Mechanical Strain-Enabled Reconstitution of Dynamic Environment in Organ-on-a-Chip Platforms: A Review
Abstract
:1. Introduction
2. A General Overview of Organ-on-a-Chip
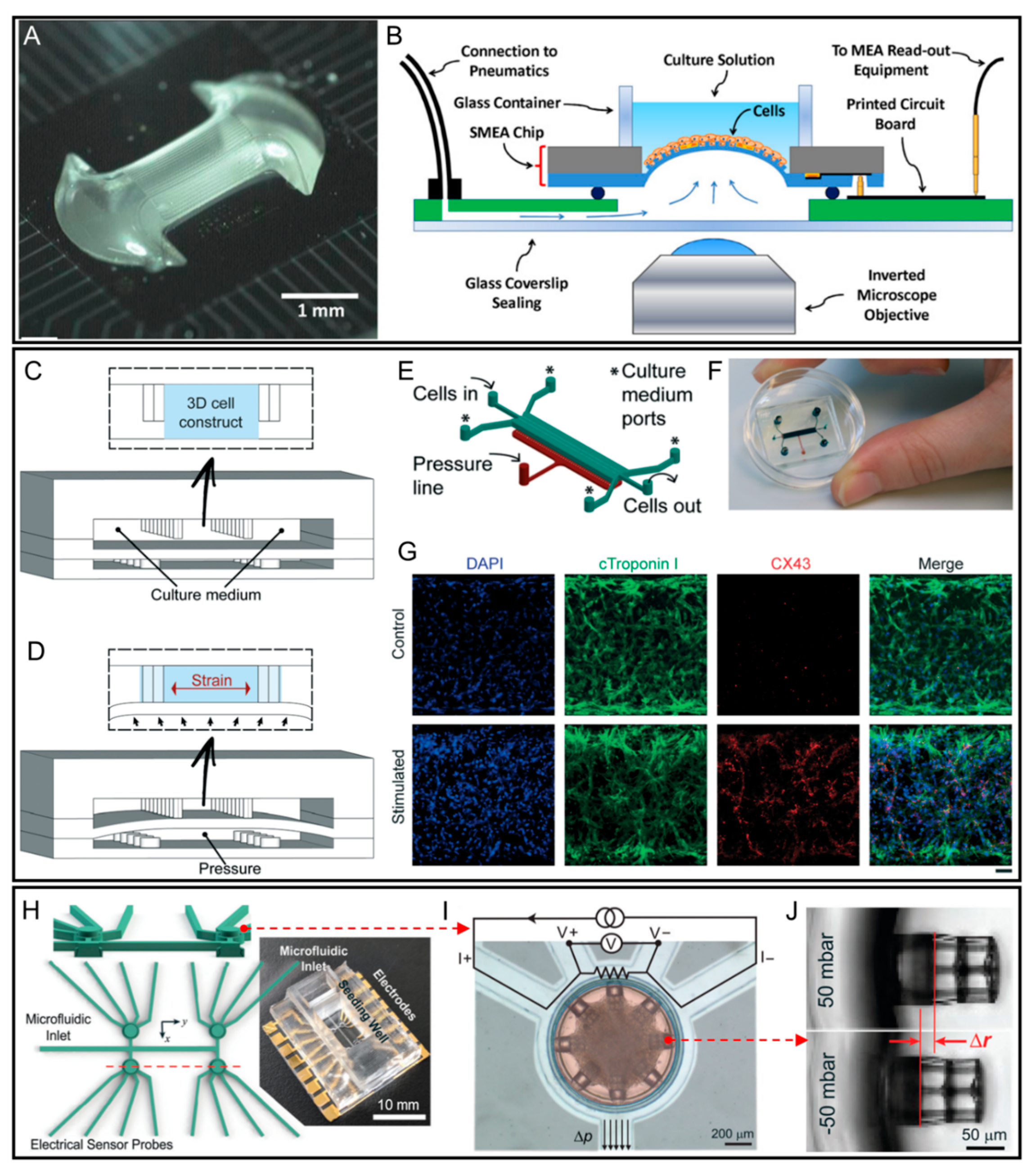
3. Heart-on-a-Chip
4. Kidney-on-a-Chip
5. Lung-on-a-Chip
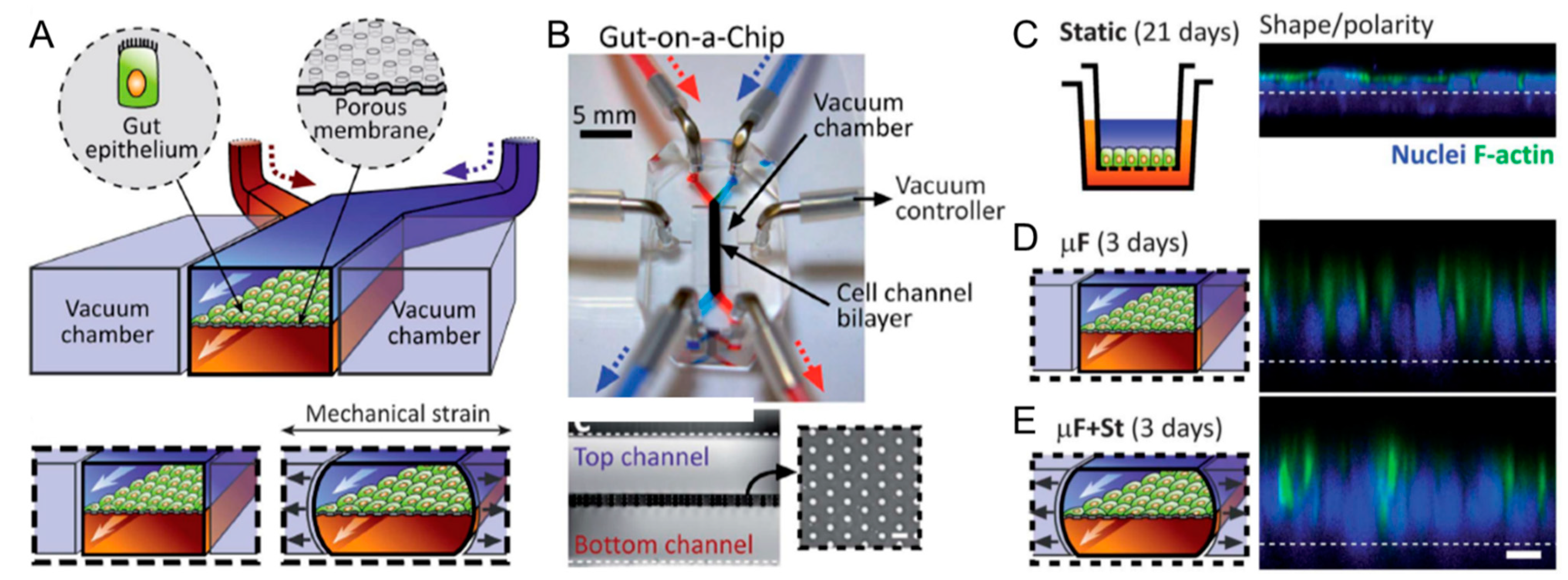
6. Gut-on-a-Chip
7. Vessel-on-a-Chip
8. Other Organ-on-a-Chip Platforms
9. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Shehab, N.; Lovegrove, M.C.; Geller, A.I.; Rose, K.O.; Weidle, N.J.; Budnitz, D.S. US emergency department visits for outpatient adverse drug events, 2013–2014. JAMA 2016, 316, 2115–2125. [Google Scholar] [CrossRef]
- Olson, H.; Betton, G.; Robinson, D.; Thomas, K.; Monro, A.; Kolaja, G.; Lilly, P.; Sanders, J.; Sipes, G.; Bracken, W. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000, 32, 56–67. [Google Scholar] [CrossRef]
- Cao, X.; Gibbs, S.T.; Fang, L.; Miller, H.A.; Landowski, C.P.; Shin, H.-C.; Lennernas, H.; Zhong, Y.; Amidon, G.L.; Lawrence, X.Y. Why is it challenging to predict intestinal drug absorption and oral bioavailability in human using rat model. Pharm. Res. 2006, 23, 1675–1686. [Google Scholar] [CrossRef] [PubMed]
- Davila, J.C.; Rodriguez, R.J.; Melchert, R.B.; Acosta, D., Jr. Predictive value of in vitro model systems in toxicology. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 63–96. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Liu, J.R.; Patel, B.; Solomon, D.E.; Vaidya, B.; Gupta, V. Microfluidics-based 3D cell culture models: Utility in novel drug discovery and delivery research. Bioeng. Transl. Med. 2016, 1, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Woo, F.W.; Castro, C.S.; Cohen, M.A.; Karanxha, J.; Mittal, J.; Chhibber, T.; Jhaveri, V.M. Organ-on-chip models: Implications in drug discovery and clinical applications. J. Cell. Physiol. 2019, 234, 8352–8380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polacheck, W.J.; Li, R.; Uzel, S.G.; Kamm, R.D. Microfluidic platforms for mechanobiology. Lab Chip 2013, 13, 2252–2267. [Google Scholar] [CrossRef] [Green Version]
- Ergir, E.; Bachmann, B.; Redl, H.; Forte, G.; Ertl, P. Small force, big impact: Next generation organ-on-a-chip systems incorporating biomechanical cues. Front. Physiol. 2018, 9, 1417. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-Chip Systems: Microengineering to biomimic living systems. Small 2016, 12, 2253–2282. [Google Scholar] [CrossRef]
- Richardson, L.; Kim, S.; Menon, R.; Han, A. Organ-on-chip technology: The future of feto-maternal interface research? Front. Physiol. 2020, 11, 715. [Google Scholar] [CrossRef]
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of biochip technology: A review from lab-on-a-chip to organ-on-a-chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef]
- Viravaidya, K.; Sin, A.; Shuler, M.L. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol. Prog. 2004, 20, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Sin, A.; Chin, K.C.; Jamil, M.F.; Kostov, Y.; Rao, G.; Shuler, M.L. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol. Prog. 2004, 20, 338–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffith, L.G.; Naughton, G. Tissue engineering--current challenges and expanding opportunities. Science 2002, 295, 1009–1014. [Google Scholar] [CrossRef]
- Khademhosseini, A.; Langer, R.; Borenstein, J.; Vacanti, J.P. Microscale technologies for tissue engineering and biology. Proc. Natl. Acad. Sci. USA 2006, 103, 2480–2487. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Van Der Meer, A.D.; Van Den Berg, A. Organs-on-chips: Breaking the in vitro impasse. Integr. Biol. 2012, 4, 461–470. [Google Scholar] [CrossRef]
- Verpoorte, E.; De Rooij, N.F. Microfluidics meets MEMS. Proc. IEEE 2003, 91, 930–953. [Google Scholar] [CrossRef] [Green Version]
- Manz, A.; Graber, N.; Widmer, H.Á. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Abgrall, P.; Gue, A. Lab-on-chip technologies: Making a microfluidic network and coupling it into a complete microsystem—A review. J. Micromech. Microeng. 2007, 17, R15. [Google Scholar] [CrossRef]
- Hutmacher, D.W. Scaffold design and fabrication technologies for engineering tissues—State of the art and future perspectives. J. Biomater. Sci. Polym. Ed. 2001, 12, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Tsang, V.L.; Bhatia, S.N. Three-dimensional tissue fabrication. Adv. drug Deliv. Rev. 2004, 56, 1635–1647. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Yu, J.; Luo, D.; Shuler, M.L.; March, J.C. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011, 11, 389–392. [Google Scholar] [CrossRef]
- Andersson, H.; Van Den Berg, A. Microfabrication and microfluidics for tissue engineering: State of the art and future opportunities. Lab Chip 2004, 4, 98–103. [Google Scholar] [CrossRef]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef]
- Kim, D.-H.; Wong, P.K.; Park, J.; Levchenko, A.; Sun, Y. Microengineered platforms for cell mechanobiology. Annu. Rev. Biomed. Eng. 2009, 11, 203–233. [Google Scholar] [CrossRef] [Green Version]
- Ingber, D.E. Cellular mechanotransduction: Putting all the pieces together again. FASEB J. 2006, 20, 811–827. [Google Scholar] [CrossRef]
- Tulloch, N.L.; Muskheli, V.; Razumova, M.V.; Korte, F.S.; Regnier, M.; Hauch, K.D.; Pabon, L.; Reinecke, H.; Murry, C.E. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ. Res. 2011, 109, 47–59. [Google Scholar] [CrossRef]
- Zheng, W.; Jiang, B.; Wang, D.; Zhang, W.; Wang, Z.; Jiang, X. A microfluidic flow-stretch chip for investigating blood vessel biomechanics. Lab Chip 2012, 12, 3441–3450. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Chen, C.S. Mechanotransduction in development: A growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009, 10, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Zaman, M.H.; Trapani, L.M.; Sieminski, A.L.; MacKellar, D.; Gong, H.; Kamm, R.D.; Wells, A.; Lauffenburger, D.A.; Matsudaira, P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 10889–10894. [Google Scholar] [CrossRef] [Green Version]
- Saez, A.; Ghibaudo, M.; Buguin, A.; Silberzan, P.; Ladoux, B. Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc. Natl. Acad. Sci. USA 2007, 104, 8281–8286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, A.I.; Ilkhanizadeh, S.; Wigenius, J.A.; Duckworth, J.K.; Inganäs, O.; Hermanson, O. The promotion of neuronal maturation on soft substrates. Biomaterials 2009, 30, 4567–4572. [Google Scholar] [CrossRef]
- Ongaro, A.E.; Di Giuseppe, D.; Kermanizadeh, A.; Miguelez Crespo, A.; Mencattini, A.; Ghibelli, L.; Mancini, V.; Wlodarczyk, K.L.; Hand, D.P.; Martinelli, E. Polylactic is a sustainable, low absorption, low autofluorescence alternative to other plastics for microfluidic and organ-on-chip applications. Anal. Chem. 2020, 92, 6693–6701. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Jung, S.H.; Lee, M.S.; Park, T.-E.; Ahn, S.-k.; Kang, J.H. Robust chemical bonding of PMMA microfluidic devices to porous PETE membranes for reliable cytotoxicity testing of drugs. Lab Chip 2019, 19, 3706–3713. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, J.W.; Yu, J.; Park, D.; Ha, J.; Son, K.; Lee, S.; Chung, M.; Kim, H.-Y.; Jeon, N.L. Microfluidics within a well: An injection-molded plastic array 3D culture platform. Lab Chip 2018, 18, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, B.; de Vries, H.; Firth, K.; van Weerd, J.; Tertoolen, L.; Karperien, H.; Jonkheijm, P.; Denning, C.; IJzerman, A.; Mummery, C. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem. Biophys. Res. Commun. 2017, 482, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Christoffersson, J.; Aronsson, C.; Jury, M.; Selegård, R.; Aili, D.; Mandenius, C.-F. Fabrication of modular hyaluronan-PEG hydrogels to support 3D cultures of hepatocytes in a perfused liver-on-a-chip device. Biofabrication 2018, 11, 015013. [Google Scholar] [CrossRef]
- Zhu, L.; Fan, X.; Wang, B.; Liu, L.; Yan, X.; Zhou, L.; Zeng, Y.; Poznansky, M.C.; Wang, L.; Chen, H. Biomechanically primed liver microtumor array as a high-throughput mechanopharmacological screening platform for stroma-reprogrammed combinatorial therapy. Biomaterials 2017, 124, 12–24. [Google Scholar] [CrossRef]
- Van Der Helm, M.W.; Van Der Meer, A.D.; Eijkel, J.C.; van den Berg, A.; Segerink, L.I. Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 2016, 4, e1142493. [Google Scholar] [CrossRef] [Green Version]
- Caballero, D.; Kaushik, S.; Correlo, V.; Oliveira, J.M.; Reis, R.; Kundu, S. Organ-on-chip models of cancer metastasis for future personalized medicine: From chip to the patient. Biomaterials 2017, 149, 98–115. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. Biomed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef] [Green Version]
- Rothbauer, M.; Rosser, J.M.; Zirath, H.; Ertl, P. Tomorrow today: Organ-on-a-chip advances towards clinically relevant pharmaceutical and medical in vitro models. Curr. Opin. Biotechnol. 2019, 55, 81–86. [Google Scholar] [CrossRef]
- Ghaemmaghami, A.M.; Hancock, M.J.; Harrington, H.; Kaji, H.; Khademhosseini, A. Biomimetic tissues on a chip for drug discovery. Drug Discov. Today 2012, 17, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Kaarj, K.; Yoon, J.-Y. Methods of delivering mechanical stimuli to organ-on-a-chip. Micromachines 2019, 10, 700. [Google Scholar] [CrossRef] [Green Version]
- Laverty, H.; Benson, C.; Cartwright, E.; Cross, M.; Garland, C.; Hammond, T.; Holloway, C.; McMahon, N.; Milligan, J.; Park, B. How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br. J. Pharmacol. 2011, 163, 675–693. [Google Scholar] [CrossRef]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.G.; Marks, N.; Mandegar, M.; Conklin, B.R.; Lee, L.P. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Pakazad, S.K.; Savov, A.; Van de Stolpe, A.; Dekker, R. A novel stretchable micro-electrode array (SMEA) design for directional stretching of cells. J. Micromech. Microeng. 2014, 24, 034003. [Google Scholar] [CrossRef]
- Gaio, N.; Van Meer, B.; Quirós Solano, W.; Bergers, L.; Van de Stolpe, A.; Mummery, C.; Sarro, P.M.; Dekker, R. Cytostretch, an organ-on-chip platform. Micromachines 2016, 7, 120. [Google Scholar] [CrossRef] [Green Version]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayne, R.K.; Karakan, M.Ç.; Zhang, K.; Pierce, N.; Michas, C.; Bishop, D.J.; Chen, C.S.; Ekinci, K.L.; White, A.E. Direct laser writing for cardiac tissue engineering: A microfluidic heart on a chip with integrated transducers. Lab Chip 2021, 21, 1724–1737. [Google Scholar] [CrossRef]
- Morrissey, K.M.; Stocker, S.L.; Wittwer, M.B.; Xu, L.; Giacomini, K.M. Renal transporters in drug development. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 503–529. [Google Scholar] [CrossRef] [Green Version]
- Peired, A.J.; Mazzinghi, B.; De Chiara, L.; Guzzi, F.; Lasagni, L.; Romagnani, P.; Lazzeri, E. Bioengineering strategies for nephrologists: Kidney was not built in a day. Expert Opin. Biol. Ther. 2020, 20, 467–480. [Google Scholar] [CrossRef] [Green Version]
- Homan, K.A.; Gupta, N.; Kroll, K.T.; Kolesky, D.B.; Skylar-Scott, M.; Miyoshi, T.; Mau, D.; Valerius, M.T.; Ferrante, T.; Bonventre, J.V. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 2019, 16, 255–262. [Google Scholar] [CrossRef]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef]
- Musah, S.; Mammoto, A.; Ferrante, T.C.; Jeanty, S.S.; Hirano-Kobayashi, M.; Mammoto, T.; Roberts, K.; Chung, S.; Novak, R.; Ingram, M. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng. 2017, 1, 1–12. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, X.; Wen, X.; Wu, T.; Wang, W.; Yang, M.; Wang, J.; Fang, M.; Lin, B.; Lin, H. Development of a functional glomerulus at the organ level on a chip to mimic hypertensive nephropathy. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Humayun, M.; Chow, C.-W.; Young, E.W. Microfluidic lung airway-on-a-chip with arrayable suspended gels for studying epithelial and smooth muscle cell interactions. Lab Chip 2018, 18, 1298–1309. [Google Scholar] [CrossRef]
- Douville, N.J.; Zamankhan, P.; Tung, Y.-C.; Li, R.; Vaughan, B.L.; Tai, C.-F.; White, J.; Christensen, P.J.; Grotberg, J.B.; Takayama, S. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip 2011, 11, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Bai, H.; Si, L.; Jiang, A.; Belgur, C.; Plebani, R.; Oh, C.Y.; Rodas, M.; Nurani, A.; Gilpin, S.E.; Powers, R.K. Mechanical control of innate immune responses against viral infection revealed in a human Lung Alveolus Chip. bioRxiv 2021, 2026, 441498. [Google Scholar]
- Stucki, A.O.; Stucki, J.D.; Hall, S.R.; Felder, M.; Mermoud, Y.; Schmid, R.A.; Geiser, T.; Guenat, O.T. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip 2015, 15, 1302–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stucki, J.D.; Hobi, N.; Galimov, A.; Stucki, A.O.; Schneider-Daum, N.; Lehr, C.-M.; Huwer, H.; Frick, M.; Funke-Chambour, M.; Geiser, T. Medium throughput breathing human primary cell alveolus-on-chip model. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Felder, M.; Trueeb, B.; Stucki, A.O.; Borcard, S.; Stucki, J.D.; Schnyder, B.; Geiser, T.; Guenat, O.T. Impaired wound healing of alveolar lung epithelial cells in a breathing lung-on-a-chip. Front. Bioeng. Biotechnol. 2019, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Radiom, M.; He, Y.; Peng-Wang, J.; Baeza-Squiban, A.; Berret, J.F.; Chen, Y. Alveolar mimics with periodic strain and its effect on the cell layer formation. Biotechnol. Bioeng. 2020, 117, 2827–2841. [Google Scholar] [CrossRef] [PubMed]
- Zamprogno, P.; Wüthrich, S.; Achenbach, S.; Thoma, G.; Stucki, J.D.; Hobi, N.; Schneider-Daum, N.; Lehr, C.-M.; Huwer, H.; Geiser, T. Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef]
- Shanks, N.; Greek, R.; Greek, J. Are animal models predictive for humans? Philos. Ethics Humanit. Med. 2009, 4, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Nasiri, R.; De Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseni, A. Gut-on-a-chip: Current progress and future opportunities. Biomaterials 2020, 225, 120196. [Google Scholar] [CrossRef] [PubMed]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadabad, M.S.; Pinheiro, I.; Possemiers, S.; Van den Abbeele, P.; Derycke, L.; Bracke, M.; Pieters, J. The HMI™ module: A new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Shim, K.-Y.; Lee, D.; Han, J.; Nguyen, N.-T.; Park, S.; Sung, J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 2017, 19, 37. [Google Scholar] [CrossRef] [Green Version]
- Verhulsel, M.; Simon, A.; Bernheim-Dennery, M.; Gannavarapu, V.R.; Gérémie, L.; Ferraro, D.; Krndija, D.; Talini, L.; Viovy, J.-L.; Vignjevic, D.M. Developing an advanced gut on chip model enabling the study of epithelial cell/fibroblast interactions. Lab Chip 2021, 21, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Basson, M.D.; Di Li, G.; Hong, F.; Han, O.; Sumpio, B.E. Amplitude-dependent modulation of brush border enzymes and proliferation by cyclic strain in human intestinal Caco-2 monolayers. J. Cell. Physiol. 1996, 168, 476–488. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef]
- Villenave, R.; Wales, S.Q.; Hamkins-Indik, T.; Papafragkou, E.; Weaver, J.C.; Ferrante, T.C.; Bahinski, A.; Elkins, C.A.; Kulka, M.; Ingber, D.E. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS ONE 2017, 12, e0169412. [Google Scholar] [CrossRef]
- Nelson, M.T.; Charbonneau, M.R.; Coia, H.G.; Castillo, M.J.; Holt, C.; Greenwood, E.S.; Robinson, P.J.; Merrill, E.A.; Lubkowicz, D.; Mauzy, C.A. Characterization of an engineered live bacterial therapeutic for the treatment of phenylketonuria in a human gut-on-a-chip. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [Green Version]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folkman, J.; D’Amore, P.A. Blood vessel formation: What is its molecular basis? Cell 1996, 87, 1153–1155. [Google Scholar] [CrossRef] [Green Version]
- Maron, B.J.; Wolfson, J.K.; Epstein, S.E.; Roberts, W.C. Intramural (“small vessel”) coronary artery disease in hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 1986, 8, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Sinha, R.; Le Gac, S.; Verdonschot, N.; Van Den Berg, A.; Koopman, B.; Rouwkema, J. Endothelial cell alignment as a result of anisotropic strain and flow induced shear stress combinations. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Van Engeland, N.C.; Pollet, A.M.; den Toonder, J.M.; Bouten, C.V.; Stassen, O.M.; Sahlgren, C.M. A biomimetic microfluidic model to study signalling between endothelial and vascular smooth muscle cells under hemodynamic conditions. Lab Chip 2018, 18, 1607–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Niklason, L.E. Microfluidic artificial “vessels” for dynamic mechanical stimulation of mesenchymal stem cells. Integr. Biol. 2012, 4, 1487–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribas, J.; Zhang, Y.S.; Pitrez, P.R.; Leijten, J.; Miscuglio, M.; Rouwkema, J.; Dokmeci, M.R.; Nissan, X.; Ferreira, L.; Khademhosseini, A. Biomechanical strain exacerbates inflammation on a progeria-on-a-Chip model. Small 2017, 13, 1603737. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.H.; Liu, Y.L.; Fan, W.T.; Huang, W.H. Integrating flexible electrochemical sensor into microfluidic chip for simulating and monitoring vascular mechanotransduction. Small 2020, 16, 1903204. [Google Scholar] [CrossRef]
- Dessalles, C.A.; Ramón-Lozano, C.; Babataheri, A.; Barakat, A.I. Luminal Flow Actuation Generates Coupled Shear and Strain in a Microvessel-on-Chip. bioRxiv 2021, 2010, 439271. [Google Scholar]
- Peng, Z.; Zhou, L.; Wong, J.K.W.; Chan, Y.K. Eye-on-a-chip (EOC) models and their role in the future of ophthalmic drug discovery. Expert Rev. Ophthalmol. 2020, 15, 259–261. [Google Scholar] [CrossRef]
- Seo, J.; Byun, W.Y.; Alisafaei, F.; Georgescu, A.; Yi, Y.-S.; Massaro-Giordano, M.; Shenoy, V.B.; Lee, V.; Bunya, V.Y.; Huh, D. Multiscale reverse engineering of the human ocular surface. Nat. Med. 2019, 25, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Dollé, J.-P.; Morrison, B., III; Schloss, R.S.; Yarmush, M.L. Brain-on-a-chip microsystem for investigating traumatic brain injury: Axon diameter and mitochondrial membrane changes play a significant role in axonal response to strain injuries. Technology 2014, 2, 106–117. [Google Scholar] [CrossRef]
- Rigat-Brugarolas, L.; Elizalde-Torrent, A.; Bernabeu, M.; De Niz, M.; Martin-Jaular, L.; Fernandez-Becerra, C.; Homs-Corbera, A.; Samitier, J.; Del Portillo, H. A functional microengineered model of the human splenon-on-a-chip. Lab Chip 2014, 14, 1715–1724. [Google Scholar] [CrossRef]
- Park, S.-H.; Sim, W.Y.; Min, B.-H.; Yang, S.S.; Khademhosseini, A.; Kaplan, D.L. Chip-based comparison of the osteogenesis of human bone marrow-and adipose tissue-derived mesenchymal stem cells under mechanical stimulation. PLoS ONE 2012, 7, e46689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, S.; Pu, H.; Xia, S.; Sun, W. A minimized valveless electromagnetic micropump for microfluidic actuation on organ chips. Sens. Actuators A Phys. 2020, 301, 111704. [Google Scholar] [CrossRef]
- Tang, S.Y.; Zhang, X.; Sun, S.; Yuan, D.; Zhao, Q.; Yan, S.; Deng, L.; Yun, G.; Zhang, J.; Zhang, S. Versatile microfluidic platforms enabled by novel magnetorheological elastomer microactuators. Adv. Funct. Mater. 2018, 28, 1705484. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Cole, T.; Yun, G.; Li, Y.; Zhao, Q.; Lu, H.; Zheng, J.; Li, W.; Tang, S.-Y. Modular and Self-Contained Microfluidic Analytical Platforms Enabled by Magnetorheological Elastomer Microactuators. Micromachines 2021, 12, 604. [Google Scholar] [CrossRef]
- Harmon, M.E.; Tang, M.; Frank, C.W. A microfluidic actuator based on thermoresponsive hydrogels. Polymer 2003, 44, 4547–4556. [Google Scholar] [CrossRef]
- Kwon, G.H.; Choi, Y.Y.; Park, J.Y.; Woo, D.H.; Lee, K.B.; Kim, J.H.; Lee, S.-H. Electrically-driven hydrogel actuators in microfluidic channels: Fabrication, characterization, and biological application. Lab Chip 2010, 10, 1604–1610. [Google Scholar] [CrossRef]
- Ha, J.H.; Shin, H.H.; Choi, H.W.; Lim, J.H.; Mo, S.J.; Ahrberg, C.D.; Lee, J.M.; Chung, B.G. Electro-responsive hydrogel-based microfluidic actuator platform for photothermal therapy. Lab Chip 2020, 20, 3354–3364. [Google Scholar] [CrossRef]
- Eddington, D.T.; Liu, R.H.; Moore, J.S.; Beebe, D.J. An organic self-regulating microfluidic system. Lab Chip 2001, 1, 96–99. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Q.; Cole, T.; Zhang, Y.; Tang, S.-Y. Mechanical Strain-Enabled Reconstitution of Dynamic Environment in Organ-on-a-Chip Platforms: A Review. Micromachines 2021, 12, 765. https://doi.org/10.3390/mi12070765
Zhao Q, Cole T, Zhang Y, Tang S-Y. Mechanical Strain-Enabled Reconstitution of Dynamic Environment in Organ-on-a-Chip Platforms: A Review. Micromachines. 2021; 12(7):765. https://doi.org/10.3390/mi12070765
Chicago/Turabian StyleZhao, Qianbin, Tim Cole, Yuxin Zhang, and Shi-Yang Tang. 2021. "Mechanical Strain-Enabled Reconstitution of Dynamic Environment in Organ-on-a-Chip Platforms: A Review" Micromachines 12, no. 7: 765. https://doi.org/10.3390/mi12070765
APA StyleZhao, Q., Cole, T., Zhang, Y., & Tang, S.-Y. (2021). Mechanical Strain-Enabled Reconstitution of Dynamic Environment in Organ-on-a-Chip Platforms: A Review. Micromachines, 12(7), 765. https://doi.org/10.3390/mi12070765






