A Novel 3D Helical Microelectrode Array for In Vitro Extracellular Action Potential Recording
Abstract
1. Introduction
2. Materials and Methods
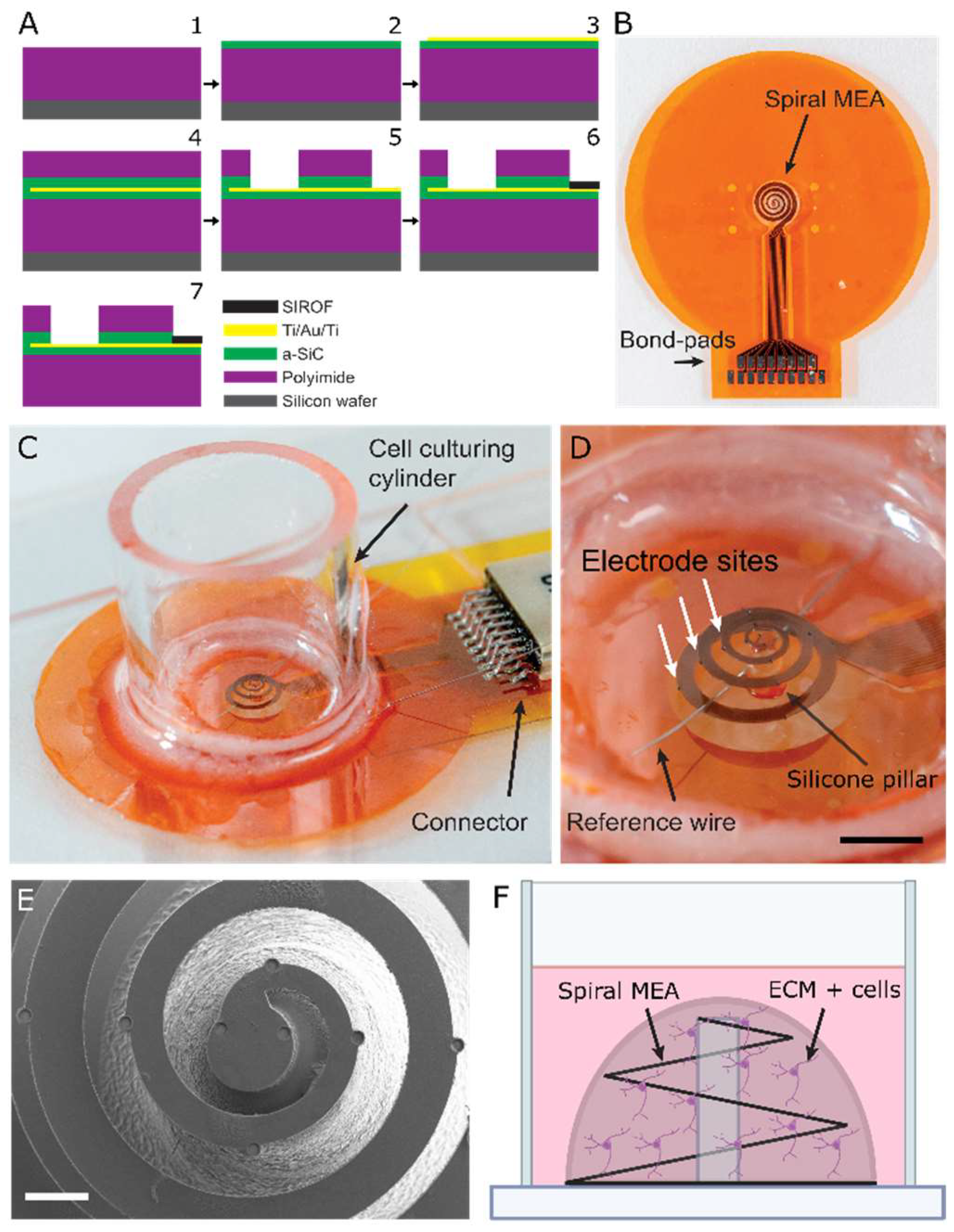
2.1. MEA Fabrication
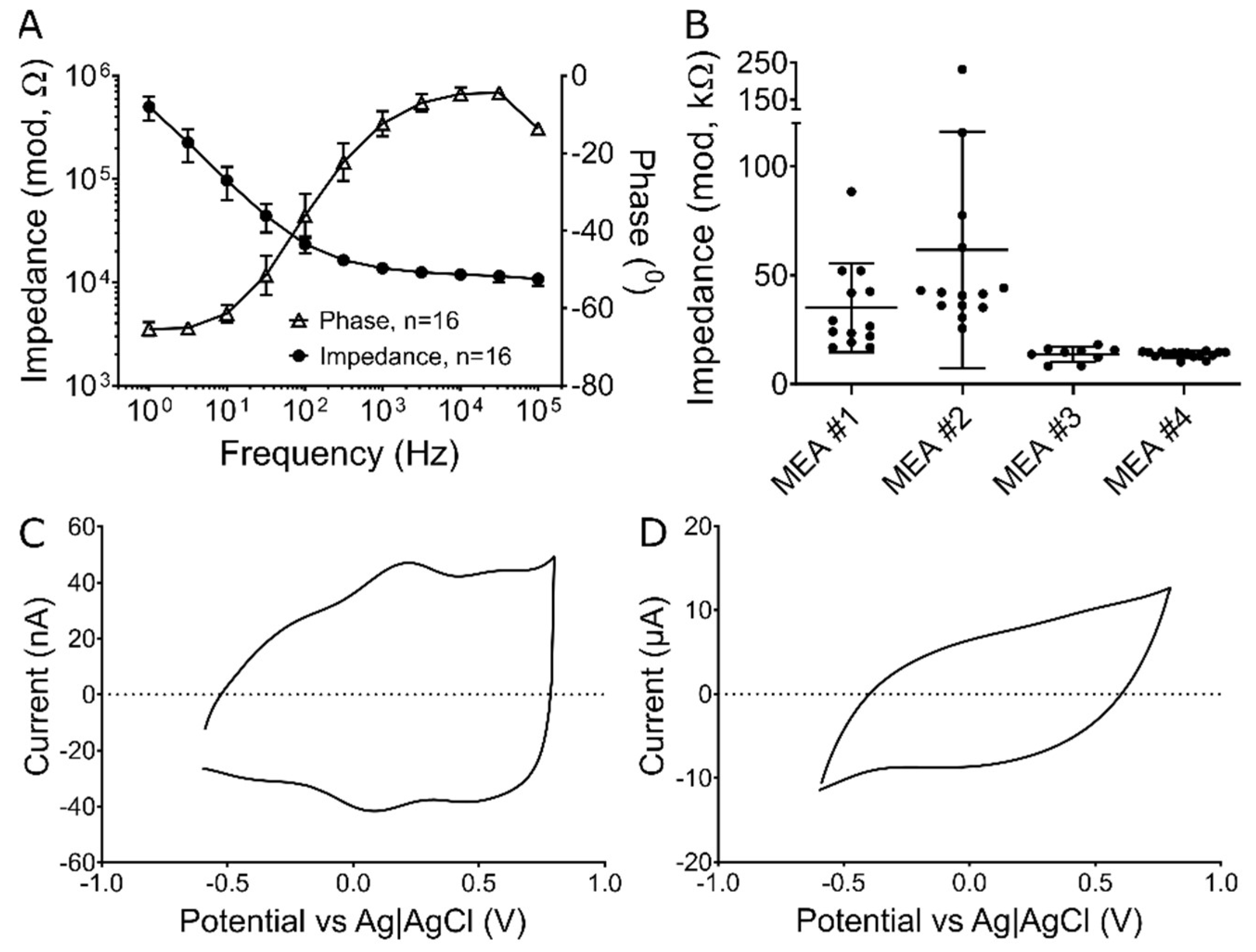
2.2. Electrochemical Testing
2.3. D iPSC Culture and Pharmacology
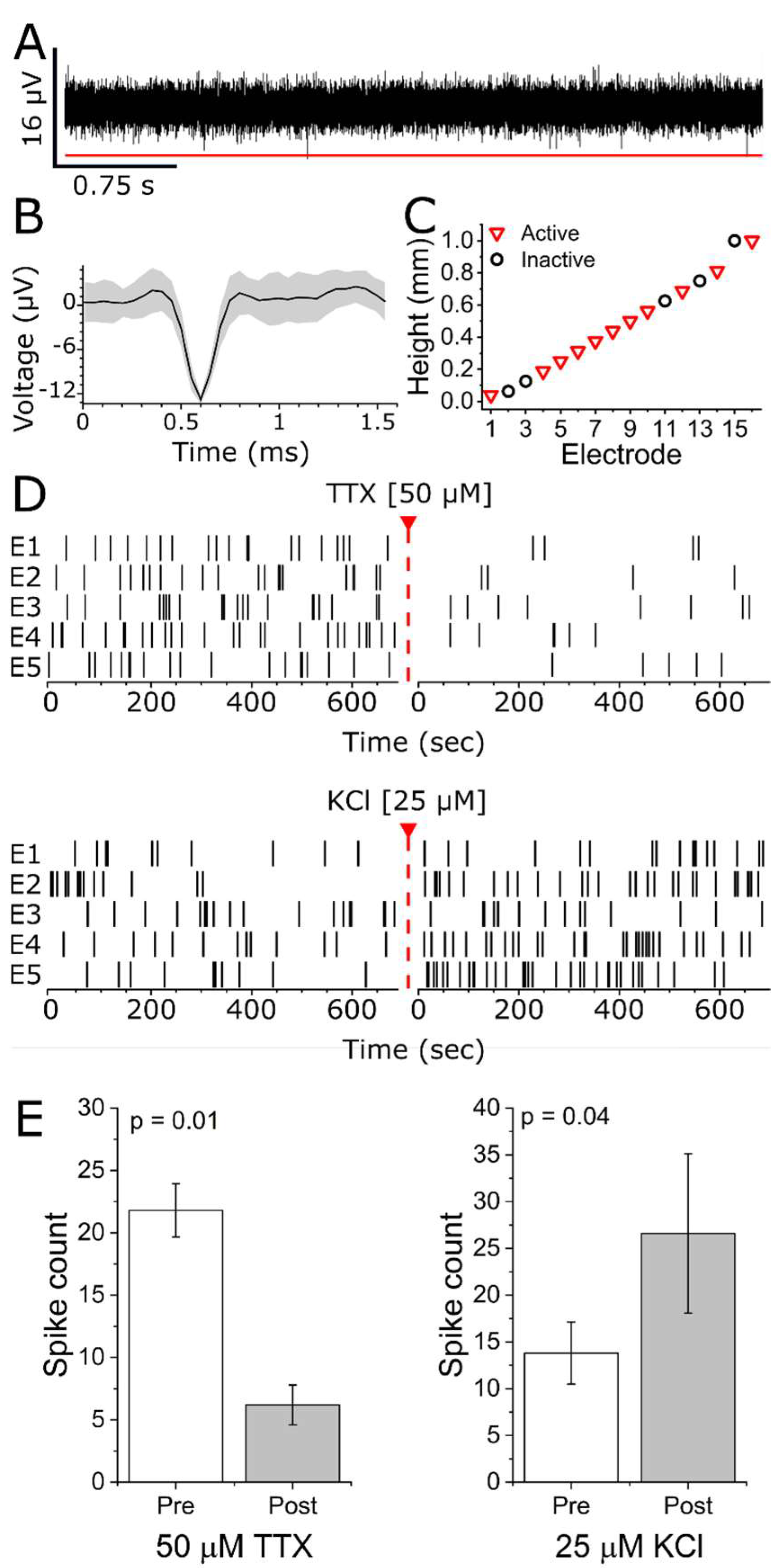
2.4. MEA Recordings and Data Analysis
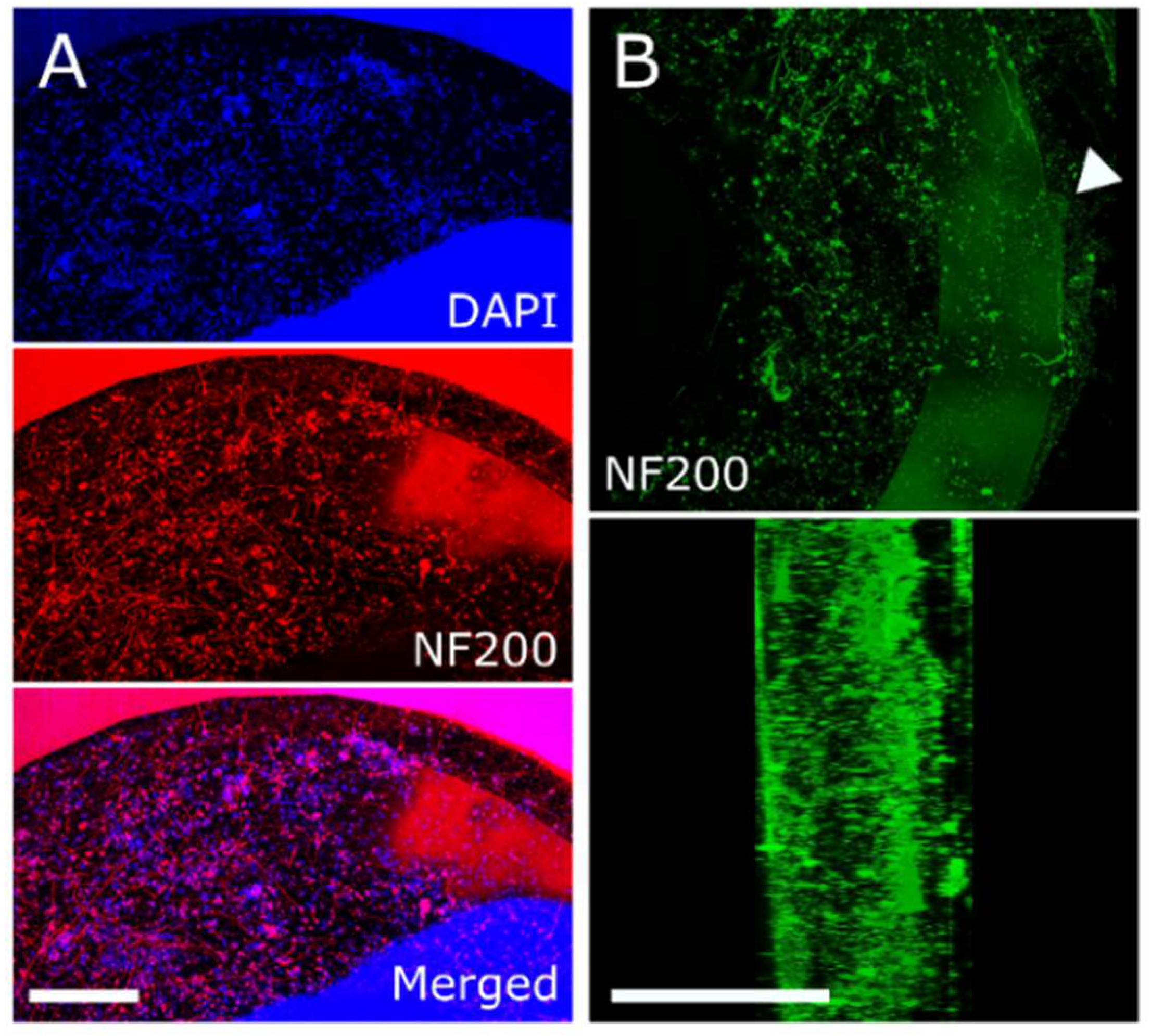
2.5. Immunocytochemistry and Imaging
3. Results
3.1. Preliminary Electrochemistry
3.2. Extracellular Action Potential Recordings
4. Discussion
4.1. Comparison with Other In Vitro 3D Electrophysiological Recording Approaches
4.2. Limitations and Potential Challenges
4.3. Advantages of 3D In Vitro Culture/Recordings
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Black, B.J.; Atmaramani, R.; Plagens, S.; Campbell, Z.; Dussor, G.; Price, T.J.; Pancrazio, J.J. Emerging neurotechnology for antinoceptive mechanisms and therapeutics discovery. Biosens. Bioelectron. 2018, 126, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Kamaly, N.; Xiao, Z.; Valencia, P.M.; Radovic-Moreno, A.F.; Farokhzad, O.C. Targeted polymeric therapeutic nanoparticles: Design, development and clinical translation. Biomaterial. 2011, 32, 4267–4274. [Google Scholar] [CrossRef] [PubMed]
- Accardi, M.V.; Pugsley, M.K.; Forster, R.; Troncy, E.; Huang, H.; Authier, S. The emerging role of in vitro electrophysiological methods in CNS safety pharmacology. J. Pharmacol. Toxicol. Methods 2016, 81, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Clements, M.; Thomas, N. High-Throughput Multi-Parameter Profiling of Electrophysiological Drug Effects in Human Embryonic Stem Cell Derived Cardiomyocytes Using Multi-Electrode Arrays. Toxicol. Sci. 2014, 140, 445–461. [Google Scholar] [CrossRef]
- Pancrazio, J.J.; Gopal, K.; Keefer, E.W.; Gross, G.W. Botulinum Toxin Suppression of CNS Network Activity In Vitro. J. Toxicol. 2014, 2014, 732913. [Google Scholar] [CrossRef]
- Valdivia, P.; Martin, M.; LeFew, W.R.; Ross, J.; Houck, K.A.; Shafer, T.J. Multi-well microelectrode array recordings detect neuroactivity of ToxCast compounds. NeuroToxicology 2014, 44, 204–217. [Google Scholar] [CrossRef]
- Manz, K.M.; Siemann, J.K.; McMahon, D.G.; Grueter, B.A. Patch-clamp and multi-electrode array electrophysiological analysis in acute mouse brain slices. STAR Protoc. 2021, 2, 100442. [Google Scholar] [CrossRef]
- Jäckel, D.; Bakkum, D.J.; Russell, T.L.; Müller, J.; Radivojevic, M.; Frey, U.; Franke, F.; Hierlemann, A. Combination of High-density Microelectrode Array and Patch Clamp Recordings to Enable Studies of Multisynaptic Integration. Sci. Rep. 2017, 7, 978. [Google Scholar] [CrossRef]
- John, S.E.; Apollo, N.V.; Opie, N.L.; Rind, G.S.; Ronayne, S.M.; May, C.N.; Oxley, T.J.; Grayden, D.B. In Vivo Impedance Characterization of Cortical Recording Electrodes Shows Dependence on Electrode Location and Size. IEEE Trans. Biomed. Eng. 2019, 66, 675–681. [Google Scholar] [CrossRef]
- Tcw, J.; Wang, M.; Pimenova, A.A.; Bowles, K.R.; Hartley, B.J.; Lacin, E.; Machlovi, S.I.; Abdelaal, R.; Karch, C.M.; Phatnani, H.; et al. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 9, 600–614. [Google Scholar] [CrossRef]
- Farrukh, A.; Zhao, S.; Del Campo, A. Microenvironments Designed to Support Growth and Function of Neuronal Cells. Front. Mater. 2018, 5, 62. [Google Scholar] [CrossRef]
- Taga, A.; Dastgheyb, R.; Habela, C.; Joseph, J.; Richard, J.-P.; Gross, S.K.; Lauria, G.; Lee, G.; Haughey, N.; Maragakis, N.J. Role of Human-Induced Pluripotent Stem Cell-Derived Spinal Cord Astrocytes in the Functional Maturation of Motor Neurons in a Multielectrode Array System. Stem Cells Transl. Med. 2019, 8, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.A.; Bosak, A.; Hogberg, H.T.; Hartung, T.; Moore, M.J. Advances in 3D neuronal microphysiological systems: Towards a functional nervous system on a chip. Vitr. Cell. Dev. Biol.-Anim. 2021, 57, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Antill-O’Brien, N.; Bourke, J.; O’Connell, C.D. Layer-By-Layer: The Case for 3D Bioprinting Neurons to Create Patient-Specific Epilepsy Models. Materials 2019, 12, 3218. [Google Scholar] [CrossRef] [PubMed]
- Hasturk, O.; Jordan, K.E.; Choi, J.; Kaplan, D.L. Enzymatically crosslinked silk and silk-gelatin hydrogels with tunable gelation kinetics, mechanical properties and bioactivity for cell culture and encapsulation. Biomaterials 2020, 232, 119720. [Google Scholar] [CrossRef]
- Tasnim, K.; Liu, J. Emerging Bioelectronics for Brain Organoid Electrophysiology. J. Mol. Biol. 2022, 434, 167165. [Google Scholar] [CrossRef]
- Huang, Q.; Tang, B.; Romero, J.C.; Yang, Y.; Elsayed, S.K.; Pahapale, G.; Lee, T.-J.; Pantoja, I.E.M.; Han, F.; Berlinicke, C.; et al. Shell microelectrode arrays (MEAs) for brain organoids. Sci. Adv. 2022, 8, eabq5031. [Google Scholar] [CrossRef]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef]
- Hartfield, E.M.; Yamasaki-Mann, M.; Fernandes, H.J.R.; Vowles, J.; James, W.; Cowley, S.; Wade-Martins, R. Physiological Characterisation of Human iPS-Derived Dopaminergic Neurons. PLoS ONE 2014, 9, e87388. [Google Scholar] [CrossRef]
- Soscia, D.A.; Lam, D.; Tooker, A.C.; Enright, H.A.; Triplett, M.; Karande, P.; Peters, S.K.G.; Sales, A.P.; Wheeler, E.K.; Fischer, N.O. A flexible 3-dimensional microelectrode array for in vitro brain models. Lab Chip 2020, 20, 901–911. [Google Scholar] [CrossRef]
- Li, Q.; Nan, K.; Le Floch, P.; Lin, Z.; Sheng, H.; Blum, T.S.; Liu, J. Cyborg Organoids: Implantation of Nanoelectronics via Organogenesis for Tissue-Wide Electrophysiology. Nano Lett. 2019, 19, 5781–5789. [Google Scholar] [CrossRef] [PubMed]
- Pancrazio, J.J.; Deku, F.; Ghazavi, A.; Stiller, A.M.; Rihani, R.; Frewin, C.L.; Varner, V.D.; Gardner, T.J.; Cogan, S.F. Thinking Small: Progress on Microscale Neurostimulation Technology. Neuromodulation 2017, 20, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Deku, F.; Cohen, Y.; Joshi-Imre, A.; Kanneganti, A.; Gardner, T.J.; Cogan, S.F. Amorphous silicon carbide ultramicroelectrode arrays for neural stimulation and recording. J. Neural Eng. 2018, 15, 16007. [Google Scholar] [CrossRef]
- Deku, F.; Frewin, C.L.; Stiller, A.; Cohen, Y.; Aqeel, S.; Joshi-Imre, A.; Black, B.; Gardner, T.J.; Pancrazio, J.J.; Cogan, S.F. Amorphous Silicon Carbide Platform for Next Generation Penetrating Neural Interface Designs. Micromachines 2018, 9, 480. [Google Scholar] [CrossRef]
- Ghazavi, A.; González-González, M.A.; Romero-Ortega, M.I.; Cogan, S.F. Intraneural ultramicroelectrode arrays for function-specific interfacing to the vagus nerve. Biosens. Bioelectron. 2020, 170, 112608. [Google Scholar] [CrossRef] [PubMed]
- Cogan, S.; Plante, T.; Ehrlich, J. Sputtered iridium oxide films (SIROFs) for low-impedance neural stimulation and recording electrodes. Annu. Int. Conf. IEEE Eng. Med. Biol. -Proc. 2004, 2, 4153–4156. [Google Scholar] [CrossRef]
- Maeng, J.; Chakraborty, B.; Geramifard, N.; Kang, T.; Rihani, R.T.; Joshi-Imre, A.; Cogan, S.F. High-charge-capacity sputtered iridium oxide neural stimulation electrodes deposited using water vapor as a reactive plasma constituent. J. Biomed. Mater. Res. Part B Appl. Biomater. 2020, 108, 880–891. [Google Scholar] [CrossRef]
- Jiang, J.; Willett, F.R.; Taylor, D.M. Relationship between microelectrode array impedance and chronic recording quality of single units and local field potentials. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBC 2014, Chicago, IL, USA, 26–30 August 2014; pp. 3045–3048. [Google Scholar] [CrossRef]
- Rihani, R.T.; Kim, H.; Black, B.J.; Atmaramani, R.; Saed, M.O.; Pancrazio, J.J.; Ware, T.H. Liquid Crystal Elastomer-Based Microelectrode Array for In Vitro Neuronal Recordings. Micromachines 2018, 9, 416. [Google Scholar] [CrossRef]
- Atmaramani, R.; Chakraborty, B.; Rihani, R.T.; Usoro, J.; Hammack, A.; Abbott, J.; Nnoromele, P.; Black, B.J.; Pancrazio, J.J.; Cogan, S.F. Ruthenium oxide based microelectrode arrays for in vitro and in vivo neural recording and stimulation. Acta Biomater. 2020, 101, 565–574. [Google Scholar] [CrossRef]
- Koklu, A.; Atmaramani, R.; Hammack, A.; Beskok, A.; Pancrazio, J.J.; Gnade, B.E.; Black, B.J. Gold nanostructure microelectrode arrays for in vitro recording and stimulation from neuronal networks. Nanotechnology 2019, 30, 235501. [Google Scholar] [CrossRef]
- Didier, C.M.; Kundu, A.; DeRoo, D.; Rajaraman, S. Development of in vitro 2D and 3D microelectrode arrays and their role in advancing biomedical research. J. Micromech. Microeng. 2020, 30, 103001. [Google Scholar] [CrossRef]
- Gaio, N.; Silvestri, C.; van Meer, B.; Vollebregt, S.; Mummery, C.L.; Dekker, R. Fabrication and Characterization of an Upside-Down Carbon Nanotube Microelectrode Array. IEEE Sens. J. 2016, 16, 8685–8691. [Google Scholar] [CrossRef]
- Nick, C.; Yadav, S.; Joshi, R.; Schneider, J.J.; Thielemann, C. A three-dimensional microelectrode array composed of vertically aligned ultra-dense carbon nanotube networks. Appl. Phys. Lett. 2015, 107, 13101. [Google Scholar] [CrossRef]
- Mestre, A.L.G.; Cerquido, M.; Inácio, P.M.C.; Asgarifar, S.; Lourenço, A.S.; Cristiano, M.L.S.; Aguiar, P.; Medeiros, M.C.R.; Araújo, I.M.; Ventura, J.; et al. Ultrasensitive gold micro-structured electrodes enabling the detection of extra-cellular long-lasting potentials in astrocytes populations. Sci. Rep. 2017, 7, 14284. [Google Scholar] [CrossRef] [PubMed]
- Fofonoff, T.; Martel, S.; Hatsopoulos, N.; Donoghue, J.; Hunter, I. Microelectrode Array Fabrication by Electrical Discharge Machining and Chemical Etching. IEEE Trans. Biomed. Eng. 2004, 51, 890–895. [Google Scholar] [CrossRef]
- Musick, K.; Khatamia, D.; Wheeler, B.C. Three-dimensional micro-electrode array for recording dissociated neuronal cultures. Lab Chip 2009, 9, 2036–2042. [Google Scholar] [CrossRef]
- Atmaramani, R.; Pancrazio, J.J.; Black, B.J. Adaptation of robust Z’ factor for assay quality assessment in microelectrode array based screening using adult dorsal root ganglion neurons. J. Neurosci. Methods 2020, 339, 108699. [Google Scholar] [CrossRef]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Fang, A.; Li, D.; Hao, Z.; Wang, L.; Pan, B.; Gao, L.; Qu, X.; He, J. Effects of astrocyte on neuronal outgrowth in a layered 3D structure. Biomed. Eng. Online 2019, 18, 74. [Google Scholar] [CrossRef]
- Hopkins, A.M.; DeSimone, E.; Chwalek, K.; Kaplan, D.L. 3D in vitro modeling of the central nervous system. Prog. Neurobiol. 2014, 125, 1–25. [Google Scholar] [CrossRef]
- Muller, Q.; Beaudet, M.-J.; De Serres-Bérard, T.; Bellenfant, S.; Flacher, V.; Berthod, F. Development of an innervated tissue-engineered skin with human sensory neurons and Schwann cells differentiated from iPS cells. Acta Biomater. 2018, 82, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Momin, A.; McNaughton, P.A. Regulation of firing frequency in nociceptive neurons by pro-inflammatory mediators. Exp. Brain Res. 2009, 196, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Djouhri, L.; Koutsikou, S.; Fang, X.; McMullan, S.; Lawson, S.N. Spontaneous Pain, Both Neuropathic and Inflammatory, Is Related to Frequency of Spontaneous Firing in Intact C-Fiber Nociceptors. J. Neurosci. 2006, 26, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Jang, J.H.; Kim, S.-P.; Lee, S.H.; Chung, S.-C.; Kim, I.Y.; Jang, D.P.; Jung, S.J. Analysis of Nociceptive Information Encoded in the Temporal Discharge Patterns of Cutaneous C-Fibers. Front. Comput. Neurosci. 2016, 10, 118. [Google Scholar] [CrossRef]
- Bernal, L.; Roza, C. Hyperpolarization-activated channels shape temporal patterns of ectopic spontaneous discharge in C-nociceptors after peripheral nerve injury. Eur. J. Pain 2018, 22, 1377–1387. [Google Scholar] [CrossRef]
- Tanner, K.; Reichling, D.; Gear, R.; Paul, S.; Levine, J. Altered temporal pattern of evoked afferent activity in a rat model of vincristine-induced painful peripheral neuropathy. Neuroscience 2003, 118, 809–817. [Google Scholar] [CrossRef]
| Ref. | Electrode # | Size (µm), Material | Insulation | Impedance (kΩ) |
|---|---|---|---|---|
| 16 | 50, SIROF | Polyimide | 13–60 | |
| [20] | 80 | 50, Pt | Polyimide | 37 ± 7 to 48 ± 6 |
| [21] | 16 | 20, PEDOT | SU-8 | 40–60 (estimate) |
| [17] | 3–7 | 50, PEDOT:PSS | SU-8 | 3–80 (estimate) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geramifard, N.; Lawson, J.; Cogan, S.F.; Black, B.J. A Novel 3D Helical Microelectrode Array for In Vitro Extracellular Action Potential Recording. Micromachines 2022, 13, 1692. https://doi.org/10.3390/mi13101692
Geramifard N, Lawson J, Cogan SF, Black BJ. A Novel 3D Helical Microelectrode Array for In Vitro Extracellular Action Potential Recording. Micromachines. 2022; 13(10):1692. https://doi.org/10.3390/mi13101692
Chicago/Turabian StyleGeramifard, Negar, Jennifer Lawson, Stuart F. Cogan, and Bryan James Black. 2022. "A Novel 3D Helical Microelectrode Array for In Vitro Extracellular Action Potential Recording" Micromachines 13, no. 10: 1692. https://doi.org/10.3390/mi13101692
APA StyleGeramifard, N., Lawson, J., Cogan, S. F., & Black, B. J. (2022). A Novel 3D Helical Microelectrode Array for In Vitro Extracellular Action Potential Recording. Micromachines, 13(10), 1692. https://doi.org/10.3390/mi13101692






