A Cell Culture Chip with Transparent, Micropillar-Decorated Bottom for Live Cell Imaging and Screening of Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Micropattern Bottom Chips
2.1.1. Wafer Production
2.1.2. Production of PMMA Micropatterns
2.1.3. PMMA Chip Production
2.1.4. Chip Assembly
2.2. Characterization of PMMA Films
2.2.1. Transparency of the Micropatterned PMMA
2.2.2. Scanning Electron Microscopy (SEM)
2.3. In Vitro Studies
2.4. Deformation Analysis
2.4.1. Fluorescence Microscopy
2.4.2. Digital Analysis of Nucleus Deformation
2.5. Cell Viability and Apoptosis and Proliferation
2.5.1. Live–Dead Analysis
2.5.2. DNA Quantification
2.5.3. Caspase Assay
2.5.4. Annexin V Staining
2.5.5. Immunocytochemical Staining of Ki-67 Protein
2.6. Real-Time Polymerase Chain Reaction (qRT-PCR)
2.7. Statistical Analysis
3. Results and Discussion
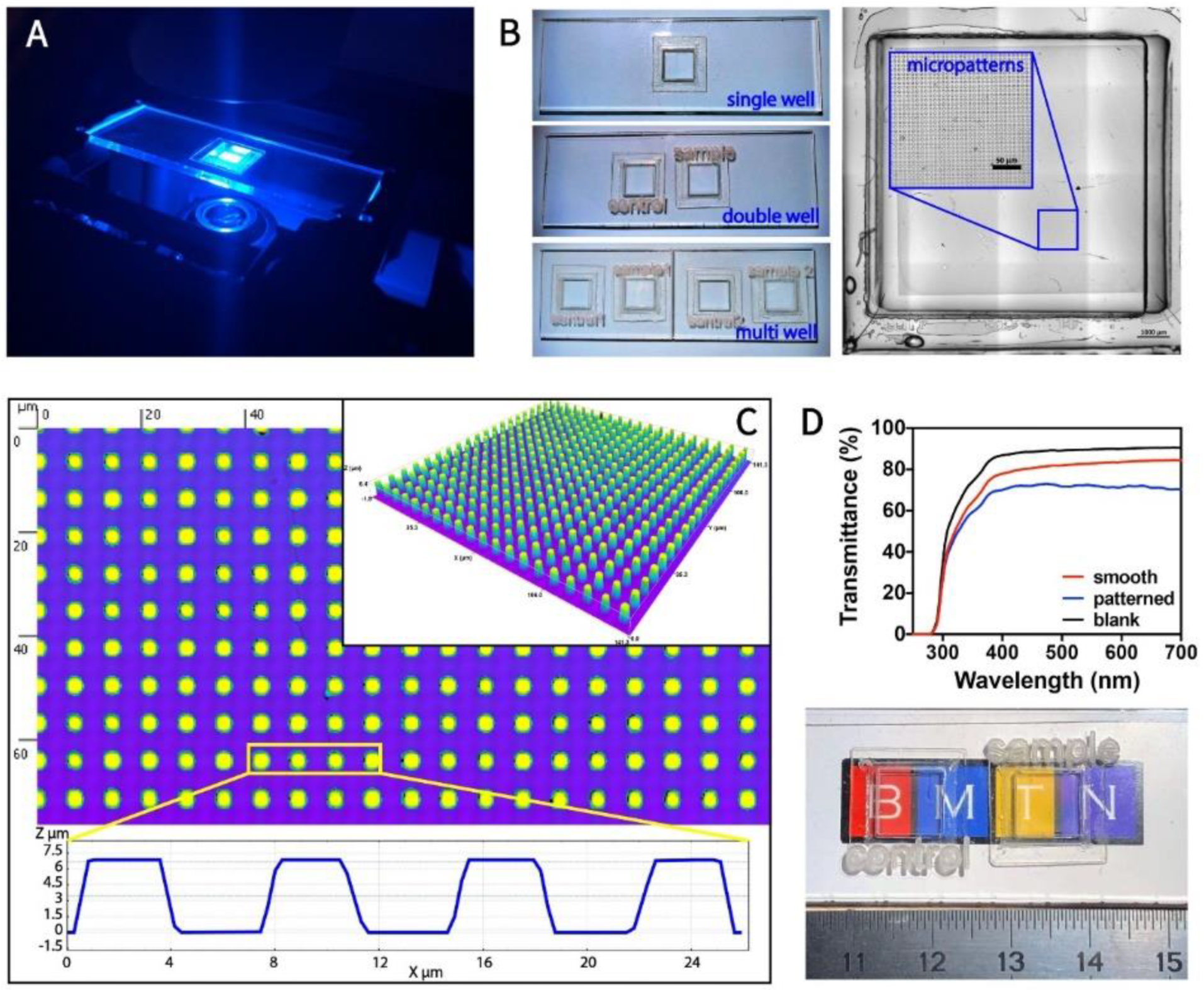
3.1. Fabrication and Characterization of Transparent, Micropillar-Bottomed Culture Chip (Ch-Pattern)
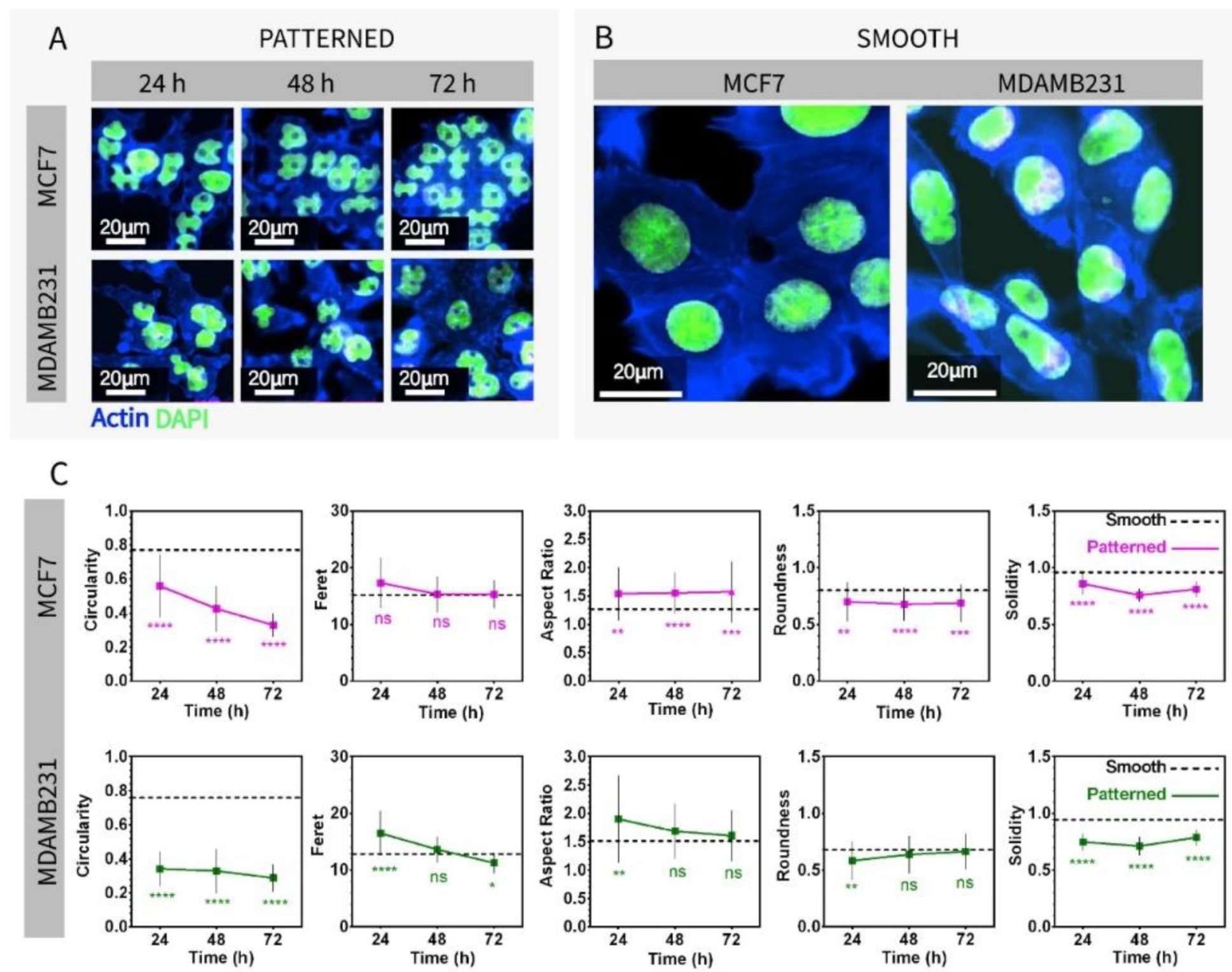
3.2. Imaging and Nuclear Shape Analysis of Breast Cancer Cells on Micropillar-Bottomed Culture Chip (Ch-Pattern)
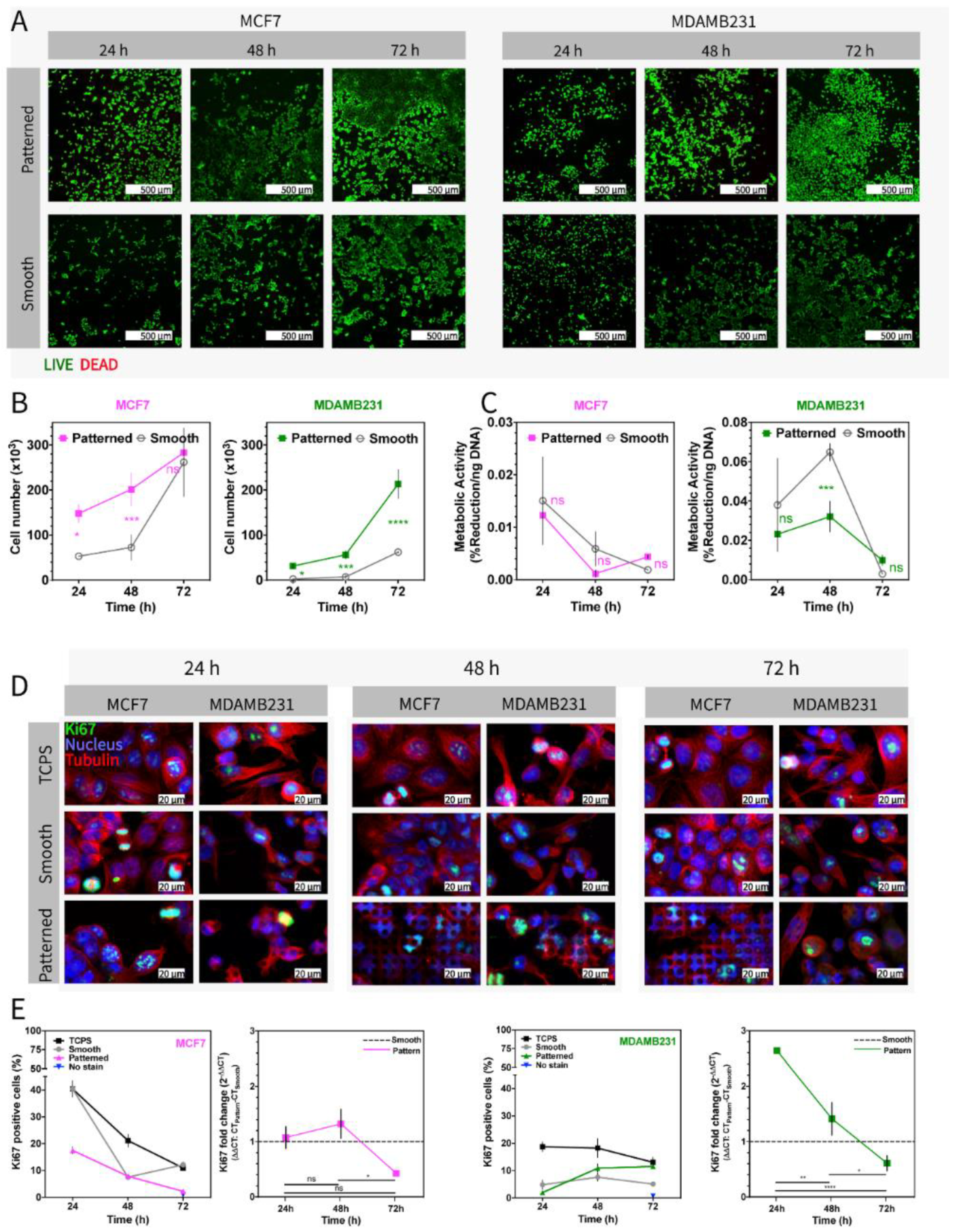
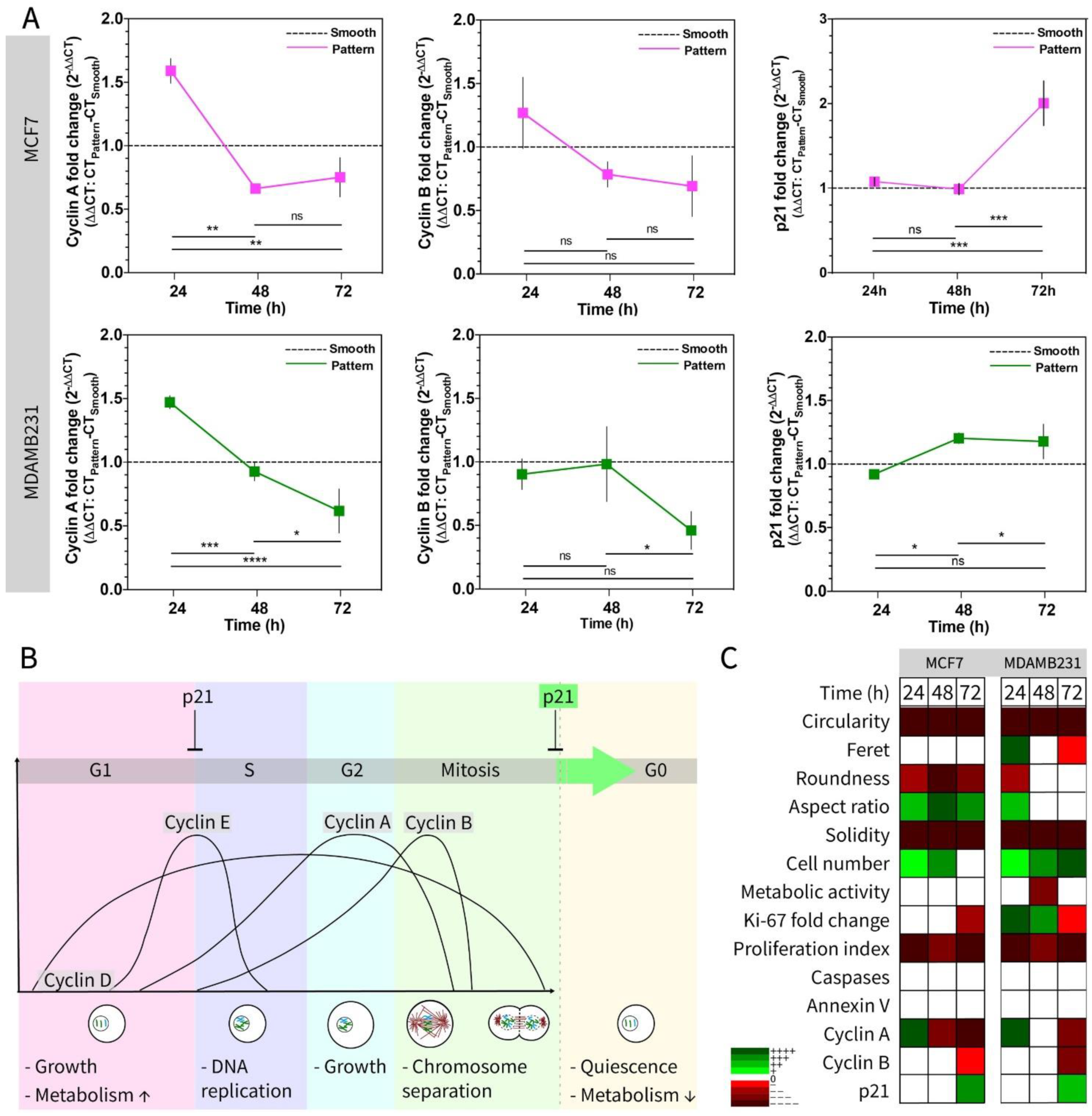
3.3. Testing the Metabolic Activity and Proliferation of Cells on Micropillar Bottom Culture Chip (Ch-Pattern)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Park, H.; Cannizzaro, C.; Vunjak-Novakovic, G.; Langer, R.; Vacanti, C.A.; Farokhzad, O.C. Nanofabrication and microfabrication of functional materials for tissue engineering. Tissue Eng. 2007, 13, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Peng, R.; Ding, J. Cell-Material Interactions Revealed Via Material Techniques of Surface Patterning. Adv. Mater. 2013, 25, 5257–5286. [Google Scholar] [CrossRef] [PubMed]
- Park, T.H.; Shuler, M.L. Integration of Cell Culture and Microfabrication Technology. Biotechnol. Prog. 2003, 19, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Lautenschläger, F.; Piel, M. Microfabricated devices for cell biology: All for one and one for all. Curr. Opin. Cell Biol. 2013, 25, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Peng, R.; Ding, J. The regulation of stem cell differentiation by cell-cell contact on micropatterned material surfaces. Biomaterials 2010, 31, 2470–2476. [Google Scholar] [CrossRef]
- García-Parra, P.; Cavaliere, F.; Maroto, M.; Bilbao, L.; Obieta, I.; de Munain, A.L.; Álava, J.I.; Izeta, A. Modeling neural differentiation on micropatterned substrates coated with neural matrix components. Front. Cell. Neurosci. 2012, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Tay, C.Y.; Pal, M.; Yu, H.; Leong, W.S.; Tan, N.S.; Ng, K.W.; Venkatraman, S.; Boey, F.; Leong, D.T.; Tan, L.P. Bio-inspired Micropatterned Platform to Steer Stem Cell Differentiation. Small 2011, 7, 1416–1421. [Google Scholar] [CrossRef]
- Sharma, A.D.; Zbarska, S.; Petersen, E.M.; Marti, M.E.; Mallapragada, S.K.; Sakaguchi, D.S. Oriented growth and transdifferentiation of mesenchymal stem cells towards a Schwann cell fate on micropatterned substrates. J. Biosci. Bioeng. 2016, 121, 325–335. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, P.; Cavanagh, B.L.; Ahearne, M. Influence of micropatterned substrates on keratocyte phenotype. Sci. Rep. 2020, 10, 6679. [Google Scholar] [CrossRef] [Green Version]
- Uzel, S.G.M.; Pavesi, A.; Kamm, R.D. Microfabrication and microfluidics for muscle tissue models. Prog. Biophys. Mol. Biol. 2014, 115, 279–293. [Google Scholar] [CrossRef]
- Takahashi, H.; Okano, T. Cell Sheet-Based Tissue Engineering for Organizing Anisotropic Tissue Constructs Produced Using Microfabricated Thermoresponsive Substrates. Adv. Healthc. Mater. 2015, 4, 2388–2407. [Google Scholar] [CrossRef]
- Vinci, B.; Cavallone, D.; Vozzi, G.; Mazzei, D.; Domenici, C.; Brunetto, M.; Ahluwalia, A. In vitro liver model using microfabricated scaffolds in a modular bioreactor. Biotechnol. J. 2010, 5, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Berthiaume, F.; Toner, M.; Yarmush, M.L.; Tilles, A.W. Microfabricated grooved substrates as platforms for bioartificial liver reactors. Biotechnol. Bioeng. 2005, 90, 632–644. [Google Scholar] [CrossRef]
- Ortega, Í.; Ryan, A.J.; Deshpande, P.; MacNeil, S.; Claeyssens, F. Combined microfabrication and electrospinning to produce 3-D architectures for corneal repair. Acta Biomater. 2013, 9, 5511–5520. [Google Scholar] [CrossRef]
- Iskandar, M.E.; Cipriano, A.F.; Lock, J.; Gott, S.C.; Rao, M.P.; Liu, H. Improved bone marrow stromal cell adhesion on micropatterned Titanium surfaces. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, San Diego, CA, USA, 28 August–1 September 2012; pp. 5666–5669. [Google Scholar]
- Schünemann, F.H.; Galárraga-Vinueza, M.E.; Magini, R.; Fredel, M.; Silva, F.; Souza, J.C.M.; Zhang, Y.; Henriques, B. Zirconia surface modifications for implant dentistry. Mater. Sci. Eng. C 2019, 98, 1294–1305. [Google Scholar] [CrossRef]
- Pelaez-Vargas, A.; Gallego-Perez, D.; Magallanes-Perdomo, M.; Fernandes, M.H.; Hansford, D.J.; De Aza, A.H.; Pena, P.; Monteiro, F.J. Isotropic micropatterned silica coatings on zirconia induce guided cell growth for dental implants. Dent. Mater. 2011, 27, 581–589. [Google Scholar] [CrossRef]
- Rabel, K.; Kohal, R.J.; Steinberg, T.; Tomakidi, P.; Rolauffs, B.; Adolfsson, E.; Palmero, P.; Fürderer, T.; Altmann, B. Controlling osteoblast morphology and proliferation via surface micro-topographies of implant biomaterials. Sci. Rep. 2020, 10, 12810. [Google Scholar] [CrossRef]
- Holthaus, M.G.; Treccani, L.; Rezwan, K. Comparison of micropatterning methods for ceramic surfaces. J. Eur. Ceram. Soc. 2011, 31, 2809–2817. [Google Scholar] [CrossRef]
- Sahlin, H.; Contreras, R.; Gaskill, D.F.; Bjursten, L.M.; Frangos, J.A. Anti-inflammatory properties of micropatterned titanium coatings. J. Biomed. Mater. Res. Part A 2006, 77, 43–49. [Google Scholar] [CrossRef]
- Persichetti, G.; Grimaldi, I.A.; Testa, G.; Bernini, R. Multifunctional optofluidic lab-on-chip platform for Raman and fluorescence spectroscopic microfluidic analysis. Lab a Chip 2017, 17, 2631–2639. [Google Scholar] [CrossRef]
- Samiei, E.; Tabrizian, M.; Hoorfar, M. A review of digital microfluidics as portable platforms for lab-on a-chip applications. Lab a Chip 2016, 16, 2376–2396. [Google Scholar] [CrossRef]
- Wongkaew, N.; Simsek, M.; Griesche, C.; Baeumner, A.J. Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-on-a-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem. Rev. 2019, 119, 120–194. [Google Scholar] [CrossRef]
- Jokerst, J.V.; McDevitt, J.T. Programmable nano-bio-chips: Multifunctional clinical tools for use at the point-of-care. Nanomedicine 2010, 5, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Neužil, P.; Giselbrecht, S.; Länge, K.; Huang, T.J.; Manz, A. Revisiting lab-on-a-chip technology for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 620–632. [Google Scholar] [CrossRef]
- Ahn, C.H.; Choi, J.W.; Beaucage, G.; Nevin, J.H.; Lee, J.B.; Puntambekar, A.; Lee, J.Y. Disposable smart lab on a chip for point-of-care clinical diagnostics. Proc. IEEE 2004, 92, 154–173. [Google Scholar] [CrossRef]
- Iandolo, D.; Pennacchio, F.A.; Mollo, V.; Rossi, D.; Dannhauser, D.; Cui, B.; Owens, R.M.; Santoro, F. Electron Microscopy for 3D Scaffolds-Cell Biointerface Characterization. Adv. Biosyst. 2019, 3, 1800103. [Google Scholar] [CrossRef]
- Kaiser, J.P.; Bruinink, A. Investigating cell-material interactions by monitoring and analysing cell migration. J. Mater. Sci. Mater. Med. 2004, 15, 429–435. [Google Scholar] [CrossRef]
- Missirlis, Y.F.; Spiliotis, A.D. Assessment of techniques used in calculating cell-material interactions. Biomol. Eng. 2002, 19, 287–294. [Google Scholar] [CrossRef]
- Punet, X.; Mauchauffé, R.; Rodríguez-Cabello, J.C.; Alonso, M.; Engel, E.; Mateos-Timoneda, M.A. Biomolecular functionalization for enhanced cell–material interactions of poly(methyl methacrylate) surfaces. Regen. Biomater. 2015, 2, 167–175. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.L.K.; Likhitpanichkul, M.; Ho, A.; Simmons, C.A. Integration of statistical modeling and high-content microscopy to systematically investigate cell-substrate interactions. Biomaterials 2010, 31, 2489–2497. [Google Scholar] [CrossRef]
- Wang, W.; Wang, S.; Liu, Q.; Wu, J.; Tao, N. Mapping single-cell-substrate interactions by surface plasmon resonance microscopy. Langmuir 2012, 28, 13373–13379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauch, K.D.; Ratner, B.D. Microscopy for Biomaterials Science. In Biomaterials Science: An Introduction to Materials, 3rd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 677–692. ISBN 9780123746269. [Google Scholar]
- Wollman, A.J.M.; Nudd, R.; Hedlund, E.G.; Leake, M.C. From animaculum to single molecules: 300 years of the light microscope. Open Biol. 2015, 5, 150019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, D.J.; Allan, V.J. Light microscopy techniques for live cell imaging. Science 2003, 300, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Thorn, K. A quick guide to light microscopy in cell biology. Mol. Biol. Cell 2016, 27, 219–222. [Google Scholar] [CrossRef]
- Ermis, M.; Akkaynak, D.; Chen, P.; Demirci, U.; Hasirci, V. A high throughput approach for analysis of cell nuclear deformability at single cell level. Sci. Rep. 2016, 6, 36917. [Google Scholar] [CrossRef] [Green Version]
- Agus, D.B. A physical sciences network characterization of non-tumorigenic and metastatic cells. Sci. Rep. 2013, 3, 1449. [Google Scholar] [CrossRef]
- Williams, D. Introduction. In Definitions of Biomaterials for the Twenty-First Century; Zhang, X., Williams, D., Eds.; Elsevier: Berkeley, CA, USA, 2019; Volume 1, pp. 1–10. ISBN 9780128182925. [Google Scholar]
- Barthes, J.; Özçelik, H.; Hindié, M.; Ndreu-Halili, A.; Hasan, A.; Vrana, N.E. Cell Microenvironment Engineering and Monitoring for Tissue Engineering and Regenerative Medicine: The Recent Advances. BioMed Res. Int. 2014, 2014, 921905. [Google Scholar] [CrossRef]
- Millet, M.; Ben Messaoud, R.; Luthold, C.; Bordeleau, F. Coupling microfluidic platforms, microfabrication, and tissue engineered scaffolds to investigate tumor cells mechanobiology. Micromachines 2019, 10, 418. [Google Scholar] [CrossRef] [Green Version]
- Hasirci, V.; Pepe-Mooney, B.J. Understanding the cell behavior on nano-/micro-patterned surfaces. Nanomedicine 2012, 7, 1375–1389. [Google Scholar] [CrossRef] [Green Version]
- Dike, L.E.; Chen, C.S.; Mrksich, M.; Tien, J.; Whitesides, G.M.; Ingber, D.E. Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. Vitr. Cell. Dev. Biol.-Anim. 1999, 35, 441–448. [Google Scholar] [CrossRef]
- Wang, W.; Itaka, K.; Ohba, S.; Nishiyama, N.; Chung, U.-i.; Yamasaki, Y.; Kataoka, K. 3D spheroid culture system on micropatterned substrates for improved differentiation efficiency of multipotent mesenchymal stem cells. Biomaterials 2009, 30, 2705–2715. [Google Scholar] [CrossRef]
- Recknor, J.B.; Sakaguchi, D.S.; Mallapragada, S.K. Directed growth and selective differentiation of neural progenitor cells on micropatterned polymer substrates. Biomaterials 2006, 27, 4098–4108. [Google Scholar] [CrossRef]
- Czöndör, K.; Garcia, M.; Argento, A.; Constals, A.; Breillat, C.; Tessier, B.; Thoumine, O. Micropatterned substrates coated with neuronal adhesion molecules for high-content study of synapse formation. Nat. Commun. 2013, 4, 2252. [Google Scholar] [CrossRef] [Green Version]
- Tay, C.Y.; Yu, H.; Pal, M.; Leong, W.S.; Tan, N.S.; Ng, K.W.; Leong, D.T.; Tan, L.P. Micropatterned matrix directs differentiation of human mesenchymal stem cells towards myocardial lineage. Exp. Cell Res. 2010, 316, 1159–1168. [Google Scholar] [CrossRef]
- Kusuma, S.; Smith, Q.; Facklam, A.; Gerecht, S. Micropattern size-dependent endothelial differentiation from a human induced pluripotent stem cell line. J. Tissue Eng. Regen. Med. 2017, 11, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Xia, L.; Zhai, D.; Zhang, N.; Liu, J.; Fang, B.; Chang, J.; Lin, K. Designing ordered micropatterned hydroxyapatite bioceramics to promote the growth and osteogenic differentiation of bone marrow stromal cells. J. Mater. Chem. B 2015, 3, 968–976. [Google Scholar] [CrossRef]
- Antmen, E.; Ermis, M.; Demirci, U.; Hasirci, V. Engineered natural and synthetic polymer surfaces induce nuclear deformation in osteosarcoma cells. J. Biomed. Mater. Res. Part B Appl. Biomater. 2019, 107, 366–376. [Google Scholar] [CrossRef]
- Davidson, P.M.; Özçelik, H.; Hasirci, V.; Reiter, G.; Anselme, K. Microstructured Surfaces Cause Severe but Non-Detrimental Deformation of the Cell Nucleus. Adv. Mater. 2009, 21, 3586–3590. [Google Scholar] [CrossRef]
- Antmen, E.; Demirci, U.; Hasirci, V. Amplification of nuclear deformation of breast cancer cells by seeding on micropatterned surfaces to better distinguish their malignancies. Colloids Surf. B Biointerfaces 2019, 183, 110402. [Google Scholar] [CrossRef]
- Liu, R.; Liu, Q.; Pan, Z.; Liu, X.; Ding, J. Cell Type and Nuclear Size Dependence of the Nuclear Deformation of Cells on a Micropillar Array. Langmuir 2019, 35, 7469–7477. [Google Scholar] [CrossRef]
- Badique, F.; Stamov, D.R.; Davidson, P.M.; Veuillet, M.; Reiter, G.; Freund, J.N.; Franz, C.M.; Anselme, K. Directing nuclear deformation on micropillared surfaces by substrate geometry and cytoskeleton organization. Biomaterials 2013, 34, 2991–3001. [Google Scholar] [CrossRef]
- Hasturk, O.; Sivas, A.; Karasozen, B.; Demirci, U.; Hasirci, N.; Hasirci, V. Quantification of Type, Timing, and Extent of Cell Body and Nucleus Deformations Caused by the Dimensions and Hydrophilicity of Square Prism Micropillars. Adv. Healthc. Mater. 2016, 5, 2972–2982. [Google Scholar] [CrossRef]
- Mallini, P.; Lennard, T.; Kirby, J.; Meeson, A. Epithelial-to-mesenchymal transition: What is the impact on breast cancer stem cells and drug resistance. Cancer Treat. Rev. 2014, 40, 341–348. [Google Scholar] [CrossRef]
- Boon Yin, K. Breast Cancer-Focusing Tumor Microenvironment, Stem Cells and Metastasis; The Mesenchymal-Like Phenotype of the MDA-MB-231 Cell Line; IntechOpen: London, UK, 2011; pp. 385–402. ISBN 978-953-307-766-6. [Google Scholar]
- Peela, N.; Sam, F.S.; Christenson, W.; Truong, D.; Watson, A.W.; Mouneimne, G.; Ros, R.; Nikkhah, M. A three dimensional micropatterned tumor model for breast cancer cell migration studies. Biomaterials 2016, 81, 72–83. [Google Scholar] [CrossRef]
- Antmen, E.; Demirci, U.; Hasirci, V. Micropatterned Surfaces: Micropatterned Surfaces Expose the Coupling between Actin Cytoskeleton-Lamin/Nesprin and Nuclear Deformability of Breast Cancer Cells with Different Malignancies. Adv. Biol. 2021, 5, 2170012. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.P.; Li, W.J.; Zhu, Y.F.; Huang, S.Y.; Fang, S.Y.; Shen, L.; Gao, Y.L. CDKN3 knockdown reduces cell proliferation, invasion and promotes apoptosis in human ovarian cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 4535–4544. [Google Scholar] [PubMed]
- Hsu, C.C.; Lee, Y.C.; Yeh, S.H.; Chen, C.H.; Wu, C.C.; Wang, T.Y.; Chen, Y.N.; Hung, L.Y.; Liu, Y.W.; Chen, H.K.; et al. 58-kDa microspherule protein (MSP58) is novel brahma-related gene 1 (BRG1)-associated protein that modulates p53/p21 senescence pathway. J. Biol. Chem. 2012, 287, 22533–22548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuokkanen, S.; Chen, B.; Ojalvo, L.; Benard, L.; Santoro, N.; Pollard, J.W. Genomic Profiling of MicroRNAs and Messenger RNAs Reveals Hormonal Regulation in MicroRNA Expression in Human Endometrium1. Biol. Reprod. 2010, 82, 791–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadikovic, B.; Thorner, P.; Chilton-MacNeill, S.; Martin, J.W.; Cervigne, N.K.; Squire, J.; Zielenska, M. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer 2010, 10, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bie, L.; Zhao, G.; Ju, Y.; Zhang, B. Integrative genomic analysis identifies CCNB1 and CDC2 as candidate genes associated with meningioma recurrence. Cancer Genet. 2011, 204, 536–540. [Google Scholar] [CrossRef]
- Ferraro, R.M.; Ginestra, P.S.; Lanzi, G.; Giliani, S.; Ceretti, E. Production of micro-patterned substrates to direct human iPSCs-derived neural stem cells orientation and interaction. Procedia Cirp. 2017, 65, 225–230. [Google Scholar] [CrossRef]
- Elacqua, J.J.; McGregor, A.L.; Lammerding, J. Automated analysis of cell migration and nuclear envelope rupture in confined environments. PLoS ONE 2018, 13, e0195664. [Google Scholar] [CrossRef] [Green Version]
- Hasturk, O.; Ermis, M.; Demirci, U.; Hasirci, N.; Hasirci, V. Square prism micropillars on poly(methyl methacrylate) surfaces modulate the morphology and differentiation of human dental pulp mesenchymal stem cells. Colloids Surf. B Biointerfaces 2019, 178, 44–55. [Google Scholar] [CrossRef]
- Hasturk, O.; Ermis, M.; Demirci, U.; Hasirci, N.; Hasirci, V. Square prism micropillars improve osteogenicity of poly(methyl methacrylate) surfaces. J. Mater. Sci. Mater. Med. 2018, 29, 53. [Google Scholar] [CrossRef]
- Seo, C.H.; Furukawa, K.; Suzuki, Y.; Kasagi, N.; Ichiki, T.; Ushida, T. A topographically optimized substrate with well-ordered lattice micropatterns for enhancing the osteogenic differentiation of murine mesenchymal stem cells. Macromol. Biosci. 2011, 11, 938–945. [Google Scholar] [CrossRef]
- Huang, Q.; Elkhooly, T.A.; Liu, X.; Zhang, R.; Yang, X.; Shen, Z.; Feng, Q. Effects of hierarchical micro/nano-topographies on the morphology, proliferation and differentiation of osteoblast-like cells. Colloids Surf. B Biointerfaces 2016, 145, 37–45. [Google Scholar] [CrossRef]
- Wang, X.; Nakamoto, T.; Dulińska-Molak, I.; Kawazoe, N.; Chen, G. Regulating the stemness of mesenchymal stem cells by tuning micropattern features. J. Mater. Chem. B 2016, 4, 37–45. [Google Scholar] [CrossRef]
- Sousa, M.P.; Caridade, S.G.; Mano, J.F. Control of Cell Alignment and Morphology by Redesigning ECM-Mimetic Nanotopography on Multilayer Membranes. Adv. Healthc. Mater. 2017, 6, 1601462. [Google Scholar] [CrossRef]
- Tusamda Wakhloo, N.; Anders, S.; Badique, F.; Eichhorn, M.; Brigaud, I.; Petithory, T.; Vassaux, M.; Milan, J.-L.; Freund, J.-N.; Rühe, J.; et al. Actomyosin, vimentin and LINC complex pull on osteosarcoma nuclei to deform on micropillar topography. Biomaterials 2020, 234, 119746. [Google Scholar] [CrossRef]
- Moroni, L.; Lee, L.P. Micropatterned hot-embossed polymeric surfaces influence cell proliferation and alignment. J. Biomed. Mater. Res. Part A 2009, 88A, 644–653. [Google Scholar] [CrossRef]
- Kim, E.J.; Boehm, C.A.; Mata, A.; Fleischman, A.J.; Muschler, G.F.; Roy, S. Post microtextures accelerate cell proliferation and osteogenesis. Acta Biomater. 2010, 6, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Nagayama, K.; Hamaji, Y.; Sato, Y.; Matsumoto, T. Mechanical trapping of the nucleus on micropillared surfaces inhibits the proliferation of vascular smooth muscle cells but not cervical cancer HeLa cells. J. Biomech. 2015, 48, 1796–1803. [Google Scholar] [CrossRef]
- Ge, J.; Yang, H.; Chen, Y.; Yan, Q.; Wu, C.; Zou, J. PMMA Bone Cement Acts on the Hippo/YAP Pathway to Regulate CTGF and Induce Intervertebral Disc Degeneration. ACS Biomater. Sci. Eng. 2019, 5, 3293–3302. [Google Scholar] [CrossRef]
- Mahadevan, G.; Valiyaveettil, S. Understanding the interactions of poly(methyl methacrylate) and poly(vinyl chloride) nanoparticles with BHK-21 cell line. Sci. Rep. 2021, 11, 2089. [Google Scholar] [CrossRef]
- Yoon, M.H.; Kang, S.-m; Lee, S.J.; Woo, T.G.; Oh, A.Y.; Park, S.; Ha, N.C.; Park, B.J. p53 induces senescence through Lamin A/C stabilization-mediated nuclear deformation. Cell Death Dis. 2019, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Shim, M.K.; Yoon, H.Y.; Lee, S.; Jo, M.K.; Park, J.; Kim, J.H.; Jeong, S.Y.; Kwon, I.C.; Kim, K. Caspase-3/-7-Specific Metabolic Precursor for Bioorthogonal Tracking of Tumor Apoptosis. Sci. Rep. 2017, 7, 16635. [Google Scholar] [CrossRef] [Green Version]
- Logue, S.E.; Elgendy, M.; Martin, S.J. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat. Protoc. 2009, 4, 1383–1395. [Google Scholar] [CrossRef]
- Lammerding, J.; Hsiao, J.; Schulze, P.C.; Kozlov, S.; Stewart, C.L.; Lee, R.T. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005, 170, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Jiao, F.; Zhao, Y.; Sun, Q.; Huo, B. Spreading area and shape regulate the apoptosis and osteogenesis of mesenchymal stem cells on circular and branched micropatterned islands. J. Biomed. Mater. Res. Part A 2020, 108, 2080–2089. [Google Scholar] [CrossRef]
- Dash, B.C.; El-Deiry, W.S. Phosphorylation of p21 in G2/M Promotes Cyclin B-Cdc2 Kinase Activity. Mol. Cell. Biol. 2005, 25, 3364–3387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ermis, M.; Antmen, E.; Kuren, O.; Demirci, U.; Hasirci, V. A Cell Culture Chip with Transparent, Micropillar-Decorated Bottom for Live Cell Imaging and Screening of Breast Cancer Cells. Micromachines 2022, 13, 93. https://doi.org/10.3390/mi13010093
Ermis M, Antmen E, Kuren O, Demirci U, Hasirci V. A Cell Culture Chip with Transparent, Micropillar-Decorated Bottom for Live Cell Imaging and Screening of Breast Cancer Cells. Micromachines. 2022; 13(1):93. https://doi.org/10.3390/mi13010093
Chicago/Turabian StyleErmis, Menekse, Ezgi Antmen, Ozgur Kuren, Utkan Demirci, and Vasif Hasirci. 2022. "A Cell Culture Chip with Transparent, Micropillar-Decorated Bottom for Live Cell Imaging and Screening of Breast Cancer Cells" Micromachines 13, no. 1: 93. https://doi.org/10.3390/mi13010093
APA StyleErmis, M., Antmen, E., Kuren, O., Demirci, U., & Hasirci, V. (2022). A Cell Culture Chip with Transparent, Micropillar-Decorated Bottom for Live Cell Imaging and Screening of Breast Cancer Cells. Micromachines, 13(1), 93. https://doi.org/10.3390/mi13010093






