Exploring the Structural Variability of Dynamic Biological Complexes by Single-Particle Cryo-Electron Microscopy
Abstract
1. Introduction
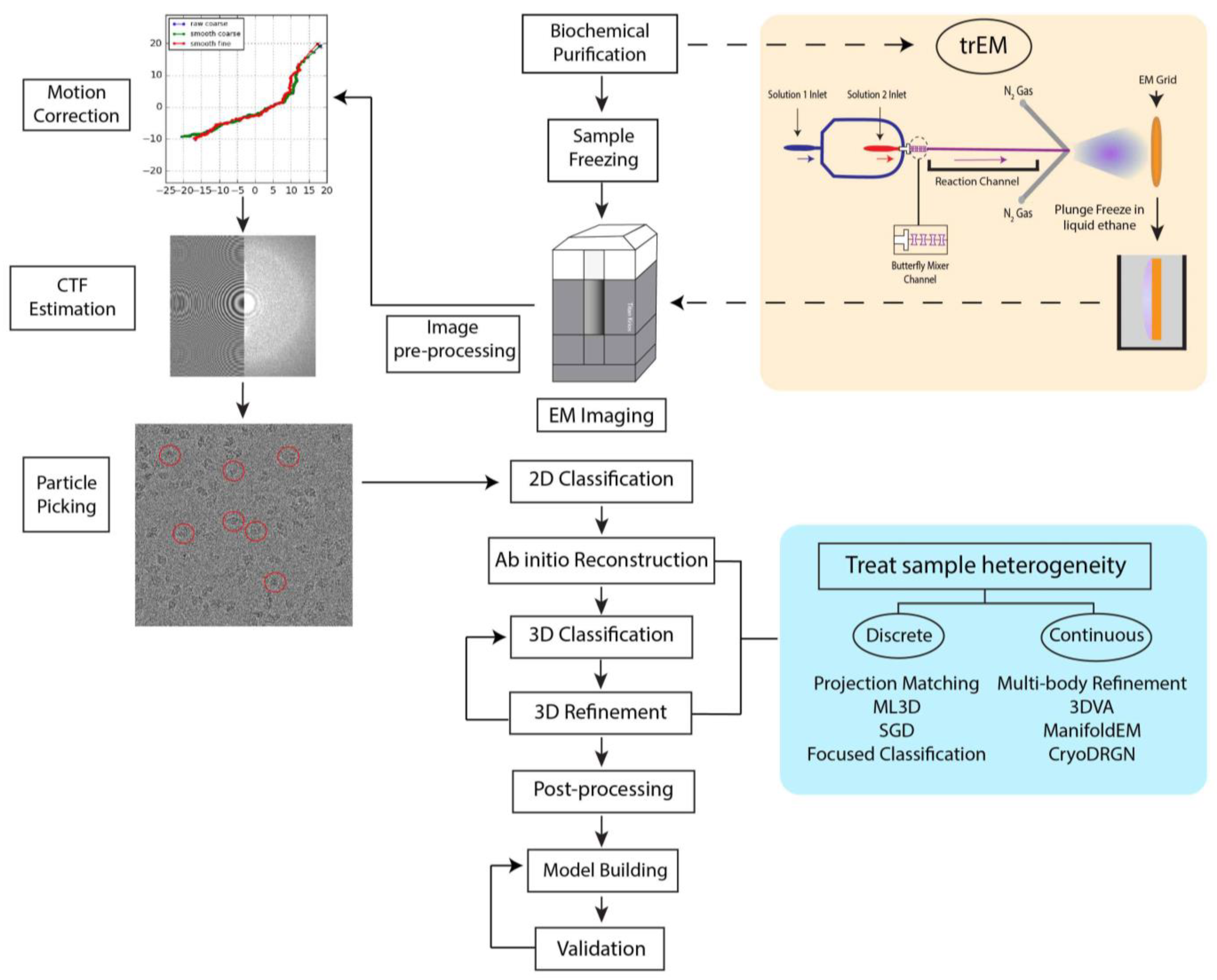
2. The Cryo-EM Workflow
3. Structural Heterogeneity in Cryo-EM Samples
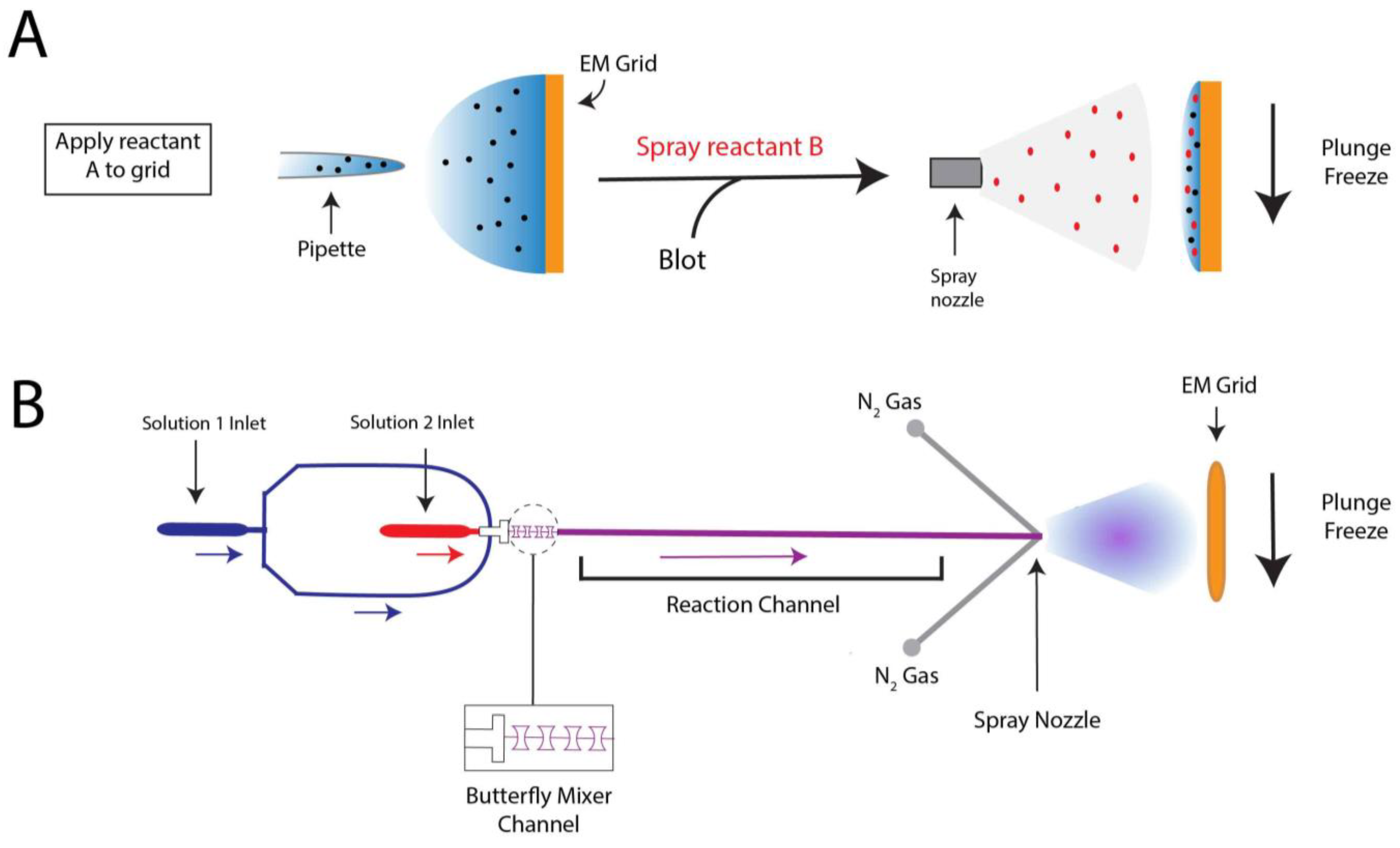
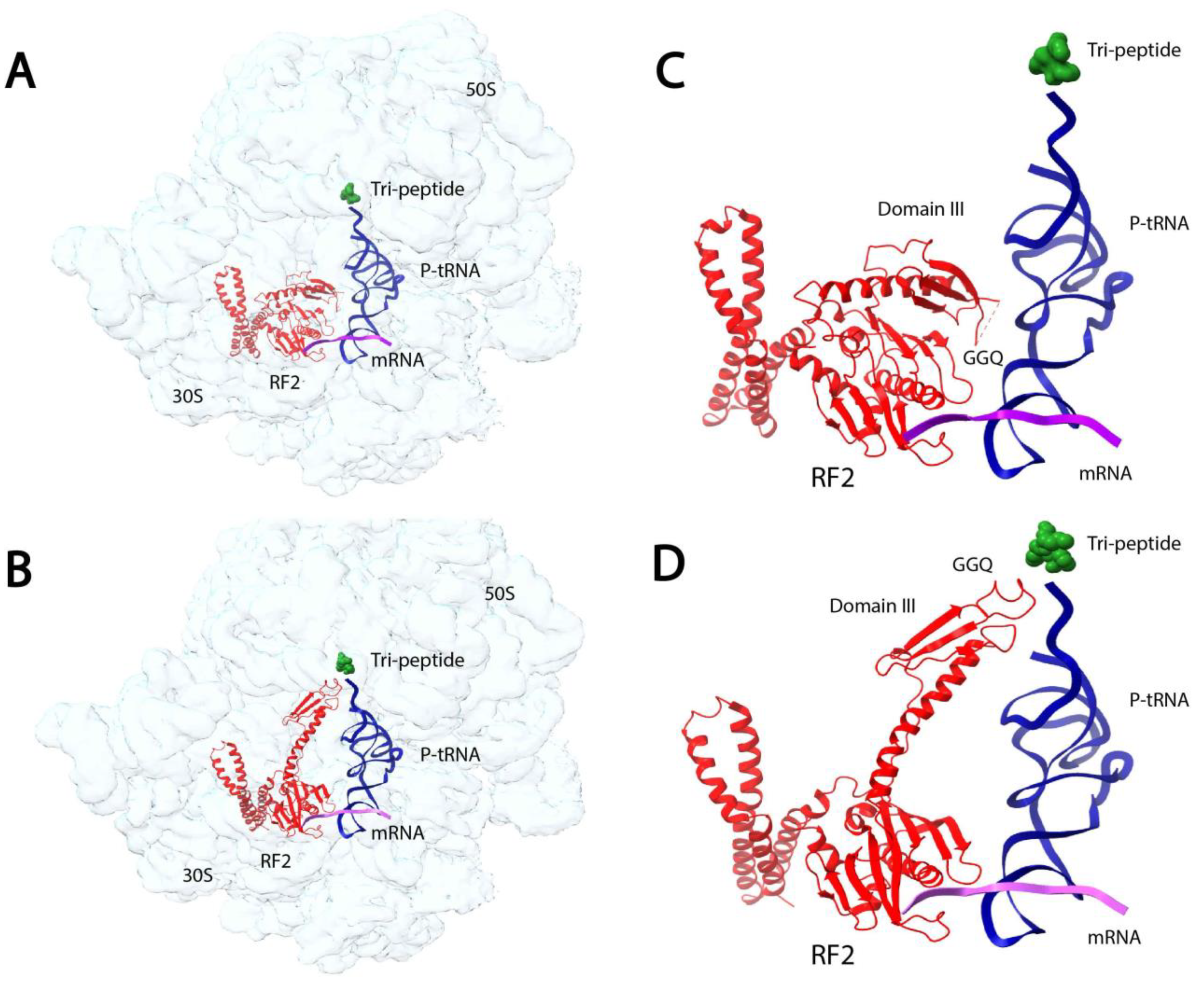
4. Probing Short-Lived Conformational Changes by Time-Resolved Cryo-EM
5. 3D Classification & Refinement: Approaches to Modeling Discrete Heterogeneity
6. Masking-Based Approaches to Resolve Discrete Structural States and Continuous Flexibility
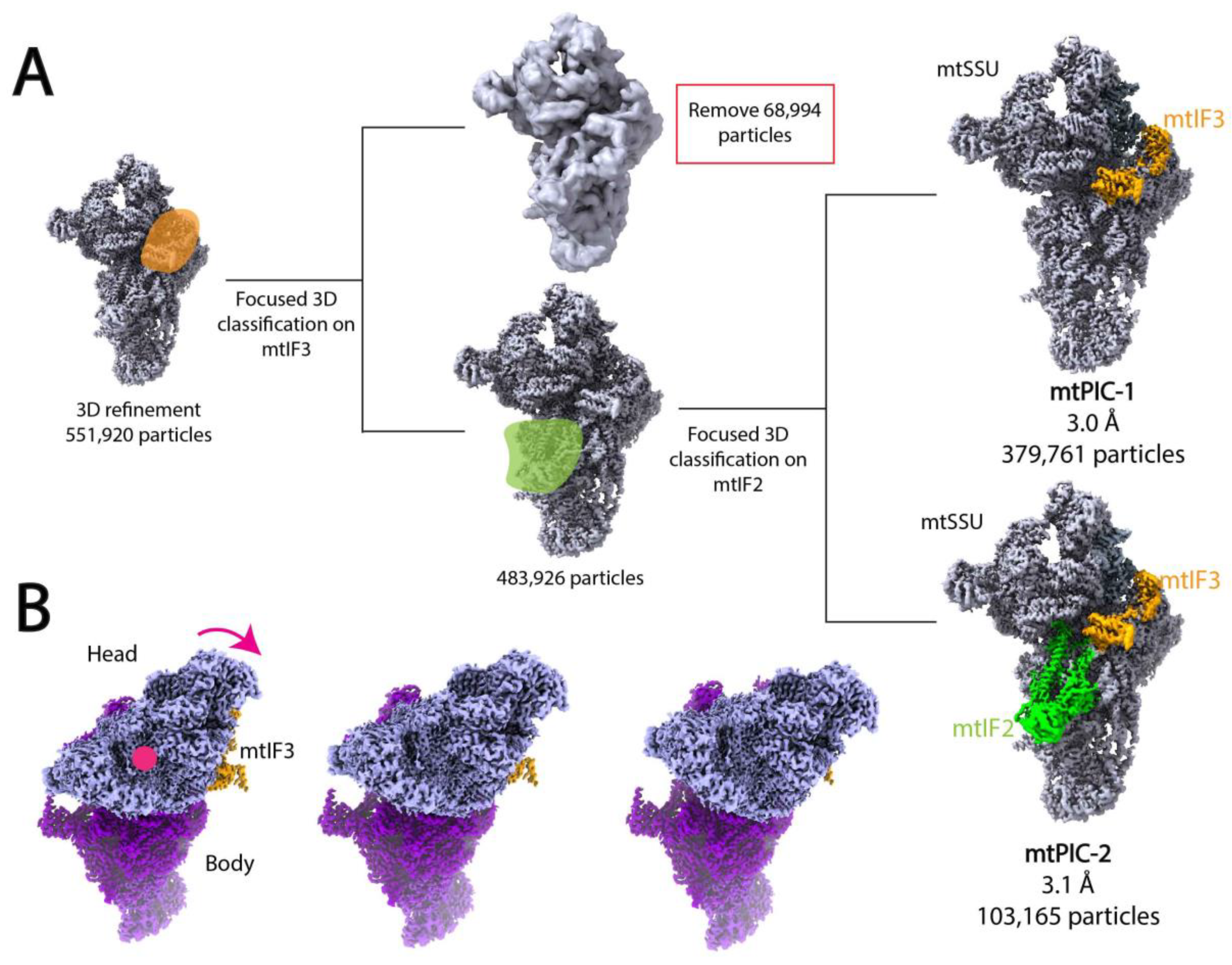
7. Focused Classification and Multi-Body Refinement of Ribosomal Complexes
8. Approaches to Modeling Continuous Heterogeneity
| Method | Advantages | Disadvantages | Reference |
|---|---|---|---|
| ManifoldEM | Generates free-energy landscape of the system | Fine tuning of hyperspace parameters | Frank & Ourmazd, 2016 [18] |
| AlphaCryo4D | Applicable to small proteins | Requires large dataset Oversamples conformational space | Wu et al., 2022 [162] |
| CryoDRGN | New version [159] does not require initial model or pose information Resolves discrete and continuous conformations | Long training time Empirical optimization of latent space | Zhong et al., 2021 [19] |
| CryoGAN | Does not require initial model or pose information Resolves discrete and continuous conformations | Limited resolution of reconstructions | Gupta et al., 2021 [154] |
| e2gmm | Reduces parameters needed to represent particles Intuitive interpretation by Gaussian parameters | Requires large amount of GPU memory Limited to small proteins for high resolution | Chen & Ludtke, 2021 [156] |
| 3DVA | Resolves discrete and continuous motion No fine-tuning of parameters Applicable to small proteins | Not applicable to systems with nonlinear geometry Artifact of appearing/disappearing densities | Punjani et al., 2021 [17] |
| 3DFlex | Models motion directly instead of 3D volume | Auto-decoder is computationally expensive | Punjani et al., 2022 [157] |
| Multi-body refinement | Automated implementation in RELION [44] Improves subdomain resolution | Interfaces between bodies poorly resolved Size limitation for densities < 150 kDa | Nakane et al., 2018 [15] |
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- RCSB PDB. PDB Statistics: Growth of Structures from X-ray Crystallography Experiments Released per Year. Available online: https://www.rcsb.org/stats/growth/growth-xray (accessed on 14 November 2022).
- Vedadi, M.; Niesen, F.H.; Allali-Hassani, A.; Fedorov, O.Y.; Finerty, P.J.; Wasney, G.A.; Yeung, R.; Arrowsmith, C.; Ball, L.J.; Berglund, H.; et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. USA 2006, 103, 15835–15840. [Google Scholar] [CrossRef] [PubMed]
- Dale, G.E.; Kostrewa, D.; Gsell, B.; Stieger, M.; D’Arcy, A. Crystal engineering: Deletion mutagenesis of the 24 kDa fragment of the DNA gyrase B subunit from Staphylococcus aureus. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 1626–1629. [Google Scholar] [CrossRef] [PubMed]
- Milburn, M.V.; Prive, G.G.; Milligan, D.L.; Scott, W.G.; Yeh, J.; Jancarik, J.; Koshland, D.E., Jr.; Kim, S.H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 1991, 254, 1342–1347. [Google Scholar] [CrossRef]
- Bai, X.C.; McMullan, G.; Scheres, S.H.W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015, 40, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Kuhlbrandt, W. The resolution revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- Earl, L.A.; Falconieri, V.; Milne, J.L.; Subramaniam, S. Cryo-EM: Beyond the microscope. Curr. Opin. Struct. Biol. 2017, 46, 71–78. [Google Scholar] [CrossRef]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.M.G.E.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef]
- Zhao, J.; Benlekbir, S.; Rubinstein, J.L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature 2015, 521, 241–245. [Google Scholar] [CrossRef]
- Fica, S.M.; Nagai, K. Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 2017, 24, 791–799. [Google Scholar] [CrossRef]
- Wong, W.; Bai, X.C.; Brown, A.; Fernandez, I.S.; Hanssen, E.; Condron, M.; Tan, Y.H.; Baum, J.; Scheres, S.H. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife 2014, 3, e03080. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. Classification of structural heterogeneity by maximum-likelihood methods. Methods Enzymol. 2010, 482, 295–320. [Google Scholar] [PubMed]
- Scheres, S.H. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016, 579, 125–157. [Google Scholar] [PubMed]
- Nakane, T.; Kimanius, D.; Lindahl, E.; Scheres, S.H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife 2018, 7, e36861. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Punjani, A.; Fleet, D.J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021, 213, 107702. [Google Scholar] [CrossRef]
- Frank, J.; Ourmazd, A. Continuous changes in structure mapped by manifold embedding of single-particle data in cryo-EM. Methods 2016, 100, 61–67. [Google Scholar] [CrossRef]
- Zhong, E.D.; Bepler, T.; Berger, B.; Davis, J.H. CryoDRGN: Reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods 2021, 18, 176–185. [Google Scholar] [CrossRef]
- Dubochet, J.; Lepault, J.; Freeman, R.; Berriman, J.A.; Homo, J.C. Electron-Microscopy of Frozen Water and Aqueous-Solutions. J. Microsc-Oxford 1982, 128, 219–237. [Google Scholar] [CrossRef]
- Frederik, P.M.; Storms, M.M. Automated, Robotic Preparation of Vitrified Samples for 2D and 3D Cryo Electron Microscopy. Microscopy Today 2005, 13, 32–39. [Google Scholar] [CrossRef][Green Version]
- Iancu, C.V.; Tivol, W.F.; Schooler, J.B.; Dias, D.P.; Henderson, G.P.; Murphy, G.E.; Wright, E.R.; Li, Z.; Yu, Z.H.; Briegel, A.; et al. Electron cryotomography sample preparation using the Vitrobot. Nat. Protoc. 2006, 1, 2813–2819. [Google Scholar] [CrossRef] [PubMed]
- Tivol, W.F.; Briegel, A.; Jensen, G.J. An improved cryogen for plunge freezing. Microsc. Microanal. 2008, 14, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Resch, G.P.; Brandstetter, M.; Konigsmaier, L.; Urban, E.; Pickl-Herk, A.M. Immersion freezing of suspended particles and cells for cryo-electron microscopy. Cold Spring Harb. Protoc. 2011, 2011, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.A.; Glaeser, R.M. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 2008, 163, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Fan, X.; Wang, H.; Zhao, F.; Tully, C.G.; Kong, J.; Yao, N.; Yan, N. High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 1009–1014. [Google Scholar] [CrossRef]
- Naydenova, K.; Peet, M.J.; Russo, C.J. Multifunctional graphene supports for electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 11718–11724. [Google Scholar] [CrossRef]
- Han, B.G.; Watson, Z.; Kang, H.; Pulk, A.; Downing, K.H.; Cate, J.; Glaeser, R.M. Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromolecules. J. Struct. Biol. 2016, 195, 238–244. [Google Scholar] [CrossRef]
- Llaguno, M.C.; Xu, H.; Shi, L.; Huang, N.; Zhang, H.; Liu, Q.; Jiang, Q.X. Chemically functionalized carbon films for single molecule imaging. J. Struct. Biol. 2014, 185, 405–417. [Google Scholar] [CrossRef]
- Yu, G.; Li, K.; Jiang, W. Antibody-based affinity cryo-EM grid. Methods 2016, 100, 16–24. [Google Scholar] [CrossRef]
- Feng, X.; Fu, Z.; Kaledhonkar, S.; Jia, Y.; Shah, B.; Jin, A.; Liu, Z.; Sun, M.; Chen, B.; Grassucci, R.A.; et al. A Fast and Effective Microfluidic Spraying-Plunging Method for High-Resolution Single-Particle Cryo-EM. Structure 2017, 25, 663–670. [Google Scholar] [CrossRef]
- Lu, Z.; Shaikh, T.R.; Barnard, D.; Meng, X.; Mohamed, H.; Yassin, A.; Mannella, C.A.; Agrawal, R.K.; Lu, T.M.; Wagenknecht, T. Monolithic microfluidic mixing-spraying devices for time-resolved cryo-electron microscopy. J. Struct. Biol. 2009, 168, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, R.B.G.; Nijpels, F.J.T.; Henderikx, R.J.M.; Weissenberger, G.; Thewessem, S.; Gijsbers, A.; Beulen, B.; Lopez-Iglesias, C.; Peters, P.J. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun. 2020, 11, 2563. [Google Scholar] [CrossRef] [PubMed]
- Jain, T.; Sheehan, P.; Crum, J.; Carragher, B.; Potter, C.S. Spotiton: A prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J. Struct. Biol. 2012, 179, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Brilot, A.F.; Chen, J.Z.; Cheng, A.; Pan, J.; Harrison, S.C.; Potter, C.S.; Carragher, B.; Henderson, R.; Grigorieff, N. Beam-induced motion of vitrified specimen on holey carbon film. J. Struct. Biol. 2012, 177, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Carragher, B.; Kisseberth, N.; Kriegman, D.; Milligan, R.A.; Potter, C.S.; Pulokas, J.; Reilein, A. Leginon: An automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 2000, 132, 33–45. [Google Scholar] [CrossRef]
- Mastronarde, D.N. Advanced Data Acquisition From Electron Microscopes With SerialEM. Microsc. Microanal. 2018, 24, 864–865. [Google Scholar] [CrossRef]
- Thompson, R.F.; Iadanza, M.G.; Hesketh, E.L.; Rawson, S.; Ranson, N.A. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 2019, 14, 100–118. [Google Scholar] [CrossRef]
- Zhang, J.; Nakamura, N.; Shimizu, Y.; Liang, N.; Liu, X.; Jakana, J.; Marsh, M.P.; Booth, C.R.; Shinkawa, T.; Nakata, M.; et al. JADAS: A customizable automated data acquisition system and its application to ice-embedded single particles. J. Struct. Biol. 2009, 165, 1–9. [Google Scholar] [CrossRef]
- Baldwin, P.R.; Tan, Y.Z.; Eng, E.T.; Rice, W.J.; Noble, A.J.; Negro, C.J.; Cianfrocco, M.A.; Potter, C.S.; Carragher, B. Big data in cryoEM: Automated collection, processing and accessibility of EM data. Curr. Opin. Microbiol. 2018, 43, 1–8. [Google Scholar] [CrossRef]
- de la Rosa-Trevin, J.M.; Oton, J.; Marabini, R.; Zaldivar, A.; Vargas, J.; Carazo, J.M.; Sorzano, C.O. Xmipp 3.0: An improved software suite for image processing in electron microscopy. J. Struct. Biol. 2013, 184, 321–328. [Google Scholar] [CrossRef]
- de la Rosa-Trevin, J.M.; Quintana, A.; Del Cano, L.; Zaldivar, A.; Foche, I.; Gutierrez, J.; Gomez-Blanco, J.; Burguet-Castell, J.; Cuenca-Alba, J.; Abrishami, V.; et al. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 2016, 195, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Grant, T.; Rohou, A.; Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. Elife 2018, 7, e35383. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, T.R.; Gao, H.; Baxter, W.T.; Asturias, F.J.; Boisset, N.; Leith, A.; Frank, J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 2008, 3, 1941–1974. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef]
- van Heel, M.; Harauz, G.; Orlova, E.V.; Schmidt, R.; Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996, 116, 17–24. [Google Scholar] [CrossRef]
- DiIorio, M.C.; Kulczyk, A.W. A Robust Single-Particle Cryo-Electron Microscopy (cryo-EM) Processing Workflow with cryoSPARC, RELION, and Scipion. J. Vis. Exp. 2022, 179, e63387. [Google Scholar] [CrossRef]
- Scheres, S.H. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 2015, 189, 114–122. [Google Scholar] [CrossRef]
- Henderson, R. Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proc. Natl. Acad. Sci. USA 2013, 110, 18037–18041. [Google Scholar] [CrossRef]
- Bepler, T.; Morin, A.; Rapp, M.; Brasch, J.; Shapiro, L.; Noble, A.J.; Berger, B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 2019, 16, 1153–1160. [Google Scholar] [CrossRef]
- Zhu, Y.; Ouyang, Q.; Mao, Y. A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy. BMC Bioinform. 2017, 18, 348. [Google Scholar] [CrossRef]
- Wagner, T.; Merino, F.; Stabrin, M.; Moriya, T.; Antoni, C.; Apelbaum, A.; Hagel, P.; Sitsel, O.; Raisch, T.; Prumbaum, D.; et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019, 2, 218. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gong, H.; Liu, G.; Li, M.; Yan, C.; Xia, T.; Li, X.; Zeng, J. DeepPicker: A deep learning approach for fully automated particle picking in cryo-EM. J. Struct. Biol. 2016, 195, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Tegunov, D.; Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 2019, 16, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Penczek, P.A.; Grassucci, R.A.; Frank, J. The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy 1994, 53, 251–270. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.W.; Gao, H.X.; Valle, M.; Herman, G.T.; Eggermont, P.P.B.; Frank, J.; Carazo, J.M. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods 2007, 4, 27–29. [Google Scholar] [CrossRef]
- Bai, X.C.; Rajendra, E.; Yang, G.; Shi, Y.; Scheres, S.H. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife 2015, 4, e11182. [Google Scholar] [CrossRef]
- Kulczyk, A.W.; Moeller, A.; Meyer, P.; Sliz, P.; Richardson, C.C. Cryo-EM structure of the replisome reveals multiple interactions coordinating DNA synthesis. Proc. Natl. Acad. Sci. USA 2017, 114, E1848–E1856. [Google Scholar] [CrossRef]
- Stark, H. GraFix: Stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010, 481, 109–126. [Google Scholar]
- Murakami, K.; Elmlund, H.; Kalisman, N.; Bushnell, D.A.; Adams, C.M.; Azubel, M.; Elmlund, D.; Levi-Kalisman, Y.; Liu, X.; Gibbons, B.J.; et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 2013, 342, 1238724. [Google Scholar] [CrossRef]
- Baretic, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Loveland, A.B.; Svidritskiy, E.; Susorov, D.; Lee, S.; Park, A.; Zvornicanin, S.; Demo, G.; Gao, F.B.; Korostelev, A.A. Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM. Nat. Commun. 2022, 13, 2776. [Google Scholar] [CrossRef] [PubMed]
- Pichkur, E.B.; Paleskava, A.; Tereshchenkov, A.G.; Kasatsky, P.; Komarova, E.S.; Shiriaev, D.I.; Bogdanov, A.A.; Dontsova, O.A.; Osterman, I.A.; Sergiev, P.V.; et al. Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome. RNA 2020, 26, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Kaledhonkar, S.; Sun, M.; Shen, B.X.; Lu, Z.H.; Barnard, D.; Lu, T.M.; Gonzalez, R.L.; Frank, J. Structural Dynamics of Ribosome Subunit Association Studied by Mixing-Spraying Time-Resolved Cryogenic Electron Microscopy. Structure 2015, 23, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, T.R.; Yassin, A.S.; Lu, Z.H.; Barnard, D.; Meng, X.; Lu, T.M.; Wagenknecht, T.; Agrawal, R.K. Initial bridges between two ribosomal subunits are formed within 9.4 milliseconds, as studied by time-resolved cryo-EM. Proc. Natl. Acad. Sci. USA 2014, 111, 9822–9827. [Google Scholar] [CrossRef]
- Unwin, N. Acetylcholine receptor channel imaged in the open state. Nature 1995, 373, 37–43. [Google Scholar] [CrossRef]
- Dandey, V.P.; Budell, W.C.; Wei, H.; Bobe, D.; Maruthi, K.; Kopylov, M.; Eng, E.T.; Kahn, P.A.; Hinshaw, J.E.; Kundu, N.; et al. Time-resolved cryo-EM using Spotiton. Nat. Methods 2020, 17, 897–900. [Google Scholar] [CrossRef]
- Maeots, M.E.; Enchev, R.I. Structural dynamics: Review of time-resolved cryo-EM. Acta Crystallogr. D 2022, 78, 927–935. [Google Scholar] [CrossRef]
- Carbone, C.E.; Loveland, A.B.; Gamper, H.B., Jr.; Hou, Y.M.; Demo, G.; Korostelev, A.A. Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP. Nat. Commun. 2021, 12, 7236. [Google Scholar] [CrossRef]
- Mulder, A.M.; Yoshioka, C.; Beck, A.H.; Bunner, A.E.; Milligan, R.A.; Potter, C.S.; Carragher, B.; Williamson, J.R. Visualizing ribosome biogenesis: Parallel assembly pathways for the 30S subunit. Science 2010, 330, 673–677. [Google Scholar] [CrossRef]
- Fischer, N.; Konevega, A.L.; Wintermeyer, W.; Rodnina, M.V.; Stark, H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 2010, 466, 329–333. [Google Scholar] [CrossRef]
- Berriman, J.; Unwin, N. Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy 1994, 56, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.; Trinick, J.; White, H. Millisecond time resolution electron cryo-microscopy of the M-ATP transient kinetic state of the acto-myosin ATPase. Biophys. J. 1995, 68, 87S–91S. [Google Scholar] [PubMed]
- White, H.D.; Walker, M.L.; Trinick, J. A computer-controlled spraying-freezing apparatus for millisecond time-resolution electron cryomicroscopy. J. Struct. Biol. 1998, 121, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Kontziampasis, D.; Klebl, D.P.; Iadanza, M.G.; Scarff, C.A.; Kopf, F.; Sobott, F.; Monteiro, D.C.F.; Trebbin, M.; Muench, S.P.; White, H.D. A cryo-EM grid preparation device for time-resolved structural studies. IUCrJ 2019, 6, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Voss, J.M.; Harder, O.F.; Olshin, P.K.; Drabbels, M.; Lorenz, U.J. Rapid melting and revitrification as an approach to microsecond time-resolved cryo-electron microscopy. Chem. Phys. Lett. 2021, 778, 138812. [Google Scholar] [CrossRef]
- Yoder, N.; Jalali-Yazdi, F.; Noreng, S.; Houser, A.; Baconguis, I.; Gouaux, E. Light-coupled cryo-plunger for time-resolved cryo-EM. J. Struct. Biol. 2020, 212, 107624. [Google Scholar] [CrossRef]
- Adams, D.J.; Kitchen, C.; Adams, S.; Furzeland, S.; Atkins, D.; Schuetz, P.; Fernyhough, C.M.; Tzokova, N.; Ryan, A.J.; Butler, M.F. On the mechanism of formation of vesicles from poly(ethylene oxide)-block-poly(caprolactone) copolymers. Soft Matter 2009, 5, 3086–3096. [Google Scholar] [CrossRef]
- Castrejon-Pita, J.R.; Kubiak, K.J.; Castrejon-Pita, A.A.; Wilson, M.C.; Hutchings, I.M. Mixing and internal dynamics of droplets impacting and coalescing on a solid surface. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013, 88, 023023. [Google Scholar] [CrossRef]
- Ashtiani, D.; Venugopal, H.; Belousoff, M.; Spicer, B.; Mak, J.; Neild, A.; de Marco, A. Delivery of femtolitre droplets using surface acoustic wave based atomisation for cryo-EM grid preparation. J. Struct. Biol. 2018, 203, 94–101. [Google Scholar] [CrossRef]
- Dandey, V.P.; Wei, H.; Zhang, Z.; Tan, Y.Z.; Acharya, P.; Eng, E.T.; Rice, W.J.; Kahn, P.A.; Potter, C.S.; Carragher, B. Spotiton: New features and applications. J. Struct. Biol. 2018, 202, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Levitz, T.S.; Weckener, M.; Fong, I.; Naismith, J.H.; Drennan, C.L.; Brignole, E.J.; Clare, D.K.; Darrow, M.C. Approaches to Using the Chameleon: Robust, Automated, Fast-Plunge cryoEM Specimen Preparation. Front Mol. Biosci. 2022, 9, 903148. [Google Scholar] [CrossRef]
- Hadimioglu, B.; Stearns, R.; Ellson, R. Moving Liquids with Sound: The Physics of Acoustic Droplet Ejection for Robust Laboratory Automation in Life Sciences. J. Lab. Autom. 2016, 21, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Razinkov, I.; Dandey, V.; Wei, H.; Zhang, Z.; Melnekoff, D.; Rice, W.J.; Wigge, C.; Potter, C.S.; Carragher, B. A new method for vitrifying samples for cryoEM. J. Struct. Biol. 2016, 195, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Frank, J. Two promising future developments of cryo-EM: Capturing short-lived states and mapping a continuum of states of a macromolecule. Microscopy 2016, 65, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Dumas, G.; Dietler, G.; Catsicas, S.; Adrian, M. Vitrification of cryoelectron microscopy specimens revealed by high-speed photographic imaging. J. Microsc. 2003, 211, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Kaledhonkar, S.; Fu, Z.; Caban, K.; Li, W.; Chen, B.; Sun, M.; Gonzalez, R.L.; Frank, J. Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature 2019, 570, 400–404. [Google Scholar] [CrossRef]
- Fu, Z.; Indrisiunaite, G.; Kaledhonkar, S.; Shah, B.; Sun, M.; Chen, B.; Grassucci, R.A.; Ehrenberg, M.; Frank, J. The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Nat. Commun. 2019, 10, 2579. [Google Scholar] [CrossRef]
- Shin, D.H.; Brandsen, J.; Jancarik, J.; Yokota, H.; Kim, R.; Kim, S.H. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J. Mol. Biol. 2004, 341, 227–239. [Google Scholar] [CrossRef]
- Vestergaard, B.; Van, L.B.; Andersen, G.R.; Nyborg, J.; Buckingham, R.H.; Kjeldgaard, M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell 2001, 8, 1375–1382. [Google Scholar] [CrossRef]
- Lanzavecchia, S.; Bellon, P.L.; Radermacher, M. Fast and Accurate Three-Dimensional Reconstruction from Projections with Random Orientations via Random Transforms. J. Struct. Biol. 1999, 128, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.F. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. II. Direct methods. Proc. R. Soc. Lond. B Biol. Sci. 1972, 182, 89–102. [Google Scholar] [PubMed]
- Valle, M.; Sengupta, J.; Swami, N.K.; Grassucci, R.A.; Burkhardt, N.; Nierhaus, K.H.; Agrawal, R.K.; Frank, J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002, 21, 3557–3567. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Valle, M.; Ehrenberg, M.; Frank, J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J. Struct. Biol. 2004, 147, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Elad, N.; Clare, D.K.; Salbil, H.R.; Orlova, E.V. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J. Struct. Biol. 2008, 162, 108–120. [Google Scholar] [CrossRef]
- Lyumkis, D.; Brilot, A.F.; Theobald, D.L.; Grigorieff, N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 2013, 183, 377–388. [Google Scholar] [CrossRef]
- Spahn, C.M.; Penczek, P.A. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr. Opin. Struct. Biol. 2009, 19, 623–631. [Google Scholar] [CrossRef]
- Zhang, W.; Kimmel, M.; Spahn, C.M.; Penczek, P.A. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure 2008, 16, 1770–1776. [Google Scholar] [CrossRef]
- Read, R.J.; Pannu, N.S. Improved Structure Refinement through Maximum Likelihood. Acta Crystallogr. A 1996, 52, 659–668. [Google Scholar] [CrossRef]
- Sigworth, F.J. A maximum-likelihood approach to single-particle image refinement. J. Struct. Biol. 1998, 122, 328–339. [Google Scholar] [CrossRef]
- DeVore, K.; Chiu, P.L. Probing Structural Perturbation of Biomolecules by Extracting Cryo-EM Data Heterogeneity. Biomolecules 2022, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Sigworth, F.J.; Doerschuk, P.C.; Carazo, J.M.; Scheres, S.H. An introduction to maximum-likelihood methods in cryo-EM. Methods Enzymol. 2010, 482, 263–294. [Google Scholar] [PubMed]
- Agirrezabala, X.; Liao, H.Y.; Schreiner, E.; Fu, J.; Ortiz-Meoz, R.F.; Schulten, K.; Green, R.; Frank, J. Structural characterization of mRNA-tRNA translocation intermediates. Proc. Natl. Acad. Sci. USA 2012, 109, 6094–6099. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Brown, A.; Toots, J.; Scheres, S.H.W.; Ramakrishnan, V. The structure of the human mitochondrial ribosome. Science 2015, 348, 95–98. [Google Scholar] [CrossRef]
- Singer, A.; Sigworth, F.J. Computational Methods for Single-Particle Electron Cryomicroscopy. Annu. Rev. Biomed. Data Sci. 2020, 3, 163–190. [Google Scholar] [CrossRef]
- Ilca, S.L.; Kotecha, A.; Sun, X.; Poranen, M.M.; Stuart, D.I.; Huiskonen, J.T. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 2015, 6, 8843. [Google Scholar] [CrossRef]
- Morais, M.C.; Kanamaru, S.; Badasso, M.O.; Koti, J.S.; Owen, B.A.L.; McMurray, C.T.; Anderson, D.L.; Rossmann, M.G. Bacteriophage phi 29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Biol. 2003, 10, 572–576. [Google Scholar] [CrossRef]
- Zhang, Y.; Kostyuchenko, V.A.; Rossmann, M.G. Structural analysis of viral nucleocapsids by subtraction of partial projections. J. Struct. Biol. 2007, 157, 356–364. [Google Scholar] [CrossRef]
- Park, E.; Menetret, J.F.; Gumbart, J.C.; Ludtke, S.J.; Li, W.; Whynot, A.; Rapoport, T.A.; Akey, C.W. Structure of the SecY channel during initiation of protein translocation. Nature 2014, 506, 102–106. [Google Scholar] [CrossRef]
- Roh, S.H.; Hryc, C.F.; Jeong, H.H.; Fei, X.; Jakana, J.; Lorimer, G.H.; Chiu, W. Subunit conformational variation within individual GroEL oligomers resolved by Cryo-EM. Proc. Natl. Acad. Sci. USA 2017, 114, 8259–8264. [Google Scholar] [CrossRef]
- Mannar, D.; Saville, J.W.; Zhu, X.; Srivastava, S.S.; Berezuk, A.M.; Zhou, S.; Tuttle, K.S.; Kim, A.; Li, W.; Dimitrov, D.S.; et al. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep. 2021, 37, 110156. [Google Scholar] [CrossRef]
- Nakane, T.; Scheres, S.H.W. Multi-body Refinement of Cryo-EM Images in RELION. Methods Mol. Biol. 2021, 2215, 145–160. [Google Scholar] [PubMed]
- Nguyen, T.H.; Galej, W.P.; Bai, X.C.; Savva, C.G.; Newman, A.J.; Scheres, S.H.; Nagai, K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 2015, 523, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Masureel, M.; Qu, Q.; Janetzko, J.; Inoue, A.; Kato, H.E.; Robertson, M.J.; Nguyen, K.C.; Glenn, J.S.; Skiniotis, G.; et al. Structure of the neurotensin receptor 1 in complex with beta-arrestin 1. Nature 2020, 579, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.A.; Leibundgut, M.; Thiel, V.; Muhlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Casanal, A.; Lohkamp, B.; Emsley, P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020, 29, 1069–1078. [Google Scholar] [CrossRef]
- Watson, Z.L.; Ward, F.R.; Meheust, R.; Ad, O.; Schepartz, A.; Banfield, J.F.; Cate, J.H.D. Structure of the bacterial ribosome at 2 angstrom resolution. Elife 2020, 9, e60482. [Google Scholar] [CrossRef]
- Poitevin, F.; Kushner, A.; Li, X.; Dao Duc, K. Structural Heterogeneities of the Ribosome: New Frontiers and Opportunities for Cryo-EM. Molecules 2020, 25, 4262. [Google Scholar] [CrossRef]
- Webster, M.W.; Takacs, M.; Zhu, C.; Vidmar, V.; Eduljee, A.; Abdelkareem, M.; Weixlbaumer, A. Structural basis of transcription-translation coupling and collision in bacteria. Science 2020, 369, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Molodtsov, V.; Firlar, E.; Kaelber, J.T.; Blaha, G.; Su, M.; Ebright, R.H. Structural basis of transcription-translation coupling. Science 2020, 369, 1359–1365. [Google Scholar] [CrossRef]
- Zhang, X.; Lai, M.; Chang, W.; Yu, I.; Ding, K.; Mrazek, J.; Ng, H.L.; Yang, O.O.; Maslov, D.A.; Zhou, Z.H. Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes. Nat. Commun. 2016, 7, 13223. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Llacer, J.L.; Wimberly, B.T.; Kieft, J.S.; Ramakrishnan, V. Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation. Cell 2016, 167, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gutierrez-Vargas, C.; Wei, J.; Grassucci, R.A.; Ramesh, M.; Espina, N.; Sun, M.; Tutuncuoglu, B.; Madison-Antenucci, S.; Woolford, J.L., Jr.; et al. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc. Natl. Acad. Sci. USA 2016, 113, 12174–12179. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Fernandez, I.S.; Scheres, S.H.; Hegde, R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef]
- Fischer, N.; Neumann, P.; Bock, L.V.; Maracci, C.; Wang, Z.; Paleskava, A.; Konevega, A.L.; Schroder, G.F.; Grubmuller, H.; Ficner, R.; et al. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature 2016, 540, 80–85. [Google Scholar] [CrossRef]
- Jomaa, A.; Boehringer, D.; Leibundgut, M.; Ban, N. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016, 7, 10471. [Google Scholar] [CrossRef]
- Fischer, N.; Neumann, P.; Konevega, A.L.; Bock, L.V.; Ficner, R.; Rodnina, M.V.; Stark, H. Structure of the E. coli ribosome-EF-Tu complex at < 3 angstrom resolution by C-s-corrected cryo-EM. Nature 2015, 520, 567–570. [Google Scholar] [PubMed]
- Natchiar, S.K.; Myasnikov, A.G.; Kratzat, H.; Hazemann, I.; Klaholz, B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017, 551, 472–477. [Google Scholar] [CrossRef]
- Shalev-Benami, M.; Zhang, Y.; Matzov, D.; Halfon, Y.; Zackay, A.; Rozenberg, H.; Zimmerman, E.; Bashan, A.; Jaffe, C.L.; Yonath, A.; et al. 2.8-angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania. Cell Rep. 2016, 16, 288–294. [Google Scholar] [CrossRef]
- Shalev-Benami, M.; Zhang, Y.; Rozenberg, H.; Nobe, Y.; Taoka, M.; Matzov, D.; Zimmerman, E.; Bashan, A.; Isobe, T.; Jaffe, C.L.; et al. Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin. Nat. Commun. 2017, 8, 1589. [Google Scholar] [CrossRef] [PubMed]
- Travin, D.Y.; Watson, Z.L.; Metelev, M.; Ward, F.R.; Osterman, I.A.; Khven, I.M.; Khabibullina, N.F.; Serebryakova, M.; Mergaert, P.; Polikanov, Y.S.; et al. Structure of ribosome-bound azole-modified peptide phazolicin rationalizes its species-specific mode of bacterial translation inhibition. Nat. Commun. 2019, 10, 4563. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.; Bai, X.C.; Sleebs, B.E.; Triglia, T.; Brown, A.; Thompson, J.K.; Jackson, K.E.; Hanssen, E.; Marapana, D.S.; Fernandez, I.S.; et al. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat. Microbiol. 2017, 2, 17031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Morgan, C.E.; Bonomo, R.A.; Yu, E.W. Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii. mBio 2021, 12, e0103121. [Google Scholar] [CrossRef]
- Tegunov, D.; Xue, L.; Dienemann, C.; Cramer, P.; Mahamid, J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 angstrom in cells. Nat. Methods 2021, 18, 186–193. [Google Scholar] [CrossRef]
- Xue, L.; Lenz, S.; Zimmermann-Kogadeeva, M.; Tegunov, D.; Cramer, P.; Bork, P.; Rappsilber, J.; Mahamid, J. Visualizing translation dynamics at atomic detail inside a bacterial cell. Nature 2022, 610, 205–211. [Google Scholar] [CrossRef]
- Khawaja, A.; Itoh, Y.; Remes, C.; Spahr, H.; Yukhnovets, O.; Hofig, H.; Amunts, A.; Rorbach, J. Distinct pre-initiation steps in human mitochondrial translation. Nat. Commun. 2020, 11, 2932. [Google Scholar] [CrossRef]
- Liu, W.; Frank, J. Estimation of variance distribution in three-dimensional reconstruction. I. Theory. J. Opt. Soc. Am. A Opt. Image. Sci. Vis. 1995, 12, 2615–2627. [Google Scholar] [CrossRef]
- Penczek, P.A.; Kimmel, M.; Spahn, C.M. Identifying conformational states of macromolecules by eigen-analysis of resampled cryo-EM images. Structure 2011, 19, 1582–1590. [Google Scholar] [CrossRef]
- Tagare, H.D.; Kucukelbir, A.; Sigworth, F.J.; Wang, H.; Rao, M. Directly reconstructing principal components of heterogeneous particles from cryo-EM images. J. Struct. Biol. 2015, 191, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Kulczyk, A.W.; Sorzano, C.O.; Przemyslaw, G.; Tchórzewski, M.; Tumer, N.E.; Li, X. Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk reveals residues involved in the binding interaction. J. Biol. Chem. 2022, 299, 102795. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.R.; Scaiola, A.; Loughran, G.; Leibundgut, M.; Kratzel, A.; Meurs, R.; Dreos, R.; O’Connor, K.M.; McMillan, A.; Bode, J.W.; et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science 2021, 372, 1306–1313. [Google Scholar] [CrossRef]
- Asarnow, D.; Wang, B.; Lee, W.H.; Hu, Y.Y.; Huang, C.W.; Faust, B.; Ng, P.M.L.; Ngoh, E.Z.X.; Bohn, M.; Bulkley, D.; et al. Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia. Cell 2021, 184, 3192–3204. [Google Scholar] [CrossRef] [PubMed]
- Pillon, M.C.; Frazier, M.N.; Dillard, L.B.; Williams, J.G.; Kocaman, S.; Krahn, J.M.; Perera, L.; Hayne, C.K.; Gordon, J.; Stewart, Z.D.; et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun. 2021, 12, 636. [Google Scholar] [CrossRef] [PubMed]
- Rujas, E.; Kucharska, I.; Tan, Y.Z.; Benlekbir, S.; Cui, H.; Zhao, T.T.; Wasney, G.A.; Budylowski, P.; Guvenc, F.; Newton, J.C.; et al. Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Nat. Commun. 2021, 12, 3661. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, G.; Guo, Y.; Liu, L.; Liu, L.; Zhang, Z.; Luo, Y.; Huang, Y.; Wang, H.H.; Ho, D.D.; Sheng, Z.; et al. Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep. 2022, 38, 110428. [Google Scholar] [CrossRef]
- Yang, T.J.; Yu, P.Y.; Chang, Y.C.; Hsu, S.D. D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation. J. Biol. Chem. 2021, 297, 101238. [Google Scholar] [CrossRef]
- Ghatak, A.; DiIorio, M.C.; Kulczyk, A.W. Novel artificial intelligence-based approaches for ab initio structure determination and atomic model building for cryo-electron microscopy; Rutgers University, Institute for Quantitative Biomedicine: New Brunswick, NJ, USA, 2023. [Google Scholar]
- Schwander, P.; Fung, R.; Ourmazd, A. Conformations of macromolecules and their complexes from heterogeneous datasets. Phil. Trans. R. Soc. B 2014, 369, 20130567. [Google Scholar] [CrossRef]
- Dashti, A.; Mashayekhi, G.; Shekhar, M.; Ben Hail, D.; Salah, S.; Schwander, P.; des Georges, A.; Singharoy, A.; Frank, J.; Ourmazd, A. Retrieving functional pathways of biomolecules from single-particle snapshots. Nat. Commun. 2020, 11, 4734. [Google Scholar] [CrossRef]
- Dashti, A.; Schwander, P.; Langlois, R.; Fung, R.; Li, W.; Hosseinizadeh, A.; Liao, H.Y.; Pallesen, J.; Sharma, G.; Stupina, V.A.; et al. Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. USA 2014, 111, 17492–17497. [Google Scholar] [CrossRef] [PubMed]
- Gupta, H.; McCann, M.T.; Donati, L.; Unser, M. CryoGAN: A New Reconstruction Paradigm for Single-Particle Cryo-EM Via Deep Adversarial Learning. IEEE Trans. Comput. Imaging 2021, 7, 759–774. [Google Scholar] [CrossRef]
- Gupta, H.; Phan, T.H.; Yoo, J.; Unser, M. Multi-CryoGAN: Reconstruction of Continuous Conformations in Cryo-EM Using Generative Adversarial Networks. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 18 August 2020; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Chen, M.; Ludtke, S.J. Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in cryo-EM. Nat. Methods 2021, 18, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Fleet, D.J. 3D Flexible Refinement: Structure and Motion of Flexible Proteins from Cryo-EM. Micrsoc. Microanal. 2022, 28, 1218. [Google Scholar] [CrossRef]
- Maji, S.; Liao, H.; Dashti, A.; Mashayekhi, G.; Ourmazd, A.; Frank, J. Propagation of Conformational Coordinates Across Angular Space in Mapping the Continuum of States from Cryo-EM Data by Manifold Embedding. J. Chem. Inf. Model. 2020, 60, 2484–2491. [Google Scholar] [CrossRef] [PubMed]
- Seitz, E.; Acosta-Reyes, F.; Maji, S.; Schawnder, P.; Frank, J. Optimization of ManifoldEM Informed by Ground Truth. IEEE Trans. Comput. Imaging. 2022, 8, 462–478. [Google Scholar] [CrossRef] [PubMed]
- Sztain, T.; Surl-Hee, A.; Bogetti, A.T.; Casalino, L.; Goldsmith, J.A.; Seitz, E.; McCool, R.S.; Kearns, F.L.; Acosta-Reyes, F.; Maji, S.; et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 2021, 13, 963–968. [Google Scholar] [CrossRef]
- Moscovich, A.; Halevi, A.; Anden, J.; Singer, A. Cryo-EM reconstruction of continuous heterogeneity by Laplacian spectral volumes. Inverse Probl. 2020, 36, 024003. [Google Scholar] [CrossRef]
- Wu, Z.; Chen, E.; Zhang, S.; Ma, Y.; Mao, Y. Visualizing Conformational Space of Functional Biomolecular Complexes by Deep Manifold Learning. Int. J. Mol. Sci. 2022, 23, 8872. [Google Scholar] [CrossRef]
- Zhong, E.D.; Lerer, A.; Davis, J.H.; Berger, B. CryoDRGN2: Ab Initio Neural Reconstruction of 3D Protein Structures From Real Cryo-EM Images. In Proceedings of the IEEE/CVF International Converence on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 4066–4075. [Google Scholar]
- Wang, J.; Li, D.; Chen, L.; Cao, W.; Kong, L.; Zhang, W.; Croll, T.; Deng, Z.; Liang, J.; Wang, Z. Catalytic trajectory of a dimeric nonribosomal peptide synthetase subunit with an inserted epimerase domain. Nat. Commun. 2022, 13, 592. [Google Scholar] [CrossRef]
- Gui, M.; Ma, M.; Sze-Tu, E.; Wang, X.; Koh, F.; Zhong, E.D.; Berger, B.; Davis, J.H.; Dutcher, S.K.; Zhang, R.; et al. Structures of radial spokes and associated complexes important for ciliary motility. Nat. Struct. Mol. Biol. 2021, 28, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Jaremko, M.J.; On, K.F.; Thomas, D.R.; Stillman, B.; Joshua-Tor, L. The dynamic nature of the human origin recognition complex revealed through five cryoEM structures. Elife 2020, 9, e58622. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DiIorio, M.C.; Kulczyk, A.W. Exploring the Structural Variability of Dynamic Biological Complexes by Single-Particle Cryo-Electron Microscopy. Micromachines 2023, 14, 118. https://doi.org/10.3390/mi14010118
DiIorio MC, Kulczyk AW. Exploring the Structural Variability of Dynamic Biological Complexes by Single-Particle Cryo-Electron Microscopy. Micromachines. 2023; 14(1):118. https://doi.org/10.3390/mi14010118
Chicago/Turabian StyleDiIorio, Megan C., and Arkadiusz W. Kulczyk. 2023. "Exploring the Structural Variability of Dynamic Biological Complexes by Single-Particle Cryo-Electron Microscopy" Micromachines 14, no. 1: 118. https://doi.org/10.3390/mi14010118
APA StyleDiIorio, M. C., & Kulczyk, A. W. (2023). Exploring the Structural Variability of Dynamic Biological Complexes by Single-Particle Cryo-Electron Microscopy. Micromachines, 14(1), 118. https://doi.org/10.3390/mi14010118






