Single Red Blood Cell Hydrodynamic Traps via the Generative Design
Abstract
1. Introduction
2. Materials and Methods
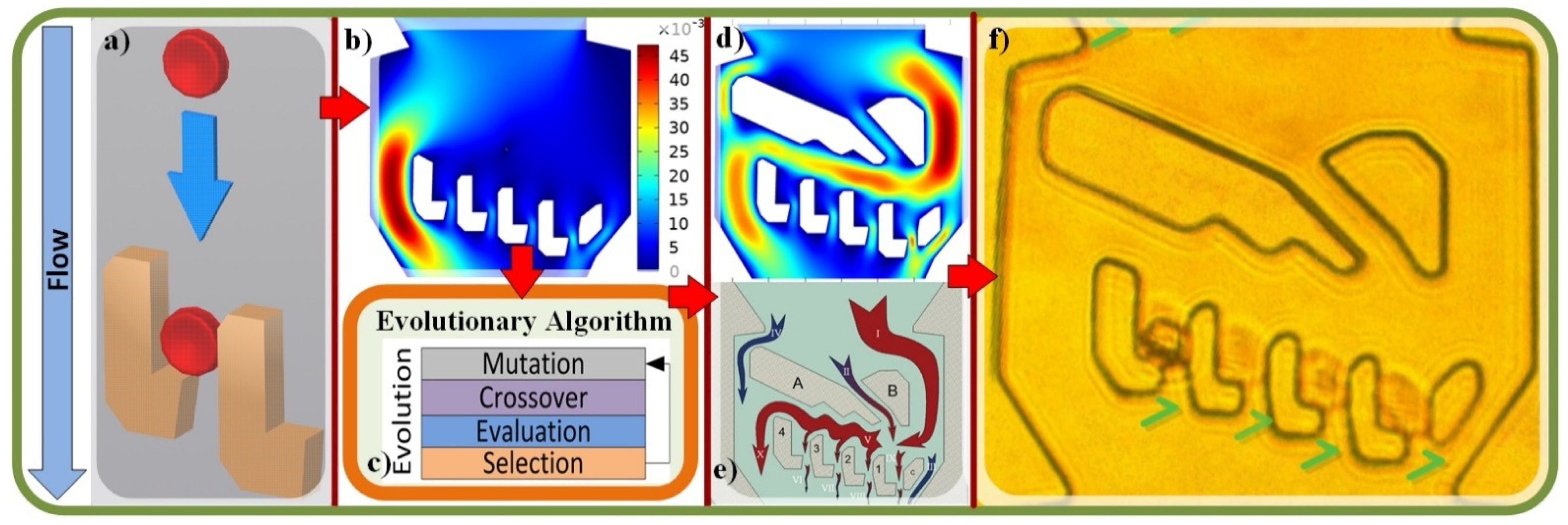
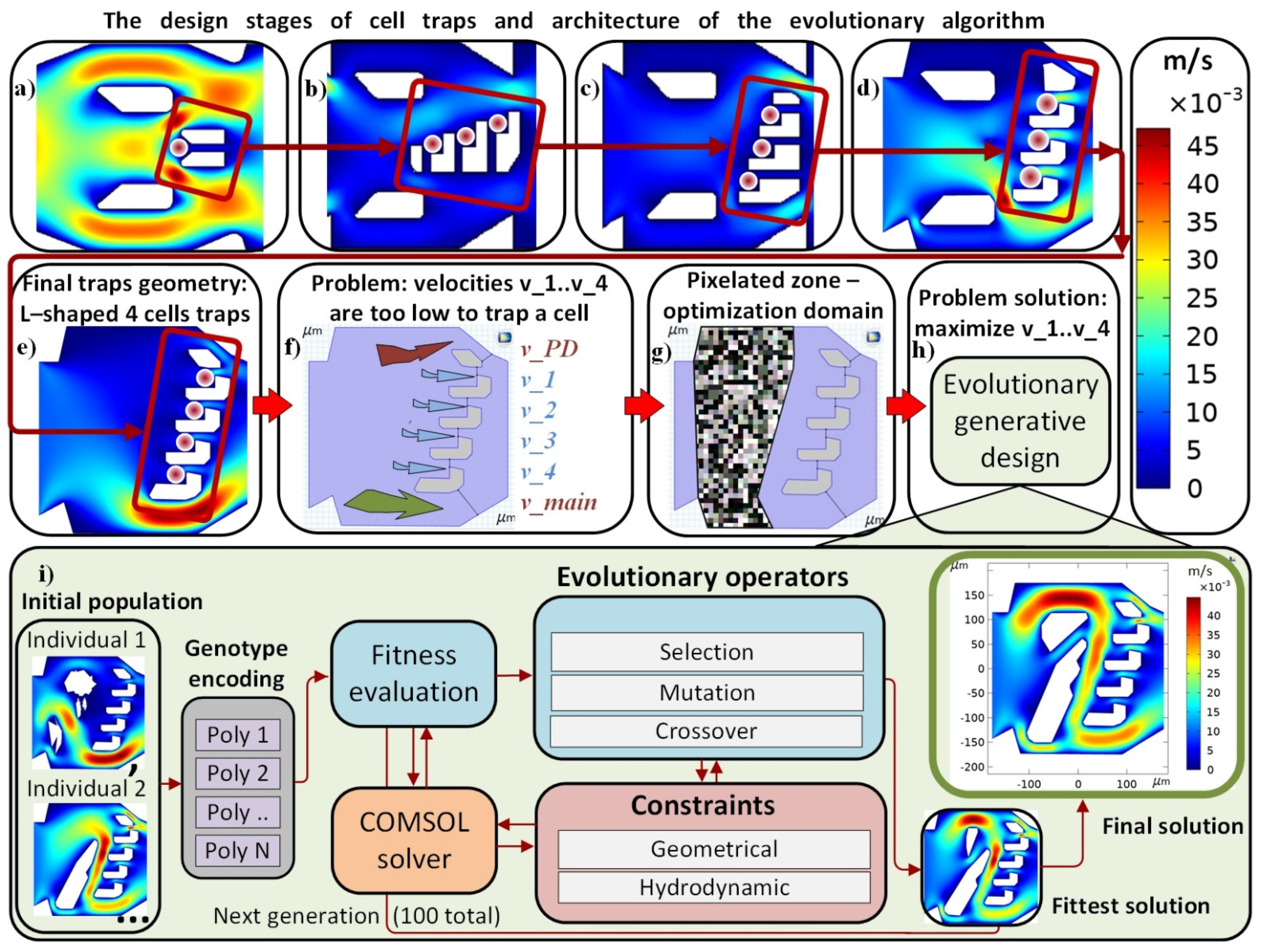
2.1. Single Cell Traps
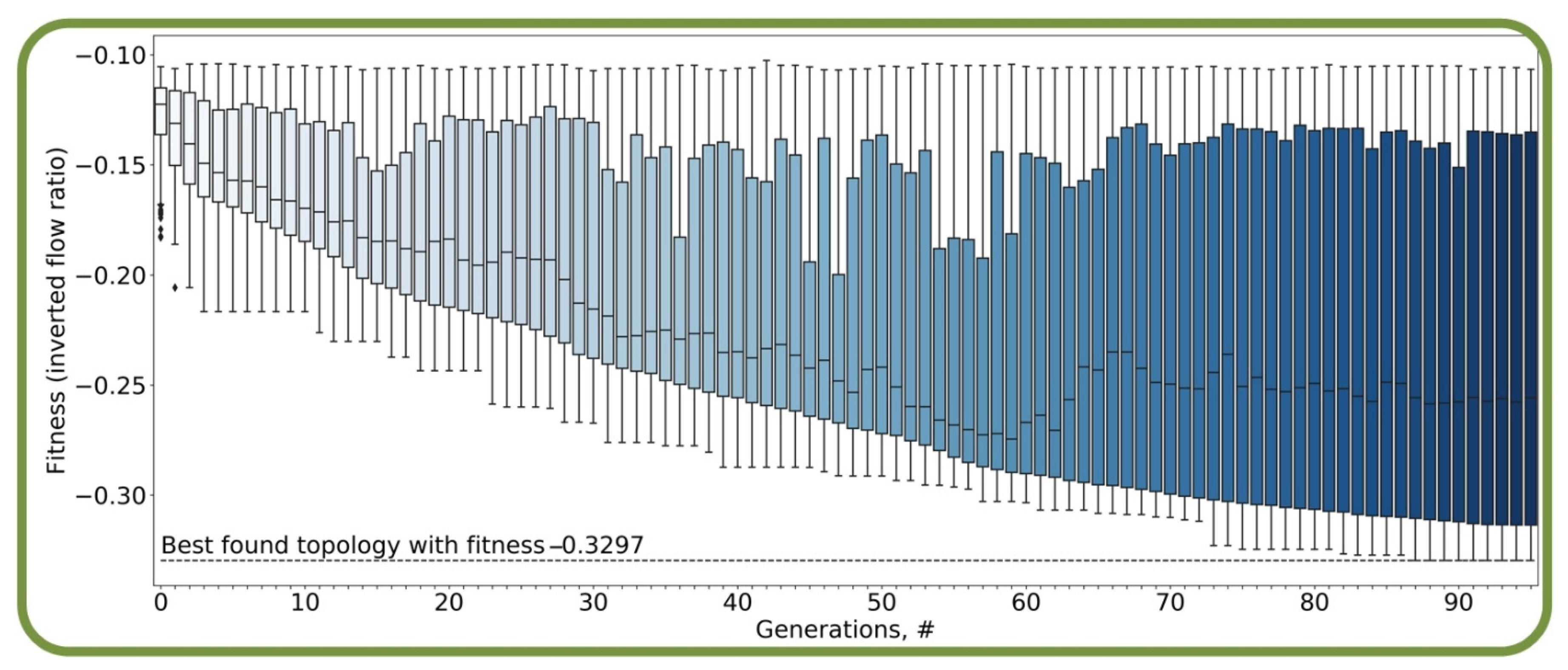
2.2. Evolutionary Algorithm
| Algorithm 1: Evolutionary algorithm for cell trap design |
| input: params = set of hyperparameters for evolutionary algorithm (population size, number of populations, etc) constraints = set of constraints for cell trap output: Best found cell trap design ▸Generate random initial population pop←InitPopulation(params.pop_size, constraints) while not IsFinished(params.num_pop) do offsprings←Reproduce(pop, constraints) pop.fitness←Fitness(pop, constraints) pop←TournamentSelection(offsprings) return Best(pop) procedure Reproduce: input: pop, constraints output: offsprings while not Validate(constraints) modify cell traps designs offsprings←Crossover (pop) offsprings←Mutation (pop) return offsprings procedure Fitness: input: pop, constraints output: fitness values for each individual fitnesses={} for ind in pop: if Validate(individual, constraints) run sim for cell trap described in genotype fitnesses[ind]←COMSOL_Sim(ind) else fitnesses[ind]←0 return fitnesses |
2.3. Fabrication
2.4. Bio-Samples Preparation
3. Results and Discussion
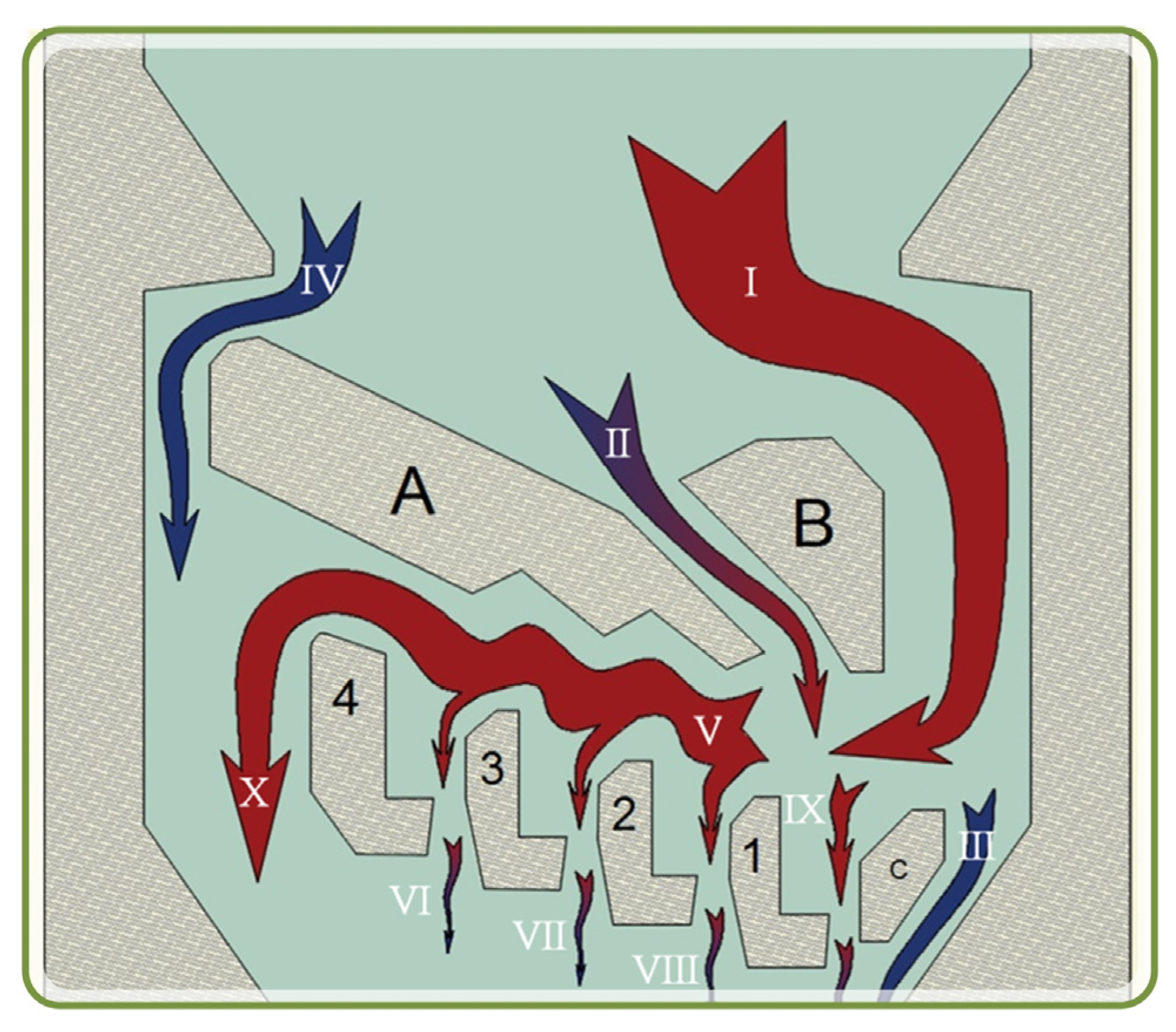
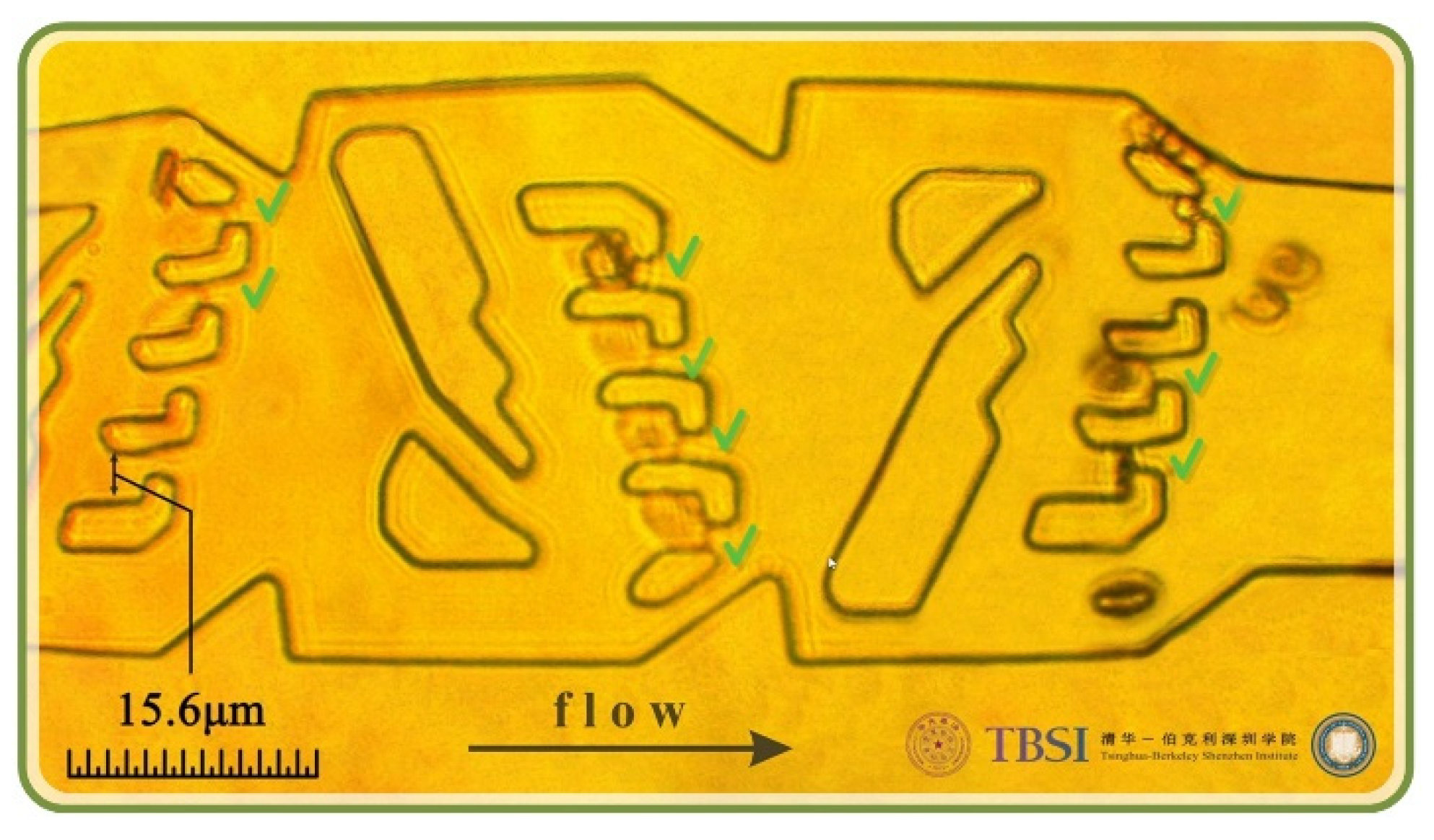
3.1. Optimal Designed Micfluidic Traps
3.2. Experimental Results
4. Conclusions
5. Code and Data Availability
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, M.G.; Park, J.; Lim, H.G.; Yoon, S.; Lee, C.; Chang, J.H.; Kirk Shung, K. Label-free analysis of the characteristics of a single cell trapped by acoustic tweezers. Sci. Rep. 2017, 7, 14092. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Franke, T. Acoustic erythrocytometer for mechanically probing cell viscoelasticity. Lab Chip 2020, 20, 1991–1998. [Google Scholar] [CrossRef] [PubMed]
- Du, E.; Dao, M.; Suresh, S. Quantitative biomechanics of healthy and diseased human red blood cells using dielectrophoresis in a microfluidic system. Extrem. Mech. Lett. 2014, 1, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.V.; Staton, S.J.R.; Hayes, M.A. Blood cell capture in a sawtooth dielectrophoretic microchannel. Anal. Bioanal. Chem. 2011, 401, 2103–2111. [Google Scholar] [CrossRef]
- Mohammadi, M.; Madadi, H.; Casals-Terré, J.; Sellarès, J. Hydrodynamic and direct-current insulator-based dielectrophoresis (H-DC-iDEP) microfluidic blood plasma separation. Anal. Bioanal. Chem. 2015, 407, 4733–4744. [Google Scholar] [CrossRef]
- Narayanamurthy, V.; Nagarajan, S.; Firus Khan, A.Y.; Samsuri, F.; Sridhar, T.M. Microfluidic hydrodynamic trapping for single cell analysis: Mechanisms, methods and applications. Anal. Methods 2017, 9, 3751–3772. [Google Scholar] [CrossRef]
- Iliescu, C.; Xu, G.; Tong, W.H.; Yu, F.; Bălan, C.M.; Tresset, G.; Yu, H. Cell patterning using a dielectrophoretic–hydrodynamic trap. Microfluid. Nanofluid. 2015, 19, 363–373. [Google Scholar] [CrossRef][Green Version]
- Tan, W.-H.; Takeuchi, S. A trap-and-release integrated microfluidic system for dynamic microarray applications. Proc. Natl. Acad. Sci. USA 2007, 104, 1146–1151. [Google Scholar] [CrossRef]
- Watarai, H.; Namba, M. Capillary magnetophoresis of human blood cells and their magnetophoretic trapping in a flow system. J. Chromatogr. A 2002, 961, 3–8. [Google Scholar] [CrossRef]
- Zborowski, M.; Ostera, G.R.; Moore, L.R.; Milliron, S.; Chalmers, J.J.; Schechter, A.N. Red blood cell magnetophoresis. Biophys. J. 2003, 84, 2638–2645. [Google Scholar] [CrossRef]
- Khokhlova, M.D. Normal and system lupus erythematosus red blood cell interactions studied by double trap optical tweezers: Direct measurements of aggregation forces. J. Biomed. Opt. 2012, 17, 25001. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.-C.; Gong, L.; Zhou, J.-H.; Wang, Z.-Q.; Li, Y.-M. Optical trapping of red blood cells in living animals with a water immersion objective. Opt. Lett. 2013, 38, 5134. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.T.F.; Aw Yong, K.M.; Fu, J. Microfluidic blood cell sorting: Now and beyond. Small 2014, 10, 1687–1703. [Google Scholar] [CrossRef] [PubMed]
- Henry, E.; Holm, S.H.; Zhang, Z.; Beech, J.P.; Tegenfeldt, J.O.; Fedosov, D.A.; Gompper, G. Sorting cells by their dynamical properties. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ye, X.; Ma, Z.; Xie, S.; Wang, W. High-throughput and clogging-free microfluidic filtration platform for on-chip cell separation from undiluted whole blood. Biomicrofluidics 2016, 10, 014118. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ju, D.; Liu, Y.; Lv, X.; Xiao, Z.; Gao, B.; Song, F.; Xu, F. Design of organ-on-a-chip to improve cell capture efficiency. Int. J. Mech. Sci. 2021, 209, 106705. [Google Scholar] [CrossRef]
- Raissi, M.; Yazdani, A.; Karniadakis, G.E. Hidden fluid mechanics: Learning velocity and pressure fields from flow visualizations. Science 2020, 367, 1026–1030. [Google Scholar] [CrossRef]
- Man, Y.; Kucukal, E.; An, R.; Watson, Q.D.; Bosch, J.; Zimmerman, P.A.; Little, J.A.; Gurkan, U.A.; Gurkan, U.A. Microfluidic assessment of red blood cell mediated microvascular occlusion. Lab Chip 2020, 20, 2086–2099. [Google Scholar] [CrossRef]
- Wu, J.; Qian, X.; Wang, M.Y. Advances in generative design. Comput.-Aided Des. 2019, 116, 102733. [Google Scholar] [CrossRef]
- Oh, S.; Jung, Y.; Kim, S.; Lee, I.; Kang, N. Deep generative design: Integration of topology optimization and generative models. J. Mech. Des. Trans. ASME 2019, 141, 111405. [Google Scholar] [CrossRef]
- Nikitin, N.O.; Polonskaia, I.S.; Kalyuzhnaya, A.V.; Boukhanovsky, A.V. The multi-objective optimisation of breakwaters using evolutionary approach. In Developments in Maritime Technology and Engineering; CRC Press: Boca Raton, FL, USA, 2021; pp. 767–774. [Google Scholar] [CrossRef]
- Kalyuzhnaya, A.V.; Nikitin, N.O.; Hvatov, A.; Maslyaev, M.; Yachmenkov, M.; Boukhanovsky, A. Towards generative design of computationally efficient mathematical models with evolutionary learning. Entropy 2021, 23, 28. [Google Scholar] [CrossRef] [PubMed]
- Toussi, H.E. The Application of Evolutionary, Generative, and Hybrid Approaches in Architecture Design Optimization. NEU J. Fac. Archit. 2020, 2, 1–20. [Google Scholar]
- Le, T.C.; Winkler, D.A. A Bright Future for Evolutionary Methods in Drug Design. Chem. Med. Chem. 2015, 10, 1296–1300. [Google Scholar] [CrossRef] [PubMed]
- Arita, T.; Fukata, M.; Maruyama, T.; Odashiro, K.; Fujino, T.; Wakana, C.; Sato, A.; Takahashi, K.; Iida, Y.; Mawatari, S.; et al. Comparison of Human Erythrocyte Filterability with Trapping Rate Obtained by Nickel Mesh Filtration Technique: Two Independent Parameters of Erythrocyte Deformability. Int. Blood Res. Rev. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Carey, T.R.; Cotner, K.L.; Li, B.; Sohn, L.L. Developments in label-free microfluidic methods for single-cell analysis and sorting. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, e1529. [Google Scholar] [CrossRef]
- Yu, X.; Gen, M. Introduction to Evolutionary Algorithms; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Nikitin, N.O.; Hvatov, A.; Polonskaia, I.S.; Kalyuzhnaya, A.V.; Grigorev, G.V.; Wang, X.; Qian, X. Generative Design of Microfluidic Channel Geometry Using Evolutionary Approach; Generative Design of Microfluidic Channel Geometry Using Evolutionary Approach. In Proceedings of the Genetic and Evolutionary Computation Conference Companion, Lille, France, 10–14 July 2021. [Google Scholar] [CrossRef]
- Kallioras, N.A.; Lagaros, N.D. DzAIℕ: Deep learning based generative design. Procedia Manuf. 2020, 44, 591–598. [Google Scholar] [CrossRef]

| Parameter | Units | Initial | Target Values | Optimized | Gain, % |
|---|---|---|---|---|---|
| vl_1 | m/s | 0.012038 | determined | 0.02308 | 92 |
| vl_2 | m/s | 0.009443 | 0.01579 | 67 | |
| vl_3 | m/s | 0.009478 | by | 0.012701 | 34 |
| vl_4 | m/s | 0.009544 | 0.010092 | 6 | |
| vl_PD | m/s | 0.005998 | TVR ratio | 0.012438 | 107 |
| vl_main | m/s | 0.027247 | 0.019577 | −28 | |
| CVR | 1/m | 70,769,000 | <7 × 107 | 17,113,000 | −76 |
| CRL | 1/s | 12,717 | <30,000 | 20,615 | 62 |
| TVR (target) | - | 1.22 | 1.22 < TVR < 2 | 1.93 | 58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigorev, G.V.; Nikitin, N.O.; Hvatov, A.; Kalyuzhnaya, A.V.; Lebedev, A.V.; Wang, X.; Qian, X.; Maksimov, G.V.; Lin, L. Single Red Blood Cell Hydrodynamic Traps via the Generative Design. Micromachines 2022, 13, 367. https://doi.org/10.3390/mi13030367
Grigorev GV, Nikitin NO, Hvatov A, Kalyuzhnaya AV, Lebedev AV, Wang X, Qian X, Maksimov GV, Lin L. Single Red Blood Cell Hydrodynamic Traps via the Generative Design. Micromachines. 2022; 13(3):367. https://doi.org/10.3390/mi13030367
Chicago/Turabian StyleGrigorev, Georgii V., Nikolay O. Nikitin, Alexander Hvatov, Anna V. Kalyuzhnaya, Alexander V. Lebedev, Xiaohao Wang, Xiang Qian, Georgii V. Maksimov, and Liwei Lin. 2022. "Single Red Blood Cell Hydrodynamic Traps via the Generative Design" Micromachines 13, no. 3: 367. https://doi.org/10.3390/mi13030367
APA StyleGrigorev, G. V., Nikitin, N. O., Hvatov, A., Kalyuzhnaya, A. V., Lebedev, A. V., Wang, X., Qian, X., Maksimov, G. V., & Lin, L. (2022). Single Red Blood Cell Hydrodynamic Traps via the Generative Design. Micromachines, 13(3), 367. https://doi.org/10.3390/mi13030367







