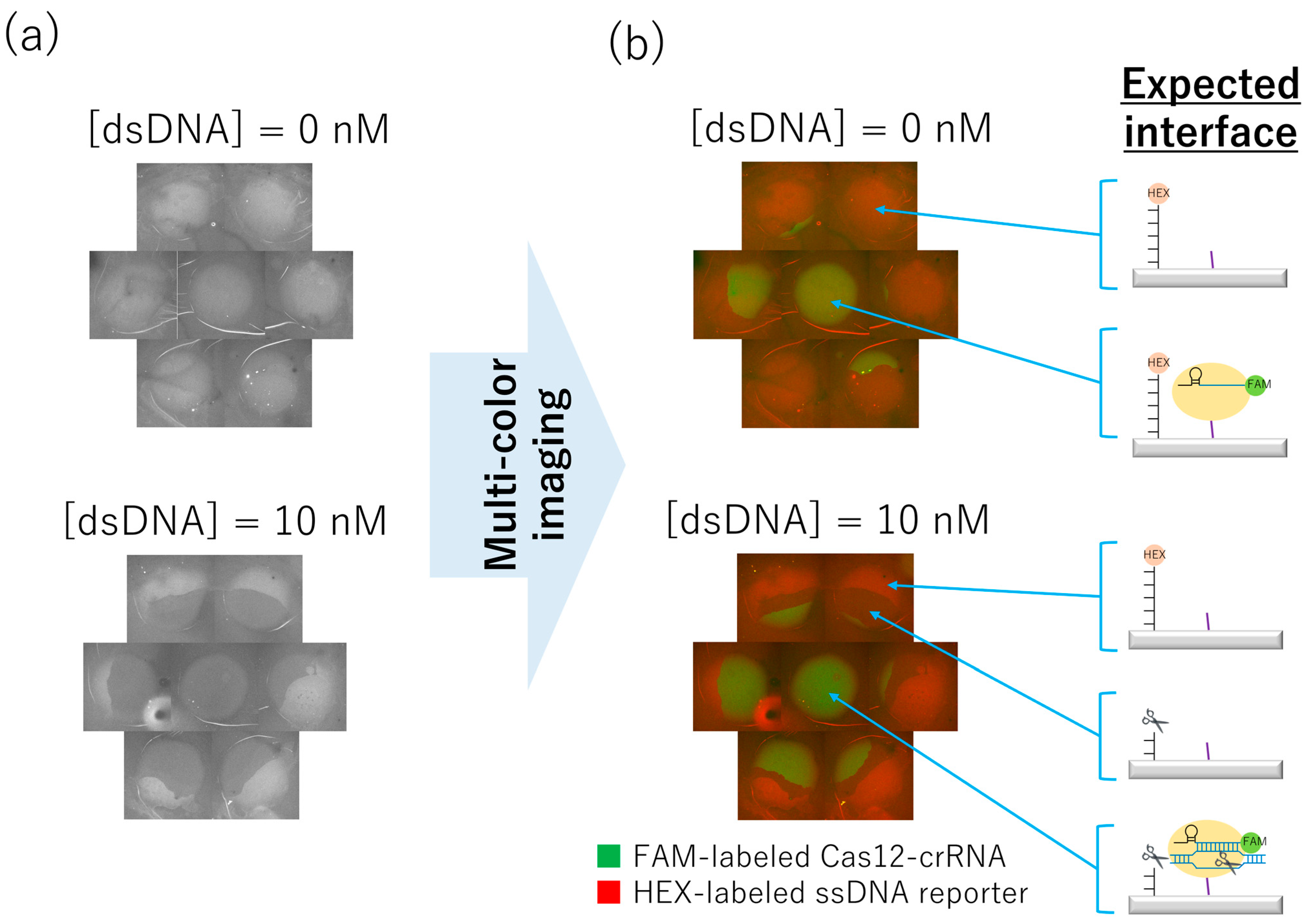
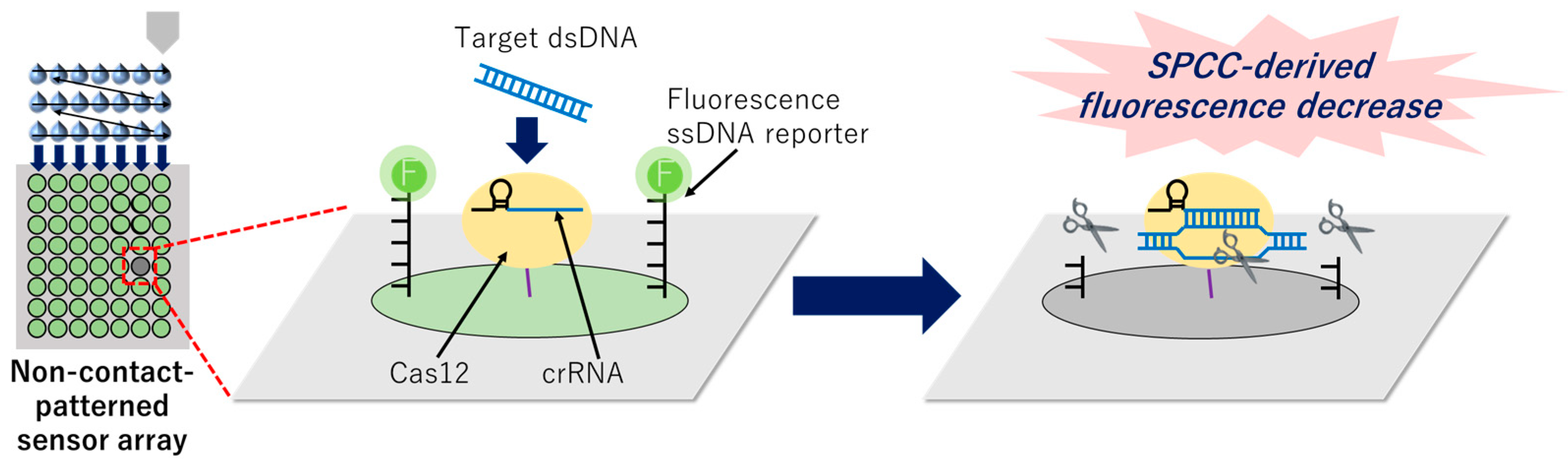
2.6. Incubation of Target dsDNA on the Non-Contact-Patterned SPCC-Based Sensor Array and Capturing Fluorescence Images
2.7. Collateral Cleavage Activity Test of Surface-Immobilized Cas12–crRNA
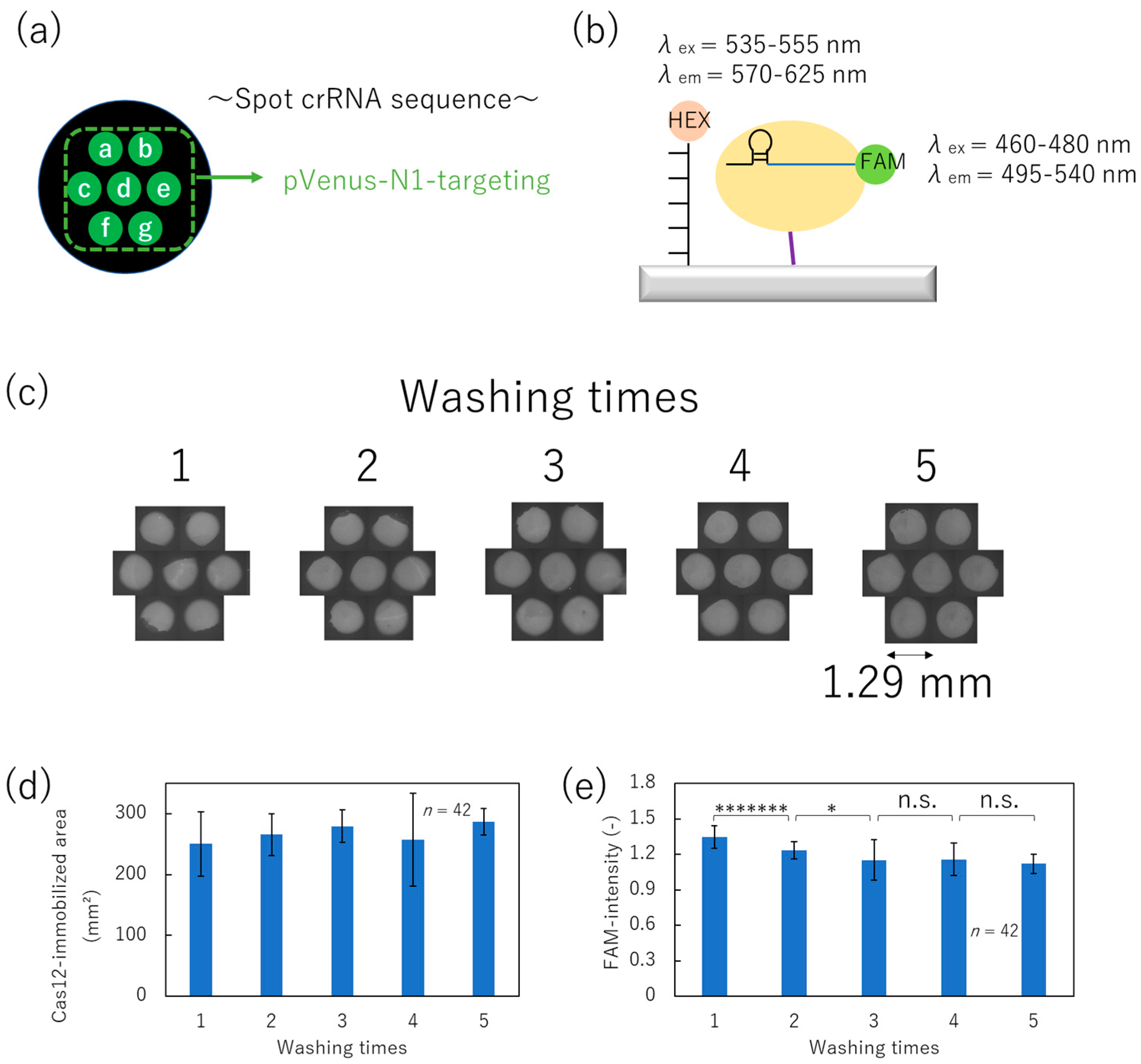
2.8. Calculating the Area of the Cas12-Immobilized Region and Its FAM-Fluorescence Intensity
3. Results
3.1. Minimization of Surface-Immobilization Time of Cas12–crRNA Complexes
3.2. Evaluation of Shear Stress from the Non-Contact Printing Nozzle against Surface-Immobilized Cas12–crRNA
3.3. Optimization of the Washing Process for Immobilization of Cas12–crRNA Complex by Non-Contact Printing
3.4. Miniaturization of the Non-Contact-Patterned SPCC-Based Sensor Array
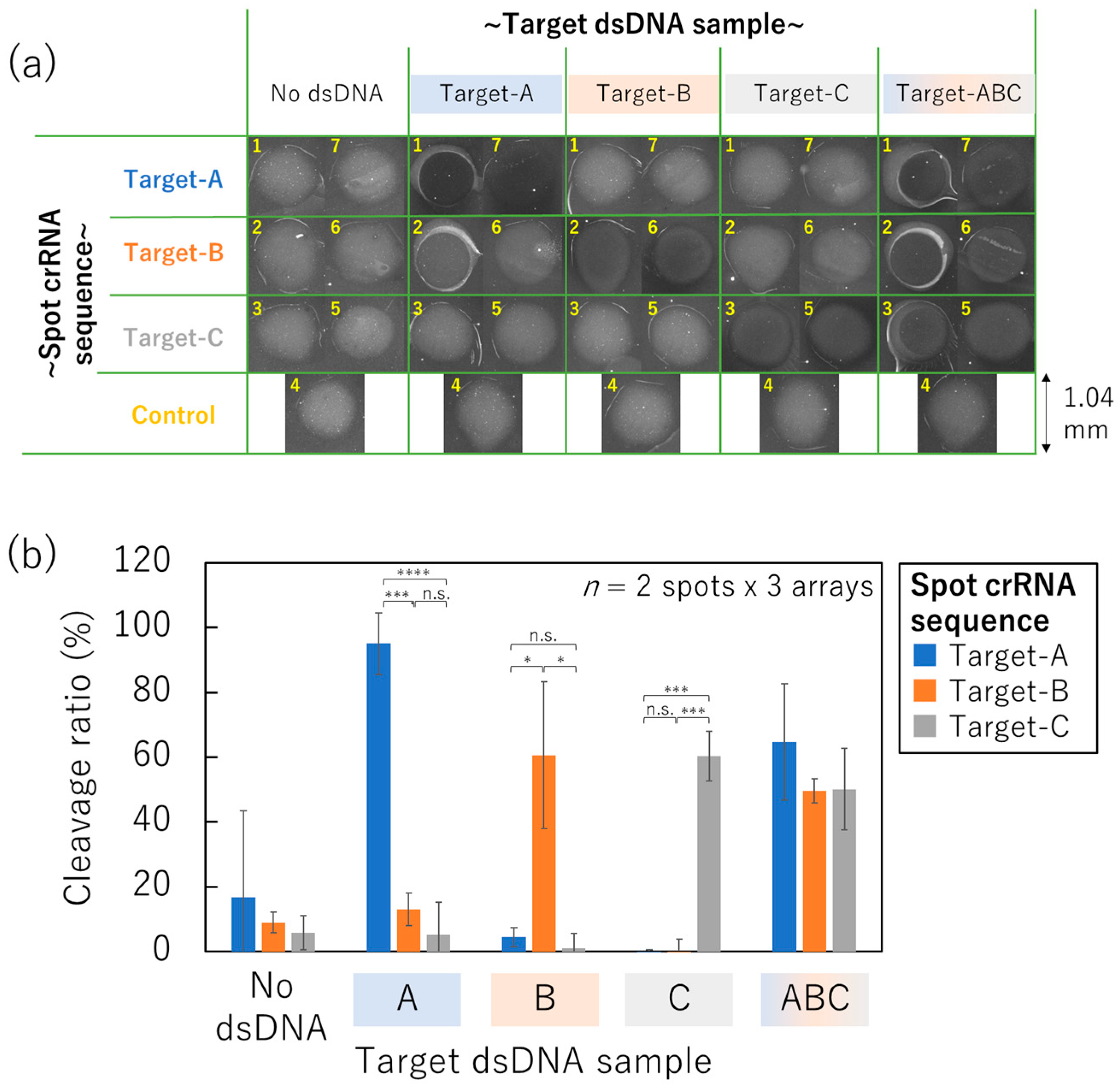
3.5. SPCC-Based Triple-Target dsDNA Detection on the Optimized Sensor Array
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Effect of the Physical Stress on the Surface-Immobilized Cas12-crRNA
References
- Japan: National Action Plan on Antimicrobial Resistance (AMR). Available online: https://www.who.int/publications/m/item/japan-national-action-plan-on-antimicrobial-resistance-(amr) (accessed on 29 November 2023).
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of Hypotension before Initiation of Effective Antimicrobial Therapy Is the Critical Determinant of Survival in Human Septic Shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Peker, N.; Couto, N.; Sinha, B.; Rossen, J.W. Diagnosis of Bloodstream Infections from Positive Blood Cultures and Directly from Blood Samples: Recent Developments in Molecular Approaches. Clin. Microbiol. Infect. 2018, 24, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Bender, A.T.; Boyle, D.S.; Drain, P.K.; Posner, J.D. Current State of Commercial Point-of-Care Nucleic Acid Tests for Infectious Diseases. Analyst 2021, 146, 2449–2462. [Google Scholar] [CrossRef] [PubMed]
- Furutani, S.; Naruishi, N.; Hagihara, Y.; Nagai, H. Development of an On-Site Rapid Real-Time Polymerase Chain Reaction System and the Characterization of Suitable DNA Polymerases for TaqMan Probe Technology. Anal. Bioanal. Chem. 2016, 408, 5641–5649. [Google Scholar] [CrossRef] [PubMed]
- Tschäpe, J.; Cobernuss-Rahn, A.; Boyle, S.; Parkin, N.; LaBrot, B.; Aslam, S.; Young, S.; Gohl, P. Multisite Performance Evaluation of the Cobas 5800 System and Comparison to the Cobas 6800/8800 Systems for Quantitative Measurement of HBV, HCV, and HIV-1 Viral Load. Microbiol. Spectr. 2022, 10, e03125-22. [Google Scholar] [CrossRef] [PubMed]
- Gaieski, D.F.; Mikkelsen, M.E.; Band, R.A.; Pines, J.M.; Massone, R.; Furia, F.F.; Shofer, F.S.; Goyal, M. Impact of Time to Antibiotics on Survival in Patients with Severe Sepsis or Septic Shock in Whom Early Goal-Directed Therapy Was Initiated in the Emergency Department. Crit. Care Med. 2010, 38, 1045. [Google Scholar] [CrossRef]
- Church, D.L. Principles of Capillary-Based Sequencing for Clinical Microbiologists. Clin. Microbiol. Newsl. 2013, 35, 11–18. [Google Scholar] [CrossRef]
- Ventimiglia, G.; Petralia, S. Recent Advances in DNA Microarray Technology: An Overview on Production Strategies and Detection Methods. BioNanoScience 2013, 3, 428–450. [Google Scholar] [CrossRef]
- Wojewoda, C.M.; Sercia, L.; Navas, M.; Tuohy, M.; Wilson, D.; Hall, G.S.; Procop, G.W.; Richter, S.S. Evaluation of the Verigene Gram-Positive Blood Culture Nucleic Acid Test for Rapid Detection of Bacteria and Resistance Determinants. J. Clin. Microbiol. 2013, 51, 2072–2076. [Google Scholar] [CrossRef]
- Ledeboer, N.A.; Lopansri, B.K.; Dhiman, N.; Cavagnolo, R.; Carroll, K.C.; Granato, P.; Thomson, R.; Butler-Wu, S.M.; Berger, H.; Samuel, L.; et al. Identification of Gram-Negative Bacteria and Genetic Resistance Determinants from Positive Blood Culture Broths by Use of the Verigene Gram-Negative Blood Culture Multiplex Microarray-Based Molecular Assay. J. Clin. Microbiol. 2015, 53, 2460–2472. [Google Scholar] [CrossRef]
- Suzuki, H.; Hitomi, S.; Yaguchi, Y.; Tamai, K.; Ueda, A.; Kamata, K.; Tokuda, Y.; Koganemaru, H.; Kurihara, Y.; Ishikawa, H.; et al. Prospective Intervention Study with a Microarray-Based, Multiplexed, Automated Molecular Diagnosis Instrument (Verigene System) for the Rapid Diagnosis of Bloodstream Infections, and Its Impact on the Clinical Outcomes. J. Infect. Chemother. 2015, 21, 849–856. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; White, C.M.; Meeder, J.; Moates, D.; Pierce, H.M.; Edwards, W.S.; Hutchinson, J.; Lee, R.A.; Leal, S.M. Analytical Performance and Potential Clinical Utility of the GenMark Dx ePlex® Blood Culture Identification Gram-Positive Panel. Diagn. Microbiol. Infect. Dis. 2022, 104, 115762. [Google Scholar] [CrossRef] [PubMed]
- Bryant, S.; Almahmoud, I.; Pierre, I.; Bardet, J.; Touati, S.; Maubon, D.; Cornet, M.; Richarme, C.; Maurin, M.; Pavese, P.; et al. Evaluation of Microbiological Performance and the Potential Clinical Impact of the ePlex® Blood Culture Identification Panels for the Rapid Diagnosis of Bacteremia and Fungemia. Front. Cell. Infect. Microbiol. 2020, 10, 594951. [Google Scholar] [CrossRef]
- Babady, N.E.; England, M.R.; Jurcic Smith, K.L.; He, T.; Wijetunge, D.S.; Tang, Y.-W.; Chamberland, R.R.; Menegus, M.; Swierkosz, E.M.; Jerris, R.C.; et al. Multicenter Evaluation of the ePlex Respiratory Pathogen Panel for the Detection of Viral and Bacterial Respiratory Tract Pathogens in Nasopharyngeal Swabs. J. Clin. Microbiol. 2018, 56, e01658-17. [Google Scholar] [CrossRef] [PubMed]
- Lipshutz, R.J.; Fodor, S.P.A.; Gingeras, T.R.; Lockhart, D.J. High Density Synthetic Oligonucleotide Arrays. Nat. Genet. 1999, 21, 20–24. [Google Scholar] [CrossRef]
- Gao, H.; Tao, S.; Wang, D.; Zhang, C.; Ma, X.; Cheng, J.; Zhou, Y. Comparison of Different Methods for Preparing Single Stranded DNA for Oligonucleotide Microarray. Anal. Lett. 2003, 36, 2849–2863. [Google Scholar] [CrossRef]
- Links to WHO TPPs and PPCs. Available online: https://www.who.int/observatories/global-observatory-on-health-research-and-development/analyses-and-syntheses/target-product-profile/links-to-who-tpps-and-ppcs (accessed on 29 September 2023).
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.E.; Berendsen, J.T.W.; Steenbergen, R.D.M.; Wolthuis, R.M.F.; Eijkel, J.C.T.; Segerink, L.I. Point-of-Care CRISPR/Cas Nucleic Acid Detection: Recent Advances, Challenges and Opportunities. Biosens. Bioelectron. 2020, 166, 112445. [Google Scholar] [CrossRef]
- de Dieu Habimana, J.; Huang, R.; Muhoza, B.; Kalisa, Y.N.; Han, X.; Deng, W.; Li, Z. Mechanistic Insights of CRISPR/Cas Nucleases for Programmable Targeting and Early-Stage Diagnosis: A Review. Biosens. Bioelectron. 2022, 203, 114033. [Google Scholar] [CrossRef]
- Wu, H.; Chen, X.; Zhang, M.; Wang, X.; Chen, Y.; Qian, C.; Wu, J.; Xu, J. Versatile Detection with CRISPR/Cas System from Applications to Challenges. TrAC Trends Anal. Chem. 2021, 135, 116150. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Nouri, R.; Dong, M.; Politza, A.J.; Guan, W. Figure of Merit for CRISPR-Based Nucleic Acid-Sensing Systems: Improvement Strategies and Performance Comparison. ACS Sens. 2022, 7, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, L.; Liu, G. CRISPR/Cas Multiplexed Biosensing: A Challenge or an Insurmountable Obstacle? Trends Biotechnol. 2019, 37, 792–795. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively Multiplexed Nucleic Acid Detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Welch, N.L.; Zhu, M.; Hua, C.; Weller, J.; Mirhashemi, M.E.; Nguyen, T.G.; Mantena, S.; Bauer, M.R.; Shaw, B.M.; Ackerman, C.M.; et al. Multiplexed CRISPR-Based Microfluidic Platform for Clinical Testing of Respiratory Viruses and Identification of SARS-CoV-2 Variants. Nat. Med. 2022, 28, 1083–1094. [Google Scholar] [CrossRef]
- Roh, Y.H.; Lee, C.Y.; Lee, S.; Kim, H.; Ly, A.; Castro, C.M.; Cheon, J.; Lee, J.; Lee, H. CRISPR-Enhanced Hydrogel Microparticles for Multiplexed Detection of Nucleic Acids. Adv. Sci. 2023, 10, 2206872. [Google Scholar] [CrossRef]
- Shigemori, H.; Fujita, S.; Tamiya, E.; Wakida, S.; Nagai, H. Solid-Phase Collateral Cleavage System Based on CRISPR/Cas12 and Its Application toward Facile One-Pot Multiplex Double-Stranded DNA Detection. Bioconjugate Chem. 2023, 34, 1754–1765. [Google Scholar] [CrossRef]
- Xing, G.; Shang, Y.; Wang, X.; Lin, H.; Chen, S.; Pu, Q.; Lin, L. Multiplexed Detection of Foodborne Pathogens Using One-Pot CRISPR/Cas12a Combined with Recombinase Aided Amplification on a Finger-Actuated Microfluidic Biosensor. Biosens. Bioelectron. 2023, 220, 114885. [Google Scholar] [CrossRef]
- Chen, Y.; Mei, Y.; Jiang, X. Universal and High-Fidelity DNA Single Nucleotide Polymorphism Detection Based on a CRISPR/Cas12a Biochip. Chem. Sci. 2021, 12, 4455–4462. [Google Scholar] [CrossRef]
- Zong, N.; Gao, Y.; Chen, Y.; Luo, X.; Jiang, X. Automated Centrifugal Microfluidic Chip Integrating Pretreatment and Molecular Diagnosis for Hepatitis B Virus Genotyping from Whole Blood. Anal. Chem. 2022, 94, 5196–5203. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, D.; Li, T.; Yan, J.; Zhu, J.; He, T.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Microfluidic Space Coding for Multiplexed Nucleic Acid Detection via CRISPR-Cas12a and Recombinase Polymerase Amplification. Nat. Commun. 2022, 13, 6480. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Ding, X.; Li, Z.; Sfeir, M.M.; Ballesteros, E.; Liu, C. Autonomous Lab-on-Paper for Multiplexed, CRISPR-Based Diagnostics of SARS-CoV-2. Lab Chip 2021, 21, 2730–2737. [Google Scholar] [CrossRef]
- Cao, H.; Mao, K.; Ran, F.; Xu, P.; Zhao, Y.; Zhang, X.; Zhou, H.; Yang, Z.; Zhang, H.; Jiang, G. Paper Device Combining CRISPR/Cas12a and Reverse-Transcription Loop-Mediated Isothermal Amplification for SARS-CoV-2 Detection in Wastewater. Environ. Sci. Technol. 2022, 56, 13245–13253. [Google Scholar] [CrossRef] [PubMed]
- Pena, J.M.; Manning, B.J.; Li, X.; Fiore, E.S.; Carlson, L.; Shytle, K.; Nguyen, P.P.; Azmi, I.; Larsen, A.; Wilson, M.K.; et al. Real-Time, Multiplexed SHERLOCK for in Vitro Diagnostics. J. Mol. Diagn. 2023, 25, 428–437. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Budach, W.; Abel, A.P.; Bruno, A.E.; Neuschäfer, D. Planar Waveguides as High-Performance Sensing Platforms for Fluorescence-Based Multiplexed Oligonucleotide Hybridization Assays. Anal. Chem. 1999, 71, 3347–3355. [Google Scholar] [CrossRef]
- Okamoto, T.; Suzuki, T.; Yamamoto, N. Microarray Fabrication with Covalent Attachment of DNA Using Bubble Jet Technology. Nat. Biotechnol. 2000, 18, 438–441. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, H.; Maejima, K.; Shibata, H.; Hiruta, Y.; Citterio, D. Evaluation of Cellophane as Platform for Colorimetric Assays on Microfluidic Analytical Devices. Microchim. Acta 2023, 190, 48. [Google Scholar] [CrossRef]
- Li, J.; Rossignol, F.; Macdonald, J. Inkjet Printing for Biosensor Fabrication: Combining Chemistry and Technology for Advanced Manufacturing. Lab Chip 2015, 15, 2538–2558. [Google Scholar] [CrossRef] [PubMed]
- Setti, L.; Piana, C.; Bonazzi, S.; Ballarin, B.; Frascaro, D.; Fraleoni-Morgera, A.; Giuliani, S. Thermal Inkjet Technology for the Microdeposition of Biological Molecules as a Viable Route for the Realization of Biosensors. Anal. Lett. 2004, 37, 1559–1570. [Google Scholar] [CrossRef]
- Derby, B. Bioprinting: Inkjet Printing Proteins and Hybrid Cell-Containing Materials and Structures. J. Mater. Chem. 2008, 18, 5717–5721. [Google Scholar] [CrossRef]
- Hu, H.; Larson, R.G. Evaporation of a Sessile Droplet on a Substrate. J. Phys. Chem. B 2002, 106, 1334–1344. [Google Scholar] [CrossRef]
- Deegan, R.D.; Bakajin, O.; Dupont, T.F.; Huber, G.; Nagel, S.R.; Witten, T.A. Capillary Flow as the Cause of Ring Stains from Dried Liquid Drops. Nature 1997, 389, 827–829. [Google Scholar] [CrossRef]
- Clancy, K.F.A.; Dery, S.; Laforte, V.; Shetty, P.; Juncker, D.; Nicolau, D.V. Protein Microarray Spots Are Modulated by Patterning Method, Surface Chemistry and Processing Conditions. Biosens. Bioelectron. 2019, 130, 397–407. [Google Scholar] [CrossRef]
- Guo, W.; Vilaplana, L.; Hansson, J.; Marco, M.-P.; van der Wijngaart, W. The Influence of Substrate Microstructures on the Fluorescent Intensity Profile, Size, Roundness, and Coffee Ring Ratio of Protein Microarray Spots. In Proceedings of the 2022 IEEE 17th International Conference on Nano/Micro Engineered and Molecular Systems (NEMS), Online, 14–17 April 2022; pp. 201–204. [Google Scholar]
- Liu, J.; Tiefenauer, L.; Tian, S.; Nielsen, P.E.; Knoll, W. PNA−DNA Hybridization Study Using Labeled Streptavidin by Voltammetry and Surface Plasmon Fluorescence Spectroscopy. Anal. Chem. 2006, 78, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Mallon, J.; Poddar, A.; Wang, Y.; Tippana, R.; Yang, O.; Bailey, S.; Ha, T. Real-Time Observation of DNA Target Interrogation and Product Release by the RNA-Guided Endonuclease CRISPR Cpf1 (Cas12a). Proc. Natl. Acad. Sci. USA 2018, 115, 5444–5449. [Google Scholar] [CrossRef]
- Vanderhoeven, J.; Pappaert, K.; Dutta, B.; Van Hummelen, P.; Desmet, G. Comparison of a Pump-around, a Diffusion-Driven, and a Shear-Driven System for the Hybridization of Mouse Lung and Testis Total RNA on Microarrays. Electrophoresis 2005, 26, 3773–3779. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Smoot, J.; McMurray, Z.; Stahl, D.A.; Yager, P. Recirculating Flow Accelerates DNA Microarray Hybridization in a Microfluidic Device. Lab Chip 2006, 6, 1163–1170. [Google Scholar] [CrossRef]
- Noerholm, M.; Bruus, H.; Jakobsen, M.H.; Telleman, P.; Ramsing, N.B. Polymer Microfluidic Chip for Online Monitoring of Microarray Hybridizations. Lab Chip 2004, 4, 28–37. [Google Scholar] [CrossRef]
- Suea-Ngam, A.; Bezinge, L.; Mateescu, B.; Howes, P.D.; deMello, A.J.; Richards, D.A. Enzyme-Assisted Nucleic Acid Detection for Infectious Disease Diagnostics: Moving toward the Point-of-Care. ACS Sens. 2020, 5, 2701–2723. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shigemori, H.; Fujita, S.; Tamiya, E.; Nagai, H. Miniaturization of CRISPR/Cas12-Based DNA Sensor Array by Non-Contact Printing. Micromachines 2024, 15, 144. https://doi.org/10.3390/mi15010144
Shigemori H, Fujita S, Tamiya E, Nagai H. Miniaturization of CRISPR/Cas12-Based DNA Sensor Array by Non-Contact Printing. Micromachines. 2024; 15(1):144. https://doi.org/10.3390/mi15010144
Chicago/Turabian StyleShigemori, Hiroki, Satoshi Fujita, Eiichi Tamiya, and Hidenori Nagai. 2024. "Miniaturization of CRISPR/Cas12-Based DNA Sensor Array by Non-Contact Printing" Micromachines 15, no. 1: 144. https://doi.org/10.3390/mi15010144
APA StyleShigemori, H., Fujita, S., Tamiya, E., & Nagai, H. (2024). Miniaturization of CRISPR/Cas12-Based DNA Sensor Array by Non-Contact Printing. Micromachines, 15(1), 144. https://doi.org/10.3390/mi15010144






