Diffusivity Measurement by Single-Molecule Recycling in a Capillary Microchannel
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecule Diffusing in a Microfluidic Channel
2.2. Single Molecule Recycling to Measure Diffusivity
2.3. Confocal Microscope System
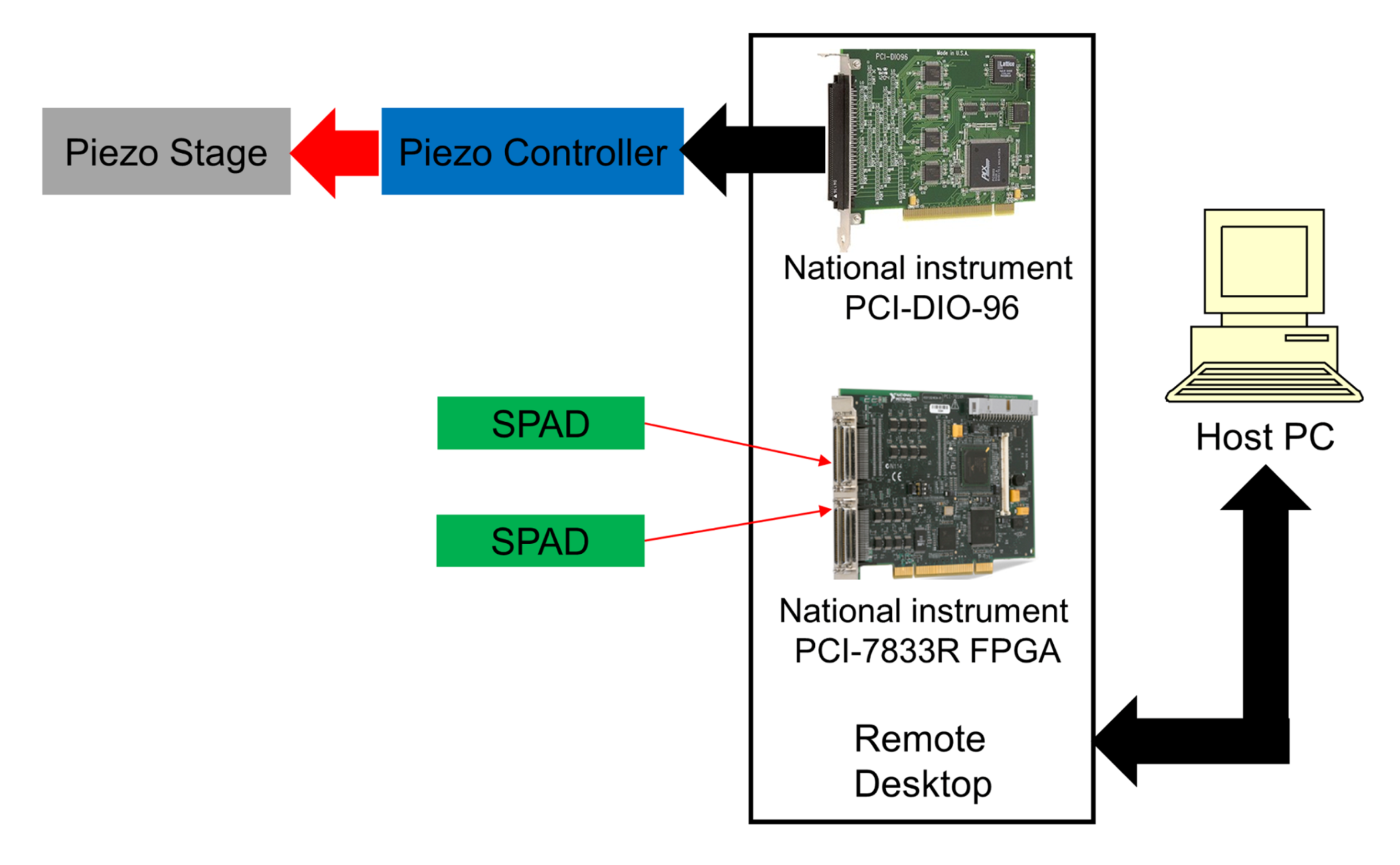
2.4. Real-Time Control System
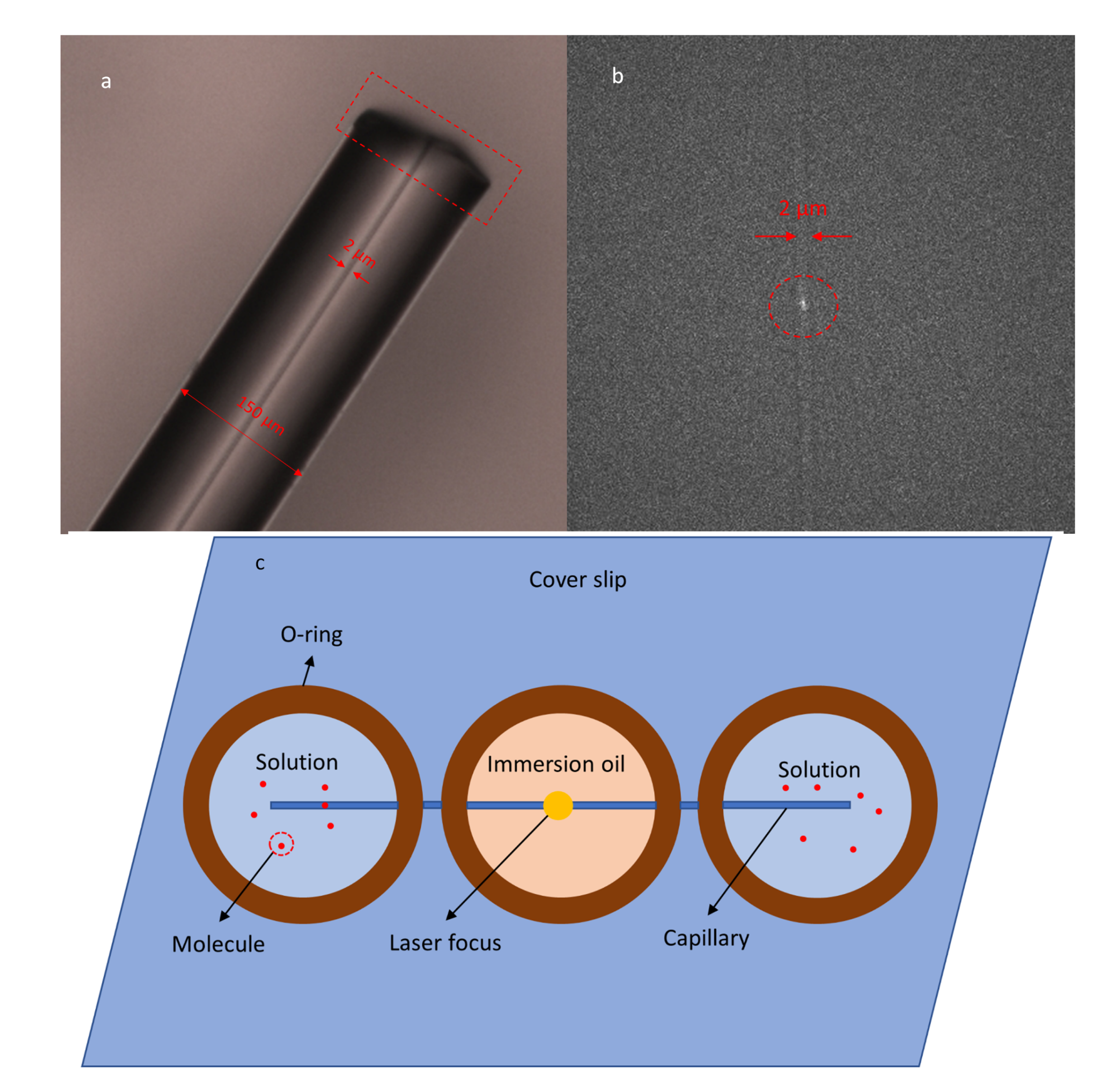
2.5. Preparation of the Capillary Microchannel
2.6. Experimental Practice
3. Results and Discussion
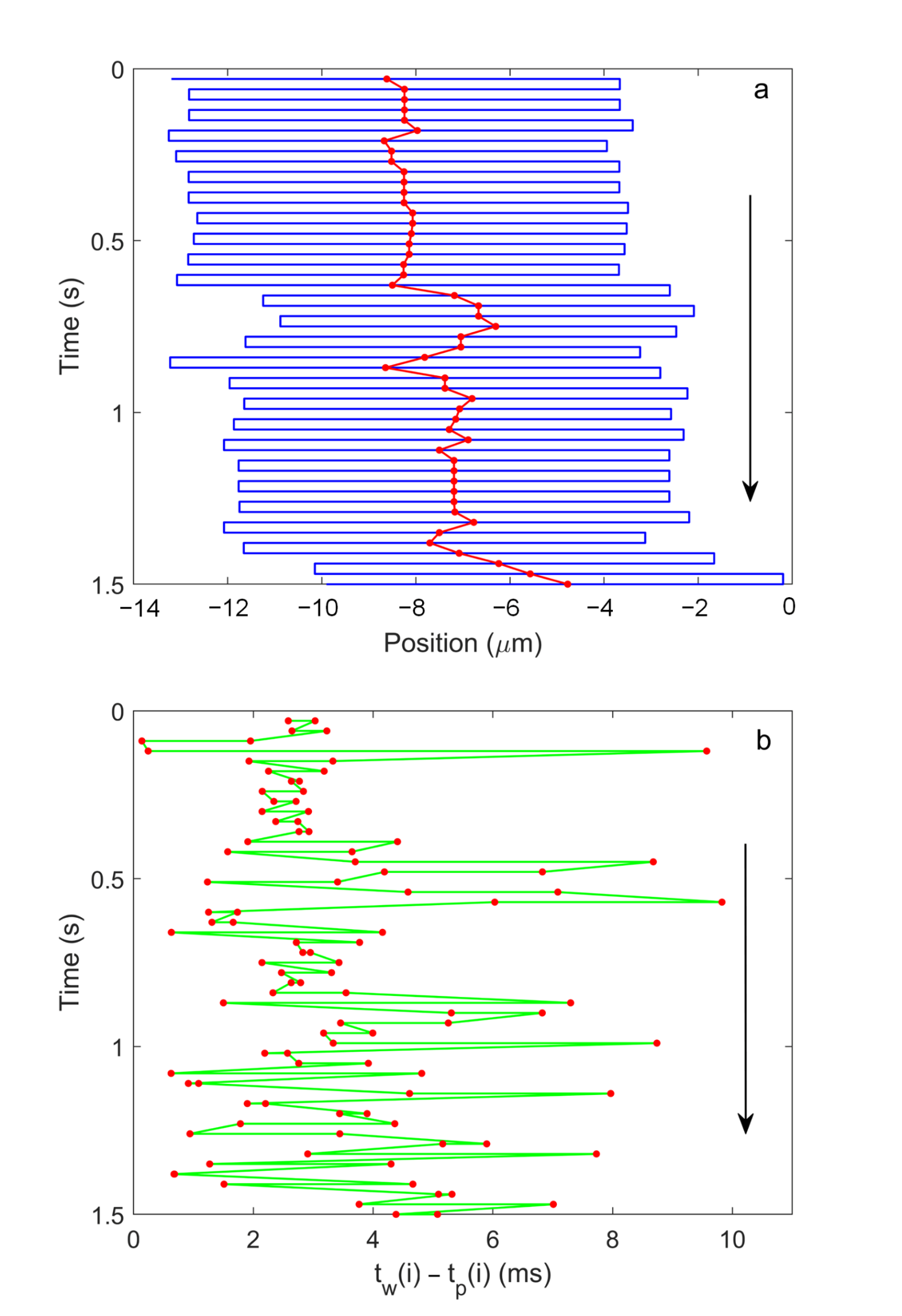
3.1. Measurement of the Translation Speed
3.2. Matched Filter for Photon Burst Detection
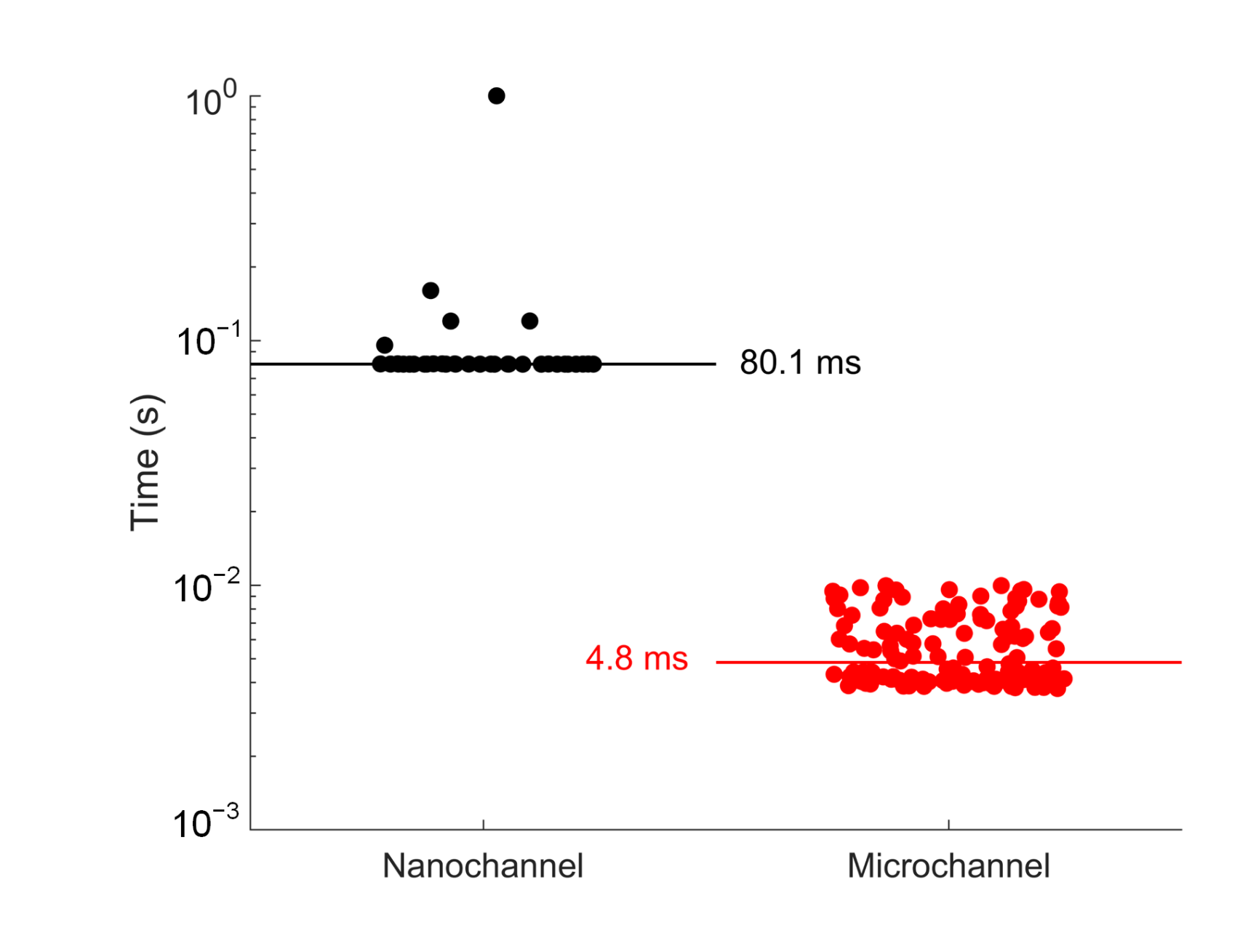
3.3. Diffusivity Measurement from Single-Molecule Recycling in a Capillary Microchannel
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barkai, E.; Jung, Y.; Silbey, R. THEORY OF SINGLE-MOLECULE SPECTROSCOPY: Beyond the Ensemble Average. Annu. Rev. Phys. Chem. 2004, 55, 457–507. [Google Scholar] [CrossRef] [Green Version]
- Moerner, W.E. Nobel Lecture: Single-molecule spectroscopy, imaging, and photocontrol: Foundations for super-resolution microscopy. Rev. Mod. Phys. 2015, 87, 1183–1212. [Google Scholar] [CrossRef] [Green Version]
- Moerner, W.E.; Fromm, D.P. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev. Sci. Instrum. 2003, 74, 3597–3619. [Google Scholar] [CrossRef]
- Goldsmith, R.H.; Moerner, W. Watching conformational- and photodynamics of single fluorescent proteins in solution. Nat. Chem. 2010, 2, 179–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Y.; Gordon, R. Optical Trapping of a Single Protein. Nano Lett. 2012, 12, 402–406. [Google Scholar] [CrossRef]
- Moerner, W.E. New directions in single-molecule imaging and analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 12596–12602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cang, H.; Xu, C.S.; Montiel, D.; Yang, H. Guiding a confocal microscope by single fluorescent nanoparticles. Opt. Lett. 2007, 32, 2729–2731. [Google Scholar] [CrossRef] [PubMed]
- Wells, N.P.; Lessard, G.A.; Werner, J.H. Confocal, Three-Dimensional Tracking of Individual Quantum Dots in High-Background Environments. Anal. Chem. 2008, 80, 9830–9834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germann, J.A.; Davis, L.M. Three-dimensional tracking of a single fluorescent nanoparticle using four-focus excitation in a confocal microscope. Opt. Express 2014, 22, 5641–5650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dissanayaka, K.D.; Canfield, B.K.; Davis, L.M. Three-dimensional feedback-driven trapping of a single nanoparticle or molecule in aqueous solution with a confocal fluorescence microscope. Opt. Express 2019, 27, 29759–29769. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. Method for trapping and manipulating nanoscale objects in solution. Appl. Phys. Lett. 2005, 86, 093109. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.E. Control of Nanoparticles with Arbitrary Two-Dimensional Force Fields. Phys. Rev. Lett. 2005, 94, 118102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, A.E.; Moerner, W.E. Controlling Brownian motion of single protein molecules and single fluorophores in aqueous buffer. Opt. Express 2008, 16, 6941–6956. [Google Scholar] [CrossRef] [PubMed]
- Lesoine, J.F.; Venkataraman, P.A.; Maloney, P.C.; Dumont, M.E.; Novotny, L. Nanochannel-Based Single Molecule Recycling. Nano Lett. 2012, 12, 3273–3278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, W.A.; Nie, S. Confinement and Detection of Single Molecules in Submicrometer Channels. Anal. Chem. 1997, 69, 3400–3405. [Google Scholar] [CrossRef]
- Wang, B.; Davis, L.M. Improved timing and diffusivity measurement in single-molecule recycling in a nanochannel. In Single Molecule Spectroscopy and Superresolution Imaging X; Enderlein, J., Gregor, I., Gryczynski, Z.K., Erdmann, R., Koberling, F., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2017; Volume 10071, pp. 20–29. [Google Scholar] [CrossRef]
- Hibara, A.; Saito, T.; Kim, H.B.; Tokeshi, M.; Ooi, T.; Nakao, M.; Kitamori, T. Nanochannels on a Fused-Silica Microchip and Liquid Properties Investigation by Time-Resolved Fluorescence Measurements. Anal. Chem. 2002, 74, 6170–6176. [Google Scholar] [CrossRef]
- Olanrewaju, A.; Beaugrand, M.; Yafia, M.; Juncker, D. Capillary microfluidics in microchannels: From microfluidic networks to capillaric circuits. Lab Chip 2018, 18, 2323–2347. [Google Scholar] [CrossRef] [Green Version]
- Magde, D.; Webb, W.W.; Elson, E.L. Fluorescence correlation spectroscopy. III. Uniform translation and laminar flow. Biopolymers 1978, 17, 361–376. [Google Scholar] [CrossRef]
- Turin, G. An introduction to matched filters. IRE Trans. Inf. Theory 1960, 6, 311–329. [Google Scholar] [CrossRef] [Green Version]
- Davis, L.; Williams, P.; Ball, D.; Swift, K.; Matayoshi, E. Data Reduction Methods for Application of Fluorescence Correlation Spectroscopy to Pharmaceutical Drug Discovery. Curr. Pharm. Biotechnol. 2004, 4, 451–462. [Google Scholar] [CrossRef] [Green Version]
- Pappaert, K.; Biesemans, J.; Clicq, D.; Vankrunkelsven, S.; Desmet, G. Measurements of diffusion coefficients in 1-D micro- and nanochannels using shear-driven flows. Lab Chip 2005, 5, 1104–1110. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. Suppressing Brownian motion of individual biomolecules in solution. Proc. Natl. Acad. Sci. USA 2006, 103, 4362–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalet, X.; Berglund, A.J. Optimal diffusion coefficient estimation in single-particle tracking. Phys. Rev. E 2012, 85, 061916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vestergaard, C.L. Optimizing experimental parameters for tracking of diffusing particles. Phys. Rev. E 2016, 94, 022401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Davis, L.M. Monte Carlo simulation and maximum-likelihood analysis of single-molecule recycling in a nanochannel. OSA Contin. 2021, 4, 212–228. [Google Scholar] [CrossRef]
- Papoulis, A.; Pillai, S.U. Probability, Random Variables, and Stochastic Processes; McGraw-Hill: New York, NY, USA, 1991; pp. 244–245. [Google Scholar]
- Elson, E.L.; Magde, D. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers 1974, 13, 1–27. [Google Scholar] [CrossRef]
- Shuming Nie, R.N.Z. Optical detection of single molecules. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 567–596. [Google Scholar] [CrossRef] [Green Version]

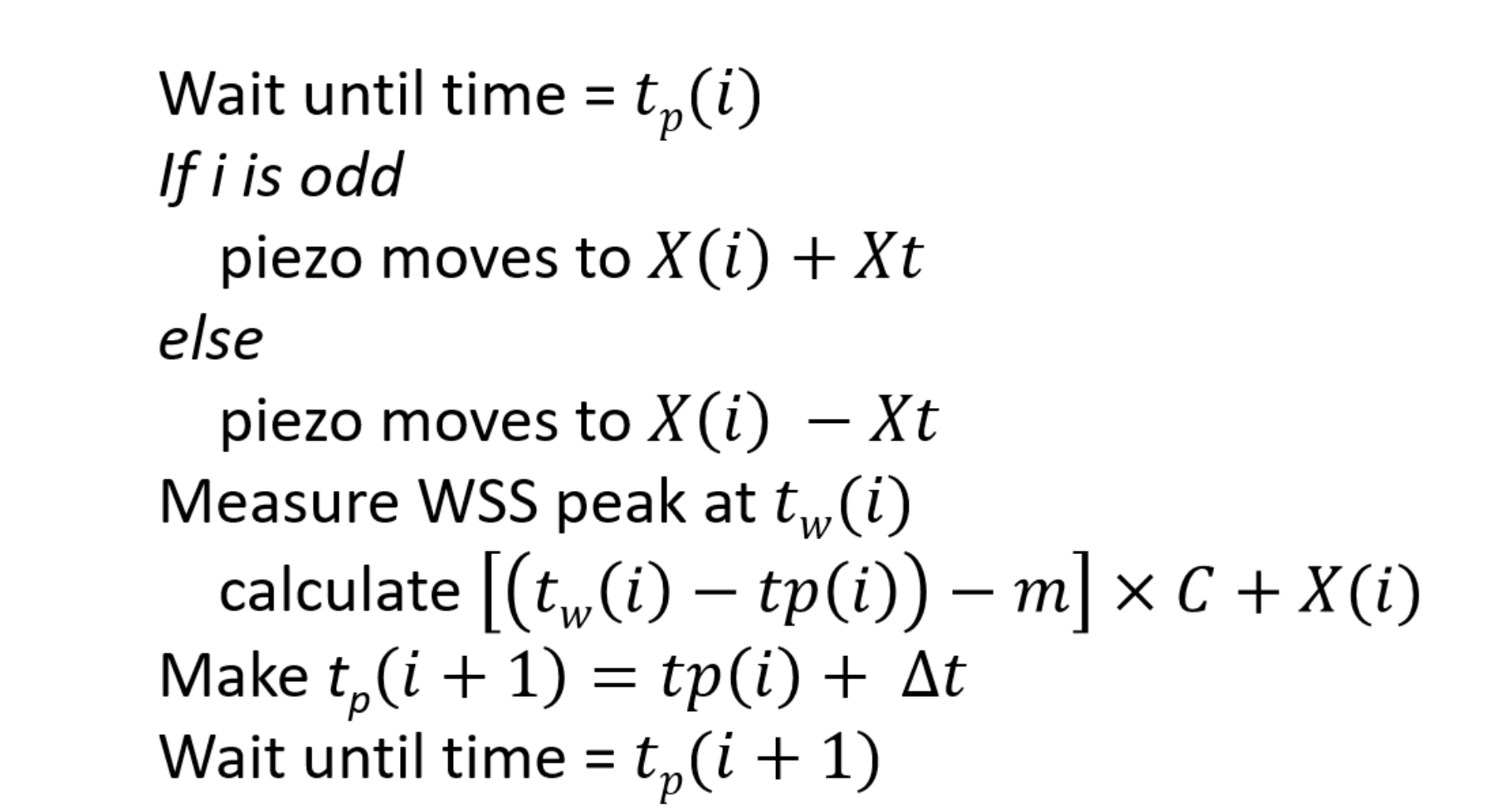



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Davis, L.M. Diffusivity Measurement by Single-Molecule Recycling in a Capillary Microchannel. Micromachines 2021, 12, 800. https://doi.org/10.3390/mi12070800
Wang B, Davis LM. Diffusivity Measurement by Single-Molecule Recycling in a Capillary Microchannel. Micromachines. 2021; 12(7):800. https://doi.org/10.3390/mi12070800
Chicago/Turabian StyleWang, Bo, and Lloyd M. Davis. 2021. "Diffusivity Measurement by Single-Molecule Recycling in a Capillary Microchannel" Micromachines 12, no. 7: 800. https://doi.org/10.3390/mi12070800
APA StyleWang, B., & Davis, L. M. (2021). Diffusivity Measurement by Single-Molecule Recycling in a Capillary Microchannel. Micromachines, 12(7), 800. https://doi.org/10.3390/mi12070800





