Rapid Fabrication by Digital Light Processing 3D Printing of a SlipChip with Movable Ports for Local Delivery to Ex Vivo Organ Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Device Design, 3D Printing, and Laser Etching
2.2. Fluorination of Resin Surface and Contact Angle Measurements
2.3. Surface Profilometry
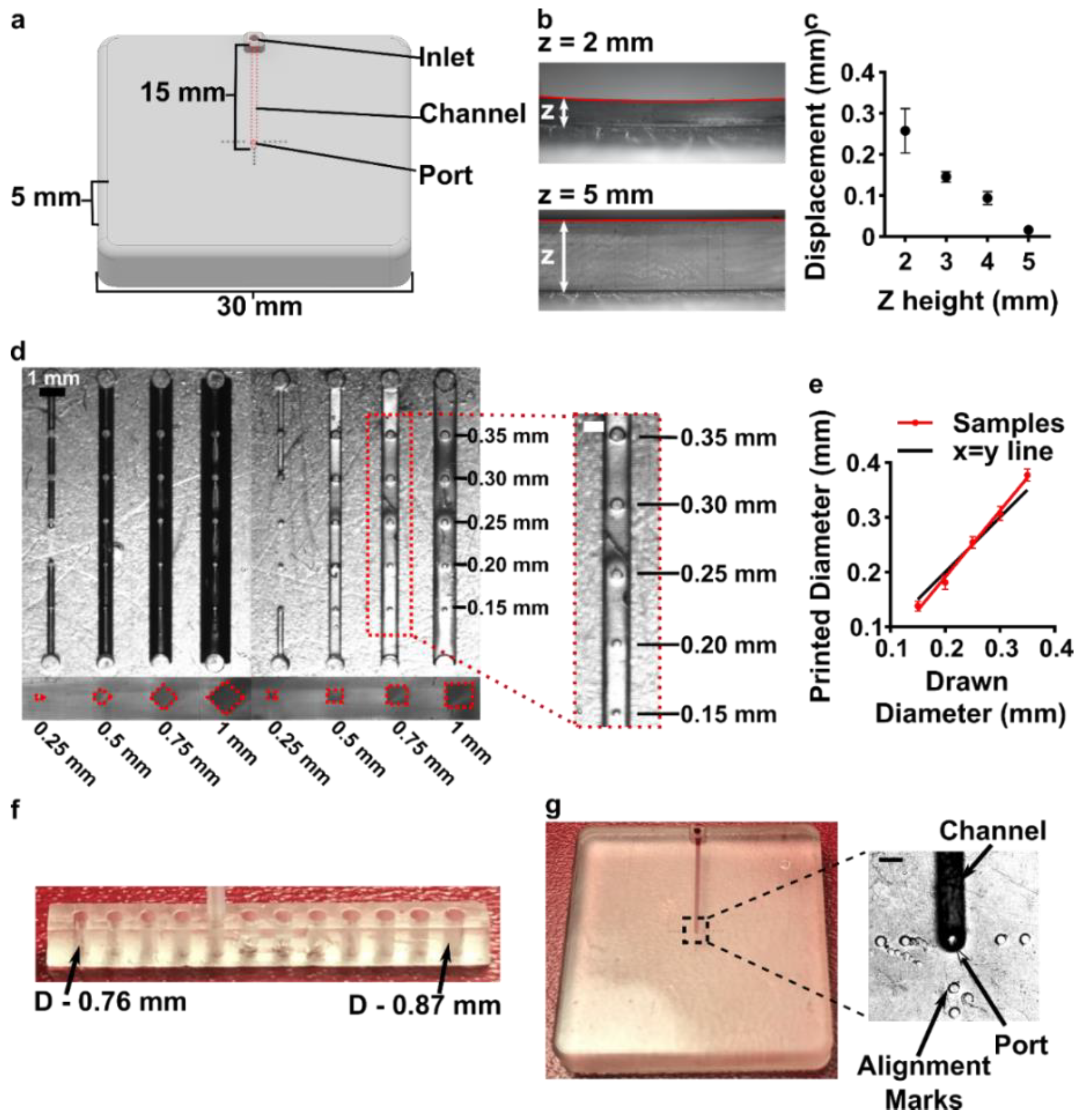
2.4. Measurement of Curvature of Printed Pieces
2.5. Animal Work and Tissue Slice Collection
2.6. Analysis of Tissue Viability
2.7. Assembly and Local Delivery with the 3D Printed Slipchip
2.8. Analysis of Delivery Widths
2.9. Delivery to Lymph Node Tissue
3. Results
3.1. Design Goals for a 3D Printed SlipChip with Movable Ports
3.2. Selection of Materials and Print Conditions for Transparency, Smoothness, Fluorination, and Cytocompatibility
3.3. Optimizing the Design and Printability of the Delivery Component
3.4. Optimization to Minimize Port Size and Preserve Optical Transparency of the Chamber Component
3.5. The Assembled 3D Printed SlipChip Delivers Fluid without Leakage into the Gap
3.6. Validation of Local Delivery to Hydrogel and Tissue Slices on the 3D Printed Chip
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shallan, A.I.; Smejkal, P.; Corban, M.; Guijt, R.M.; Breadmore, M.C. Cost-Effective Three-Dimensional Printing of Visibly Transparent Microchips within Minutes. Anal. Chem. 2014, 86, 3124–3130. [Google Scholar] [CrossRef]
- Yazdi, A.A.; Popma, A.; Wong, W.; Nguyen, T.; Pan, Y.; Xu, J. 3D Printing: An Emerging Tool for Novel Microfluidics and Lab-on-a-Chip Applications. Microfluid. Nanofluid. 2016, 20, 50. [Google Scholar] [CrossRef]
- Bhattacharjee, N.; Urrios, A.; Kang, S.; Folch, A. The Upcoming 3D-Printing Revolution in Microfluidics. Lab Chip 2016, 16, 1720–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, W.; Li, L.; Nichols, K.P.; Ismagilov, R.F. SlipChip. Lab Chip 2009, 9, 2286–2292. [Google Scholar] [CrossRef]
- Pompano, R.R.; Platt, C.E.; Karymov, M.A.; Ismagilov, R.F. Control of Initiation, Rate, and Routing of Spontaneous Capillary-Driven Flow of Liquid Droplets through Microfluidic Channels on SlipChip. Langmuir 2012, 28, 1931–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Du, W.; Ismagilov, R. User-Loaded SlipChip for Equipment-Free Multiplexed Nanoliter-Scale Experiments. J. Am. Chem. Soc. 2010, 132, 106–111. [Google Scholar] [CrossRef] [Green Version]
- Zhukov, D.V.; Khorosheva, E.M.; Khazaei, T.; Du, W.; Selck, D.A.; Shishkin, A.A.; Ismagilov, R.F. Microfluidic SlipChip Device for Multistep Multiplexed Biochemistry on a Nanoliter Scale. Lab Chip 2019, 19, 3200–3211. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-W.; Peng, C.-C.; Liao, W.-H.; Tung, Y.-C. Polydimethylsiloxane SlipChip for Mammalian Cell Culture Applications. Analyst 2015, 140, 7355–7365. [Google Scholar] [CrossRef]
- Catterton, M.A.; Dunn, A.F.; Pompano, R.R. User-Defined Local Stimulation of Live Tissue through a Movable Microfluidic Port. Lab Chip 2018, 18, 2003–2012. [Google Scholar] [CrossRef]
- Culbertson, C.T.; Sibbitts, J.; Sellens, K.; Jia, S. Fabrication of Glass Microfluidic Devices. In Microfluidic Electrophoresis: Methods and Protocols; Dutta, D., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 1–12. ISBN 978-1-4939-8964-5. [Google Scholar]
- Lyu, W.; Yu, M.; Qu, H.; Yu, Z.; Du, W.; Shen, F. Slip-Driven Microfluidic Devices for Nucleic Acid Analysis. Biomicrofluidics 2019, 13, 041502. [Google Scholar] [CrossRef]
- Yu, M.; Chen, X.; Qu, H.; Ma, L.; Xu, L.; Lv, W.; Wang, H.; Ismagilov, R.F.; Li, M.; Shen, F. Multistep SlipChip for the Generation of Serial Dilution Nanoliter Arrays and Hepatitis B Viral Load Quantification by Digital Loop Mediated Isothermal Amplification. Anal. Chem. 2019, 91, 8751–8755. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ismagilov, R.F. Protein Crystallization Using Microfluidic Technologies Based on Valves, Droplets, and SlipChip. Annu. Rev. Biophys. 2010, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Datta, S.S.; Karymov, M.A.; Pan, Q.; Begolo, S.; Ismagilov, R.F. Individually Addressable Arrays of Replica Microbial Cultures Enabled by Splitting SlipChips. Integr. Biol. 2014, 6, 796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, F.; Sun, B.; Kreutz, J.E.; Davydova, E.K.; Du, W.; Reddy, P.L.; Joseph, L.J.; Ismagilov, R.F. Multiplexed Quantification of Nucleic Acids with Large Dynamic Range Using Multivolume Digital RT-PCR on a Rotational SlipChip Tested with HIV and Hepatitis C Viral Load. J. Am. Chem. Soc. 2011, 133, 17705–17712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Scida, K.; Crooks, R.M. Detection of Hepatitis B Virus DNA with a Paper Electrochemical Sensor. Anal. Chem. 2015, 87, 9009–9015. [Google Scholar] [CrossRef]
- Banerjee, I.; Salih, T.; Ramachandraiah, H.; Erlandsson, J.; Pettersson, T.; Araújo, A.C.; Karlsson, M.; Russom, A. Slipdisc: A Versatile Sample Preparation Platform for Point of Care Diagnostics. RSC Adv. 2017, 7, 35048–35054. [Google Scholar] [CrossRef] [Green Version]
- Schoepp, N.G.; Schlappi, T.S.; Curtis, M.S.; Butkovich, S.S.; Miller, S.; Humphries, R.M.; Ismagilov, R.F. Rapid Pathogen-Specific Phenotypic Antibiotic Susceptibility Testing Using Digital LAMP Quantification in Clinical Samples. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Hull, C.W. Apparatus for Production of Three-Dimensional Objects by Stereolithography. U.S. Patent 4575330, 11 March 1986. [Google Scholar]
- Quan, H.; Zhang, T.; Xu, H.; Luo, S.; Nie, J.; Zhu, X. Photo-Curing 3D Printing Technique and Its Challenges. Bioact. Mater. 2020, 5, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Waheed, S.; Cabot, J.M.; Macdonald, N.P.; Lewis, T.; Guijt, R.M.; Paull, B.; Breadmore, M.C. 3D Printed Microfluidic Devices: Enablers and Barriers. Lab Chip 2016, 16, 1993–2013. [Google Scholar] [CrossRef] [Green Version]
- Amin, R.; Knowlton, S.; Hart, A.; Yenilmez, B.; Ghaderinezhad, F.; Katebifar, S.; Messina, M.; Khademhosseini, A.; Tasoglu, S. 3D-Printed Microfluidic Devices. Biofabrication 2016, 8, 022001. [Google Scholar] [CrossRef]
- Mehta, V.; Rath, S.N. 3D Printed Microfluidic Devices: A Review Focused on Four Fundamental Manufacturing Approaches and Implications on the Field of Healthcare. Bio-Des. Manuf. 2021, 4, 311–343. [Google Scholar] [CrossRef]
- Catterton, M.A.; Montalbine, A.N.; Pompano, R.R. Selective Fluorination of the Surface of Polymeric Materials after Stereolithography 3D Printing. Langmuir 2021. [Google Scholar] [CrossRef]
- Urrios, A.; Parra-Cabrera, C.; Bhattacharjee, N.; Gonzalez-Suarez, A.M.; Rigat-Brugarolas, L.G.; Nallapatti, U.; Samitier, J.; DeForest, C.A.; Posas, F.; Garcia-Cordero, J.L.; et al. 3D-Printing of Transparent Bio-Microfluidic Devices in PEG-DA. Lab Chip 2016, 16, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.E.; Belanger, M.C.; Woodroof, J.F.; Pompano, R.R. Spatially Resolved Microfluidic Stimulation of Lymphoid Tissue Ex Vivo. Analyst 2017, 142, 649–659. [Google Scholar] [CrossRef]
- Chang, T.C.; Mikheev, A.M.; Huynh, W.; Monnat, R.J.; Rostomily, R.C.; Folch, A. Parallel Microfluidic Chemosensitivity Testing on Individual Slice Cultures. Lab Chip 2014, 14, 4540–4551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horowitz, L.F.; Rodriguez, A.D.; Ray, T.; Folch, A. Microfluidics for Interrogating Live Intact Tissues. Microsyst. Nanoeng. 2020, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, J.S.; Caicedo, H.H.; Fall, C.P.; Eddington, D.T. Microfluidic Add-on for Standard Electrophysiology Chambers. Lab Chip 2008, 8, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.E.; Pompano, R.R. Diffusion of Cytokines in Live Lymph Node Tissue Using Microfluidic Integrated Optical Imaging. Anal. Chim. Acta 2018, 1000, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Roach, L.S.; Song, H.; Ismagilov, R.F. Controlling Nonspecific Protein Adsorption in a Plug-Based Microfluidic System by Controlling Interfacial Chemistry Using Fluorous-Phase Surfactants. Anal. Chem. 2005, 77, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Cristini, V.; Tan, Y.-C. Theory and Numerical Simulation of Droplet Dynamics in Complex Flows—a Review. Lab Chip 2004, 4, 257–264. [Google Scholar] [CrossRef]
- Belanger, M.C.; Ball, A.G.; Catterton, M.A.; Kinman, A.W.L.; Anbaei, P.; Groff, B.D.; Melchor, S.J.; Lukens, J.R.; Ross, A.E.; Pompano, R.R. Acute Lymph Node Slices Are a Functional Model System to Study Immunity Ex Vivo. ACS Pharmacol. Transl. Sci. 2021, 4, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Ball, A.G.; Belanger, M.C.; Pompano, R.R. Detergent Wash Improves Vaccinated Lymph Node Handling Ex Vivo. J. Immunol. Methods 2021, 489, 112943. [Google Scholar] [CrossRef] [PubMed]
- Karalekas, D.; Rapti, D.; Gdoutos, E.E.; Aggelopoulos, A. Investigation of Shrinkage-Induced Stresses in Stereolithography Photo-Curable Resins. Exp. Mech. 2002, 42, 439–444. [Google Scholar] [CrossRef]
- Beauchamp, M.J.; Nordin, G.P.; Woolley, A.T. Moving from Millifluidic to Truly Microfluidic Sub-100-Μm Cross-Section 3D Printed Devices. Anal. Bioanal. Chem. 2017, 409, 4311–4319. [Google Scholar] [CrossRef]
- Warr, C.; Valdoz, J.C.; Bickham, B.P.; Knight, C.J.; Franks, N.A.; Chartrand, N.; Van Ry, P.M.; Christensen, K.A.; Nordin, G.P.; Cook, A.D. Biocompatible PEGDA Resin for 3D Printing. ACS Appl. Bio Mater. 2020, 3, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Piironen, K.; Haapala, M.; Talman, V.; Järvinen, P.; Sikanen, T. Cell Adhesion and Proliferation on Common 3D Printing Materials Used in Stereolithography of Microfluidic Devices. Lab Chip 2020, 20, 2372–2382. [Google Scholar] [CrossRef] [PubMed]
- Razavi Bazaz, S.; Rouhi, O.; Raoufi, M.A.; Ejeian, F.; Asadnia, M.; Jin, D.; Ebrahimi Warkiani, M. 3D Printing of Inertial Microfluidic Devices. Sci. Rep. 2020, 10, 5929. [Google Scholar] [CrossRef] [Green Version]
- Joswig, L.; Vellekoop, M.J.; Lucklum, F. Miniature 3D-Printed Centrifugal Pump with Non-Contact Electromagnetic Actuation. Micromachines 2019, 10, 631. [Google Scholar] [CrossRef] [Green Version]
- Musgrove, H.; Catterton, M.; Pompano, R. Optimizing Stereolithographic 3D-Printed Materials for On-Chip Primary Immune Cell Culture. In Proceedings of the 25th International Conference on Miniaturized Systems for Chemistry and Life Sciences, Istanbul, Turkey, 27 September 2021. [Google Scholar]
- Rimington, R.P.; Capel, A.J.; Player, D.J.; Bibb, R.J.; Christie, S.D.R.; Lewis, M.P. Feasibility and Biocompatibility of 3D-Printed Photopolymerized and Laser Sintered Polymers for Neuronal, Myogenic, and Hepatic Cell Types. Macromol. Biosci. 2018, 18, 1800113. [Google Scholar] [CrossRef] [Green Version]
- Ukita, Y.; Takamura, Y.; Utsumi, Y. Direct Digital Manufacturing of Autonomous Centrifugal Microfluidic Device. Jpn. J. Appl. Phys. 2016, 55, 06GN02. [Google Scholar] [CrossRef]
- Bishop, G.W.; Satterwhite-Warden, J.E.; Bist, I.; Chen, E.; Rusling, J.F. Electrochemiluminescence at Bare and DNA-Coated Graphite Electrodes in 3D-Printed Fluidic Devices. ACS Sens. 2016, 1, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ligon, S.C.; Liska, R.; Stampfl, J.; Gurr, M.; Mülhaupt, R. Polymers for 3D Printing and Customized Additive Manufacturing. Chem. Rev. 2017, 117, 10212–10290. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.-M.; Lan, H.-Y. Compensation of Distortion in the Bottom Exposure of Stereolithography Process. Int. J. Adv. Manuf. Technol. 2006, 27, 1101–1112. [Google Scholar] [CrossRef]
- Hart, C.; Didier, C.M.; Sommerhage, F.; Rajaraman, S. Biocompatibility of Blank, Post-Processed and Coated 3D Printed Resin Structures with Electrogenic Cells. Biosensors 2020, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Kreß, S.; Schaller-Ammann, R.; Feiel, J.; Priedl, J.; Kasper, C.; Egger, D. 3D Printing of Cell Culture Devices: Assessment and Prevention of the Cytotoxicity of Photopolymers for Stereolithography. Materials 2020, 13, 3011. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-R.; Zhu, L.-N.; Gao, J.; Zhao, H.-X.; Zhu, Y.; Ye, S.; Fang, Q. 3D-Printed High-Density Droplet Array Chip for Miniaturized Protein Crystallization Screening under Vapor Diffusion Mode. ACS Appl. Mater. Interfaces 2017, 9, 11837–11845. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catterton, M.A.; Ball, A.G.; Pompano, R.R. Rapid Fabrication by Digital Light Processing 3D Printing of a SlipChip with Movable Ports for Local Delivery to Ex Vivo Organ Cultures. Micromachines 2021, 12, 993. https://doi.org/10.3390/mi12080993
Catterton MA, Ball AG, Pompano RR. Rapid Fabrication by Digital Light Processing 3D Printing of a SlipChip with Movable Ports for Local Delivery to Ex Vivo Organ Cultures. Micromachines. 2021; 12(8):993. https://doi.org/10.3390/mi12080993
Chicago/Turabian StyleCatterton, Megan A, Alexander G Ball, and Rebecca R Pompano. 2021. "Rapid Fabrication by Digital Light Processing 3D Printing of a SlipChip with Movable Ports for Local Delivery to Ex Vivo Organ Cultures" Micromachines 12, no. 8: 993. https://doi.org/10.3390/mi12080993
APA StyleCatterton, M. A., Ball, A. G., & Pompano, R. R. (2021). Rapid Fabrication by Digital Light Processing 3D Printing of a SlipChip with Movable Ports for Local Delivery to Ex Vivo Organ Cultures. Micromachines, 12(8), 993. https://doi.org/10.3390/mi12080993




