A Technique for Rapid Bacterial-Density Enumeration through Membrane Filtration and Differential Pressure Measurements
Abstract
1. Introduction
2. Materials and Methods
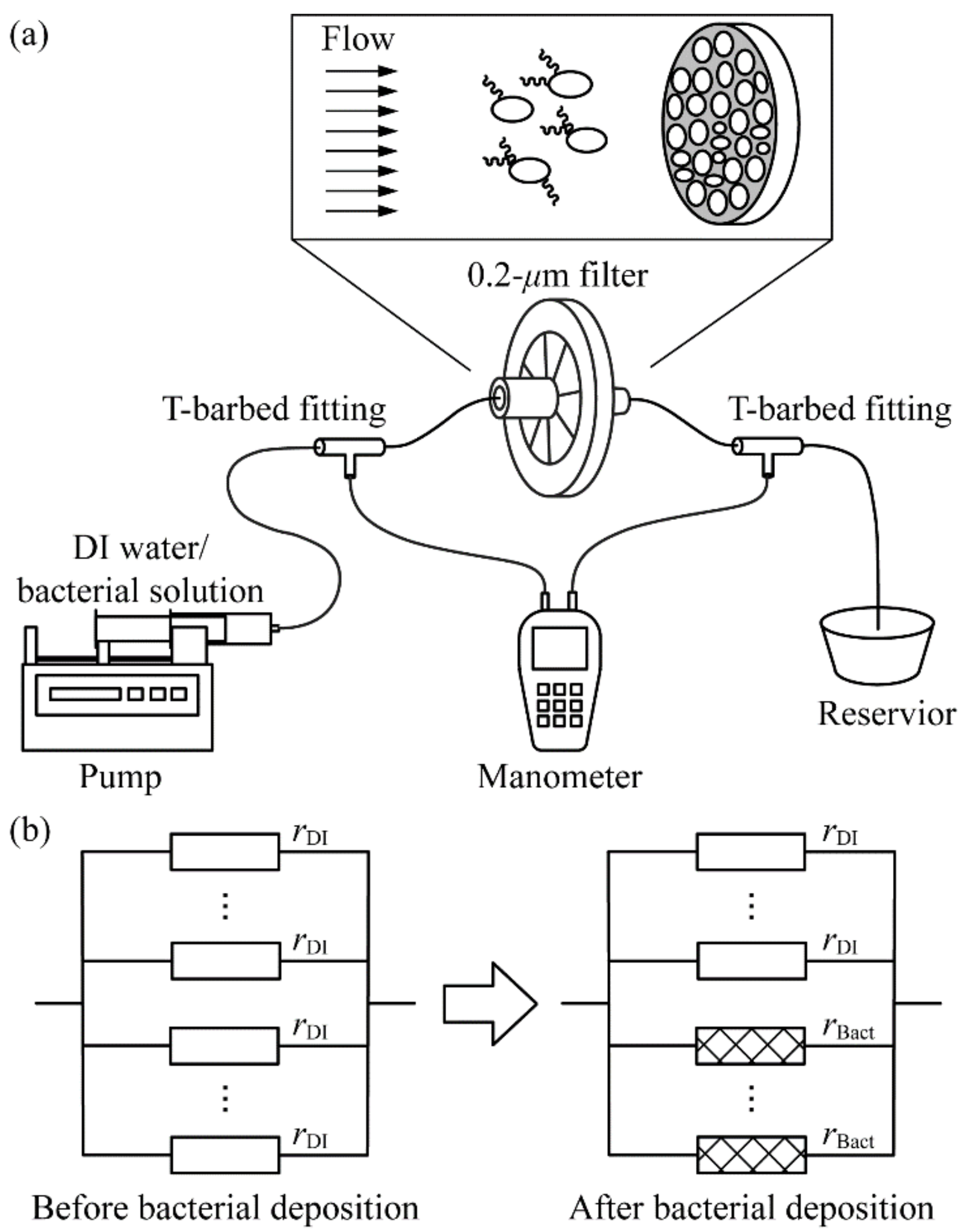
2.1. Device Schematic and Assembly
2.2. Bacteria Sample Culture and Preparation
2.3. Experiment Procedures for Obtaining the Calibration Curve for Bacteria Enumeration
3. Results and Discussions
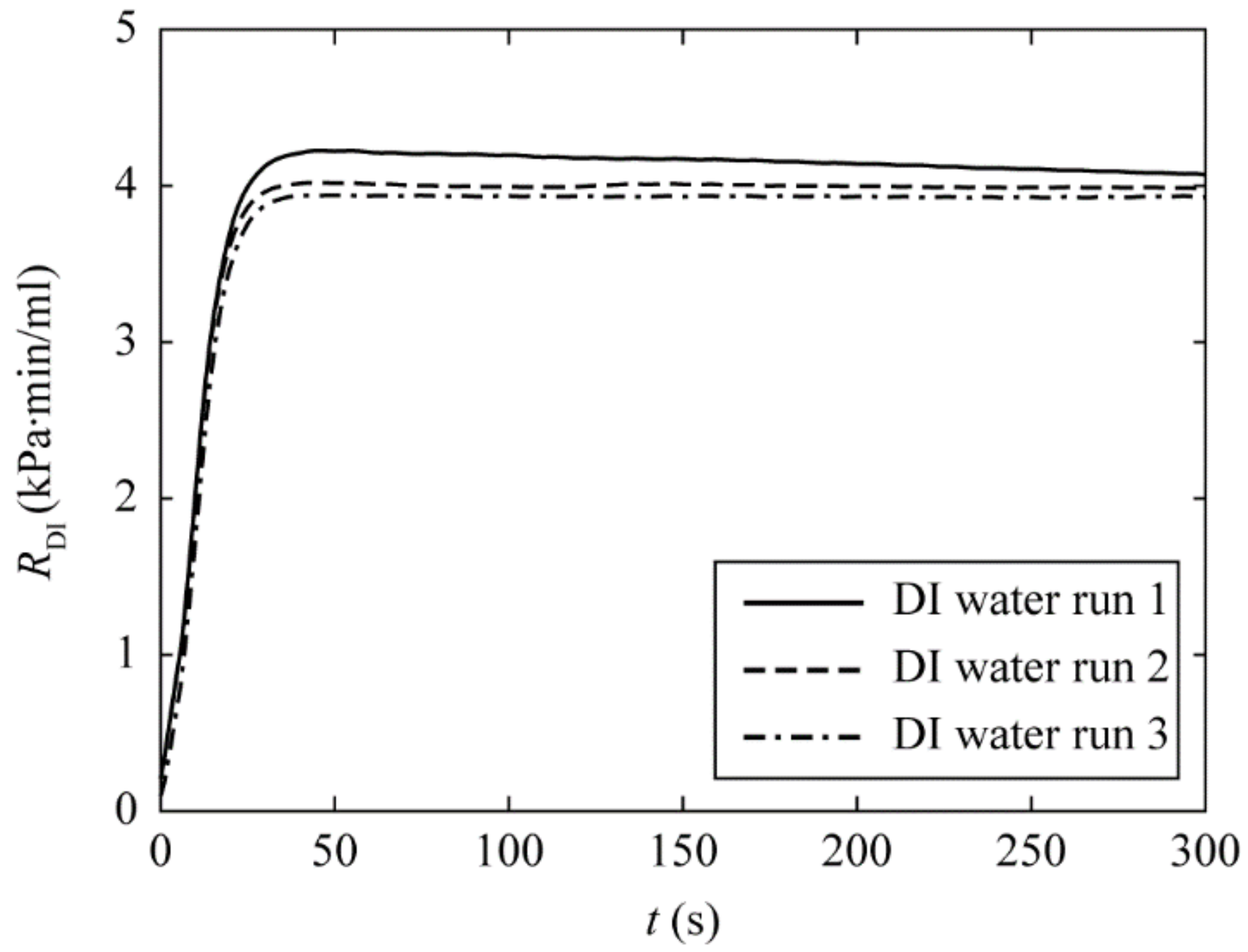
3.1. Establishing the Baseline Differential Pressure and Hydraulic Resistance of a Syringe Filter with DI Water
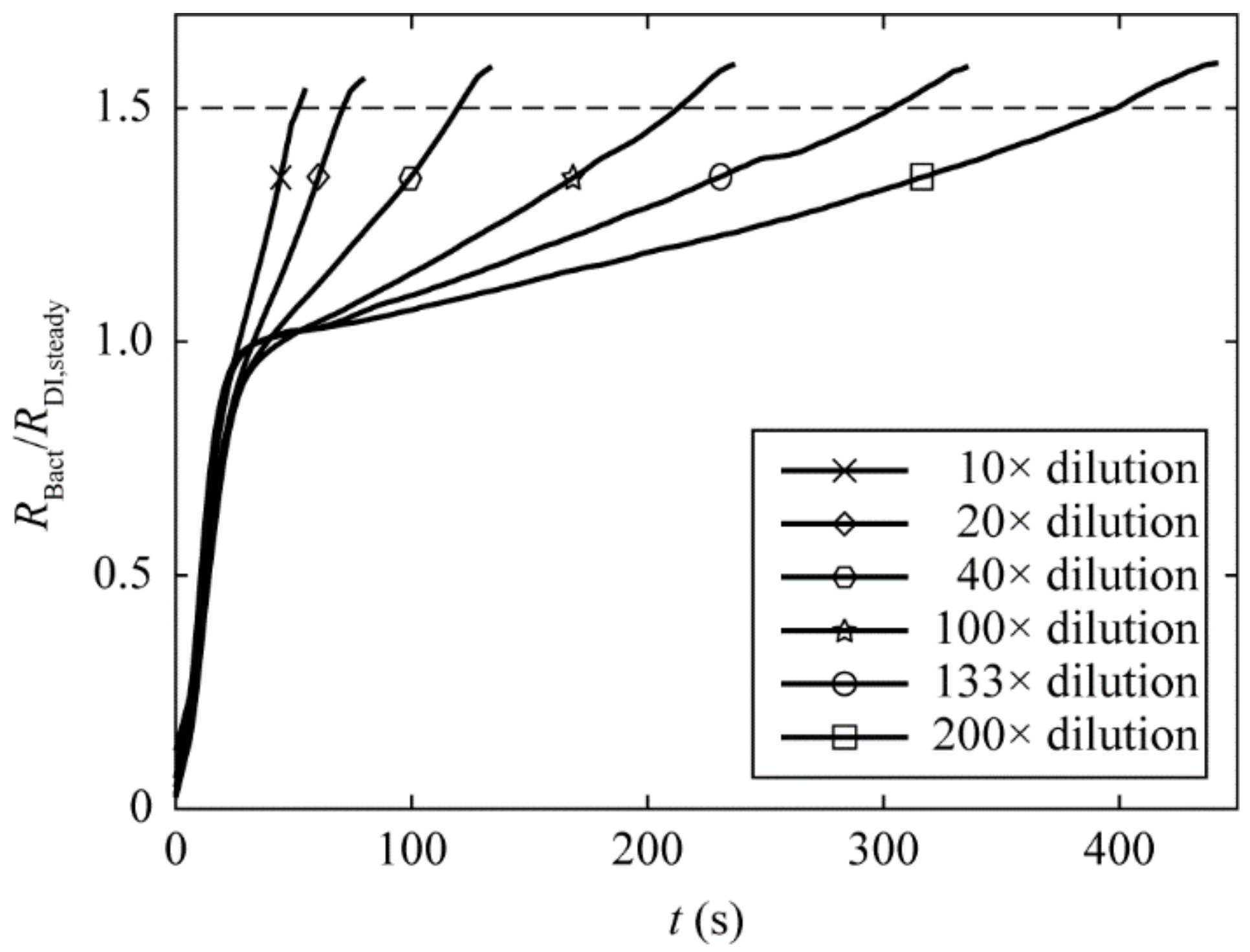
3.2. Calibration of the Differential Pressure and Hydraulic Resistance of Filtration Membrane due to Bacteria Trapping
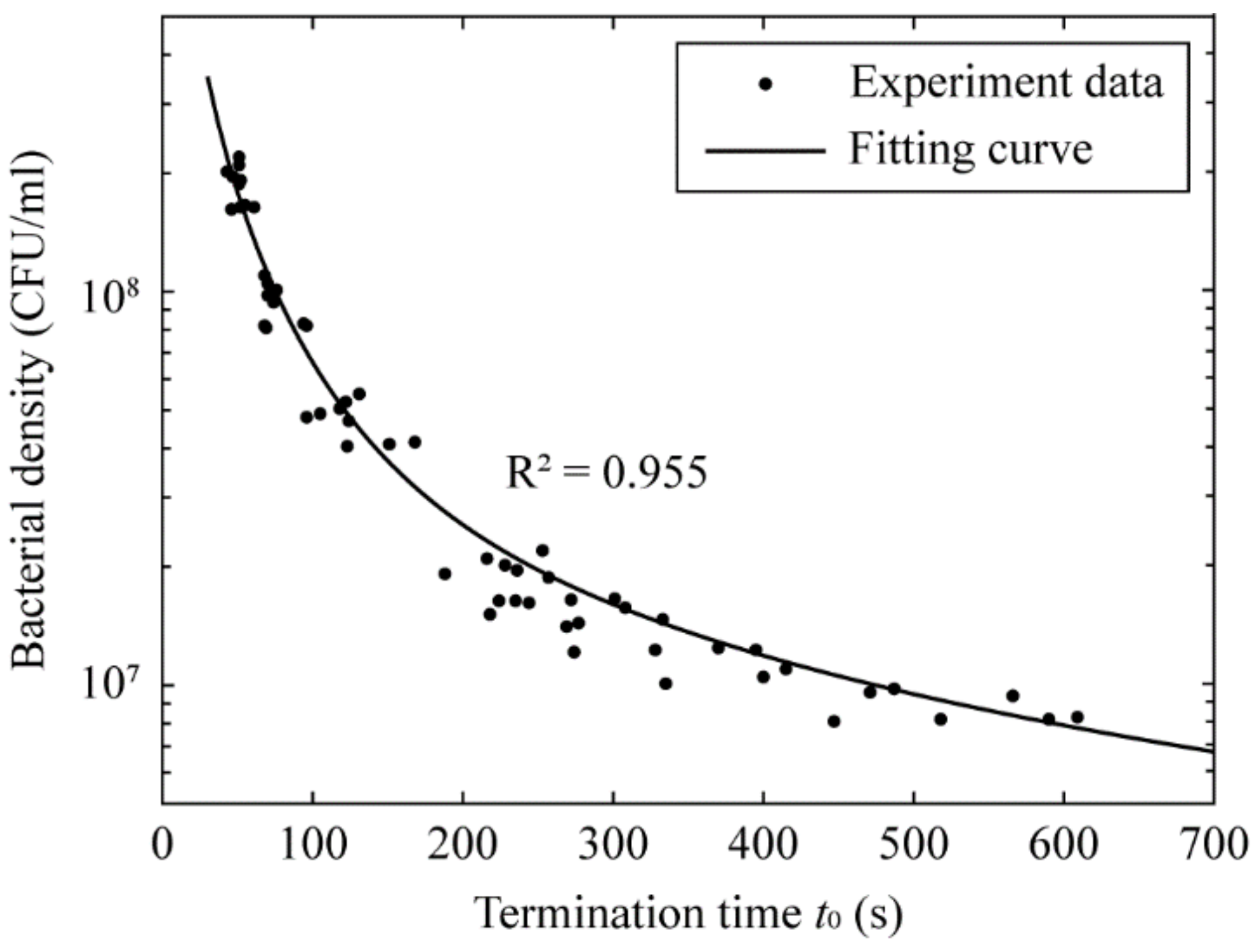
3.3. Determination of the Bacterial Density Based on Termination Time
3.4. Equivalent Electric-Circuit Model
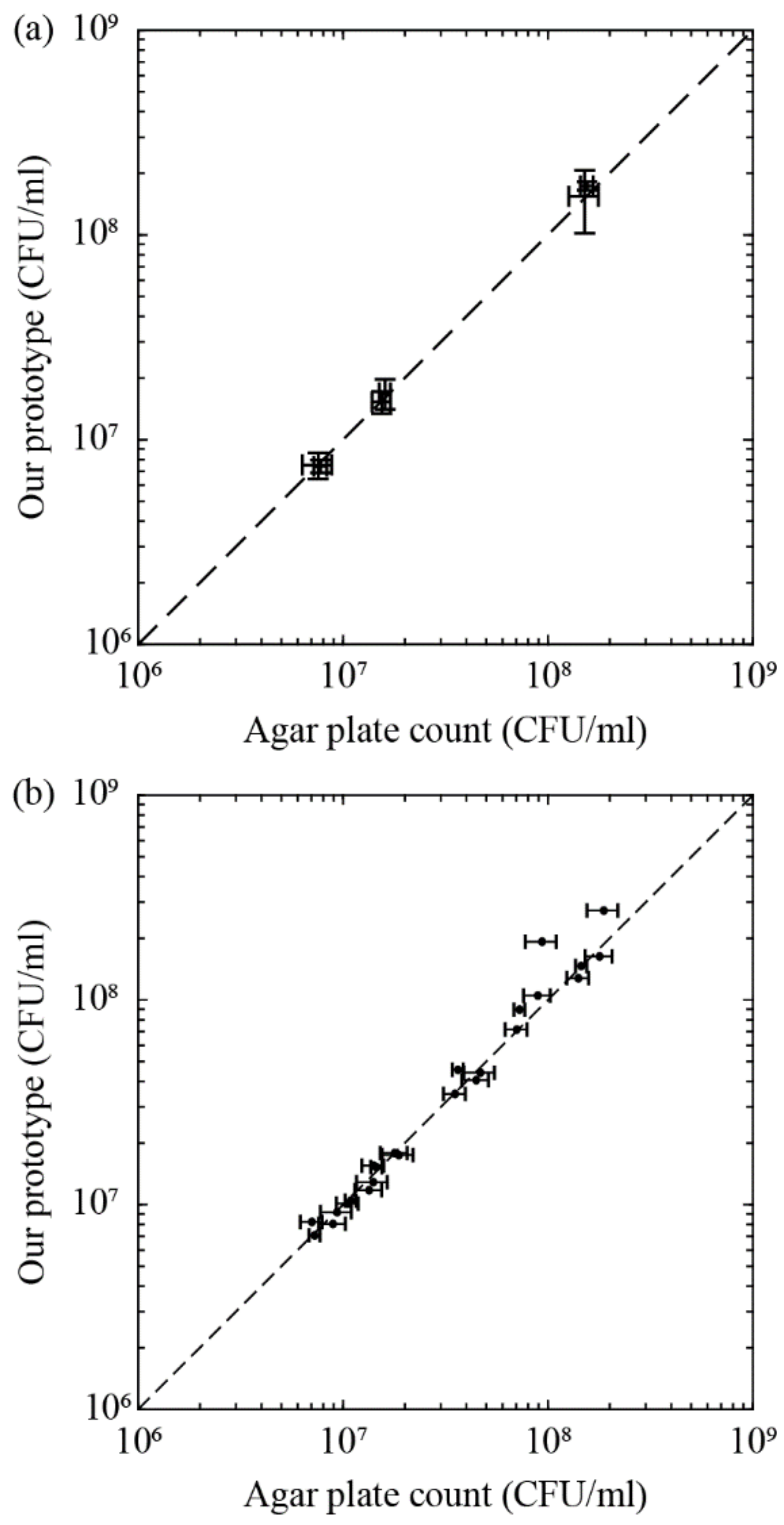
3.5. Blind Test and Proof of Concept for Bacteria Enumeration
3.6. Advantages and Improvements over Existing Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- O’Toole, D.K. Methods for the direct and indirect assessment of the bacterial content of milk. J. Appl. Bacteriol. 1983, 55, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Clais, S.; Boulet, G.; Van kerckhoven, M.; Lanckacker, E.; Delputte, P.; Maes, L.; Cos, P. Comparison of viable plate count, turbidity measurement and real-time PCR for quantification of Porphyromonas gingivalis. Lett. Appl. Microbiol. 2015, 60, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Al-Tawfiq, J.A.; Enne, V.I.; Kidd, M.; Drosten, C.; Breuer, J.; Muller, M.A.; Hui, D.; Maeurer, M.; Bates, M.; et al. Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections—Needs, advances, and future prospects. Lancet Infect. Dis. 2014, 14, 1123–1135. [Google Scholar] [CrossRef]
- Netzer, R.; Ribičić, D.; Aas, M.; Cavé, L.; Dhawan, T. Absolute quantification of priority bacteria in aquaculture using digital PCR. J. Microbiol. Methods 2021, 183, 106171. [Google Scholar] [CrossRef]
- Bej, A.K.; Mahbubani, M.H. Genetically engineered microorganisms: Monitoring and containing. In Handbook of Hazardous Materials; Corn, M., Ed.; Academic Press: Boston, MA, USA, 1993; pp. 309–319. [Google Scholar]
- Stingl, K.; Heise, J.; Thieck, M.; Wulsten, I.F.; Pacholewicz, E.; Iwobi, A.N.; Govindaswamy, J.; Zeller-Péronnet, V.; Scheuring, S.; Luu, H.Q.; et al. Challenging the “gold standard” of colony-forming units—Validation of a multiplex real-time PCR for quantification of viable Campylobacter spp. in meat rinses. Int. J. Food Microbiol. 2021, 359, 109417. [Google Scholar] [CrossRef]
- Michelutti, L.; Bulfoni, M.; Nencioni, E. A novel pharmaceutical approach for the analytical validation of probiotic bacterial count by flow cytometry. J. Microbiol. Methods 2020, 170, 105834. [Google Scholar] [CrossRef]
- Yang, X.; Zhong, Y.; Wang, D.; Lu, Z. A simple colorimetric method for viable bacteria detection based on cell counting Kit-8. Anal. Methods 2021, 13, 5211–5215. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Bacteriological Analytical Manual, 8th ed.; AOAC International: Rockville, MA, USA, 1995.
- Jett, B.D.; Hatter, K.L.; Huycke, M.M.; Gilmore, M.S. Simplified agar plate method for quantifying viable bacteria. Biotechniques 1997, 23, 648–650. [Google Scholar] [CrossRef]
- Reasoner, D.J. Heterotrophic plate count methodology in the United States. Int. J. Food Microbiol. 2004, 92, 307–315. [Google Scholar] [CrossRef]
- Fakruddin, M.; Mannan, K.S.B.; Andrews, S. Viable but nonculturable bacteria: Food safety and public health perspective. ISRN Microbiol. 2013, 2013, 703813. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Butler, M.T.; Wang, Q.; Harshey, R.M. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. USA 2010, 107, 3776–3781. [Google Scholar] [CrossRef]
- Okano, Y.; Hristova, K.R.; Leutenegger, C.M.; Jackson, L.E.; Denison, R.F.; Gebreyesus, B.; Lebauer, D.; Scow, K.M. Application of Real-Time PCR to Study Effects of Ammonium on Population Size of Ammonia-Oxidizing Bacteria in Soil. Appl. Environ. Microbiol. 2004, 70, 1008–1016. [Google Scholar] [CrossRef]
- Farrow, M.J.; Hunter, I.S.; Connolly, P. Developing a real time sensing system to monitor bacteria in wound dressings. Biosensors 2012, 2, 171–188. [Google Scholar] [CrossRef]
- Ben-David, A.; Davidson, C.E. Estimation method for serial dilution experiments. J. Microbiol. Methods 2014, 107, 214–221. [Google Scholar] [CrossRef]
- Wang, Y.; Salazar, J.K. Culture-independent rapid detection methods for bacterial pathogens and toxins in food matrices. Compr. Rev. Food Sci. Food Saf. 2016, 15, 183–205. [Google Scholar] [CrossRef]
- Hirsch, P.R.; Mauchline, T.H.; Clark, I.M. Culture-independent molecular techniques for soil microbial ecology. Soil Biol. Biochem. 2010, 42, 878–887. [Google Scholar] [CrossRef]
- McFarland, J. The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J. Am. Med. Assoc. 1907, 49, 1176–1178. [Google Scholar] [CrossRef]
- Roessler, W.G.; Brewer, C.R. Permanent turbidity standards. Appl. Microbiol. 1967, 15, 1114–1121. [Google Scholar] [CrossRef]
- Lahuerta Zamora, L.; Pérez-Gracia, M.T. Using digital photography to implement the McFarland method. J. R. Soc. Interfaces 2012, 9, 1892–1897. [Google Scholar] [CrossRef]
- Toennies, G.; Gallant, D.L. The relation between photometric turbidity and bacterial concentration. Growth 1949, 13, 7–20. [Google Scholar]
- Korukawa, M.; Hatano, M.; Kashiwagi, N.; Saito, T.; Ishida, S.; Homma, R. A new method for the turbidimetric measurement of bacterial density. J. Bacteriol. 1962, 83, 14–19. [Google Scholar] [CrossRef]
- Koch, A.L. Turbidity measurements of bacterial cultures in some available commercial instruments. Anal. Biochem. 1970, 38, 252–259. [Google Scholar] [CrossRef]
- Sutton, S. Measurement of microbial cells by optical density. J. Valid. Technol. 2011, 17, 47–49. [Google Scholar]
- Stevenson, K.; McVey, A.F.; Clark, I.B.N.; Swain, P.S.; Pilizota, T. General calibration of microbial growth in microplate readers. Sci. Rep. 2016, 6, 38828. [Google Scholar] [CrossRef]
- Jannasch, H.W. Studies on Planktonic Bacteria by Means of a Direct Membrane Filter Method. Microbiology 1958, 18, 609–620. [Google Scholar] [CrossRef]
- Mulvany, J.G. Chapter VII Membrane Filter Techniques in Microbiology. In Methods in Microbiol; Norris, J.R., Ribbons, D.W., Eds.; Academic Press: Cambridge, MA, USA, 1969; Volume 1, pp. 205–253. [Google Scholar]
- Zhao, C.; Ge, Z.; Yang, C. Microfluidic techniques foranalytes concentration. Micromachines 2017, 8, 28. [Google Scholar] [CrossRef]
- Chen, X.; Miller, A.; Cao, S.; Gan, Y.; Zhang, J.; He, Q.; Wang, R.-Q.; Yong, X.; Qin, P.; Lapizco-Encinas, B.H.; et al. Rapid Escherichia coli trapping and retrieval from bodily fluids via a three-dimensional bead-stacked nanodevice. ACS Appl. Mater. Interfaces 2020, 12, 7888–7896. [Google Scholar] [CrossRef]
- Hobbie, J.E.; Daley, R.J.; Jasper, S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microb. 1977, 33, 1225–1228. [Google Scholar] [CrossRef]
- Pettipher, G.L.; Mansell, R.; McKinnon, C.H.; Cousins, C.M. Rapid membrane filtration-epifluorescent microscopy technique for direct enumeration of bacteria in raw milk. Appl. Environ. Microbiol. 1980, 39, 423–429. [Google Scholar] [CrossRef]
- Stannard, C.J.; Wood, J.M. The rapid estimation of microbial contamination of raw meat by measurement of adenosine triphosphate (ATP). J. Appl. Bacteriol. 1983, 55, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Kuramitsu, Y.; Ookuma, A.; Trevanich, S.; Honjoh, K.; Hatano, S. Rapid detection and counting of viable bacteria in vegetables and environmental water using a photon-counting TV camera. J. Food Prot. 1998, 61, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Lin, C.W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Tortorello, M.L.; Stewart, D.S.; Raybourne, R.B. Quantitative analysis and isolation of Escherichia coli O157:H7 in a food matrix using flow cytometry and cell sorting. FEMS Immunol. Med. Microbiol. 1997, 19, 267–274. [Google Scholar] [CrossRef]
- Alvarez-Barrientos, A.; Arroyo, J.; Cantón, R.; Nombela, C.; Sánchez-Pérez, M. Applications of flow cytometry to clinical microbiology. Clin. Microbiol. Rev. 2000, 13, 167–195. [Google Scholar] [CrossRef]
- Flint, S.; Drocourt, J.-L.; Walker, K.; Stevenson, B.; Dwyer, M.; Clarke, I.; McGill, D. A rapid, two-hour method for the enumeration of total viable bacteria in samples from commercial milk powder and whey protein concentrate powder manufacturing plants. Int. Dairy J. 2006, 16, 379–384. [Google Scholar] [CrossRef]
- Liao, J.; Ren, J.; Wei, H.; Lam, R.H.W.; Chua, S.L.; Khoo, B.L. Label-free biosensor of phagocytosis for diagnosing bacterial infections. Biosens. Bioelectron. 2021, 191, 113412. [Google Scholar] [CrossRef]
- Yoon, J.Y.; Kim, B. Lab-on-a-chip pathogen sensors for food safety. Sensors 2012, 12, 10713–10741. [Google Scholar] [CrossRef]
- Li, M.; Li, W.H.; Zhang, J.; Alici, G.; Wen, W. A review of microfabrication techniques and dielectrophoretic microdevices for particle manipulation and separation. J. Phys. D Appl. Phys. 2014, 47, 063001. [Google Scholar] [CrossRef]
- Ali, A.A.; Altemimi, A.B.; Alhelfi, N.; Ibrahim, S.A. Application of biosensors for detection of pathogenic food bacteria: A review. Biosensors 2020, 10, 58. [Google Scholar] [CrossRef]
- Kong, T.F.; Shen, X.; Marcos; Yang, C.; Ibrahim, I.H. Dielectrophoretic trapping and impedance detection of Escherichia coli, Vibrio cholera, and Enterococci bacteria. Biomicrofluidics 2020, 14, 054105. [Google Scholar] [CrossRef]
- Kong, T.F.; Tan, P.Y.; Tay, B.Z.; Shen, X.; Marcos. Bacteria and cancer cell pearl chain under dielectrophoresis. Electrophroesis 2021, 42, 1070–1078. [Google Scholar] [CrossRef]
- Goldman, E.; Green, L.H. Practical Handbook of Microbiology, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Guo, R.; McGoverin, C.; Swift, S.; Vanholsbeeck, F. A rapid and low-cost estimation of bacteria counts in solution using fluorescence spectroscopy. Anal. Bioanal. Chem. 2017, 409, 3959–3967. [Google Scholar] [CrossRef]
- Kirby, B.J. Micro- and Nanoscale Fluid Mechanics: Transport in Microfluidic Devices; Cambridge University Press: New York, NY, USA, 2010. [Google Scholar]
- Zapata, A.; Ramirez-Arcos, S. A comparative study of McFarland turbidity standards and the Densimat photometer to determine bacterial cell density. Curr. Microbiol. 2015, 70, 907–909. [Google Scholar] [CrossRef]
- Eppendorf AG. OD600 Measurements Using Different Photometers; Eppendorf AG: Hamburg, Germany, 2015. [Google Scholar]
- Su, C.-C.; Li, C.-H.; Chang, N.-K.; Gao, F.; Chang, S.-H. Fabrication of high sensitivity carbon microcoil pressure sensors. Sensors 2012, 12, 10034–10041. [Google Scholar] [CrossRef]
- Kong, T.F.; Nguyen, N.-T. Liquid metal microcoils for sensing and actuation in lab-on-a-chip applications. Microsyst. Technol. 2015, 21, 519–526. [Google Scholar] [CrossRef][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Teo, T.W.; Kong, T.F.; Marcos. A Technique for Rapid Bacterial-Density Enumeration through Membrane Filtration and Differential Pressure Measurements. Micromachines 2022, 13, 1198. https://doi.org/10.3390/mi13081198
Shen X, Teo TW, Kong TF, Marcos. A Technique for Rapid Bacterial-Density Enumeration through Membrane Filtration and Differential Pressure Measurements. Micromachines. 2022; 13(8):1198. https://doi.org/10.3390/mi13081198
Chicago/Turabian StyleShen, Xinhui, Ting Wei Teo, Tian Fook Kong, and Marcos. 2022. "A Technique for Rapid Bacterial-Density Enumeration through Membrane Filtration and Differential Pressure Measurements" Micromachines 13, no. 8: 1198. https://doi.org/10.3390/mi13081198
APA StyleShen, X., Teo, T. W., Kong, T. F., & Marcos. (2022). A Technique for Rapid Bacterial-Density Enumeration through Membrane Filtration and Differential Pressure Measurements. Micromachines, 13(8), 1198. https://doi.org/10.3390/mi13081198




