Cancer Cell Direct Bioprinting: A Focused Review
Abstract
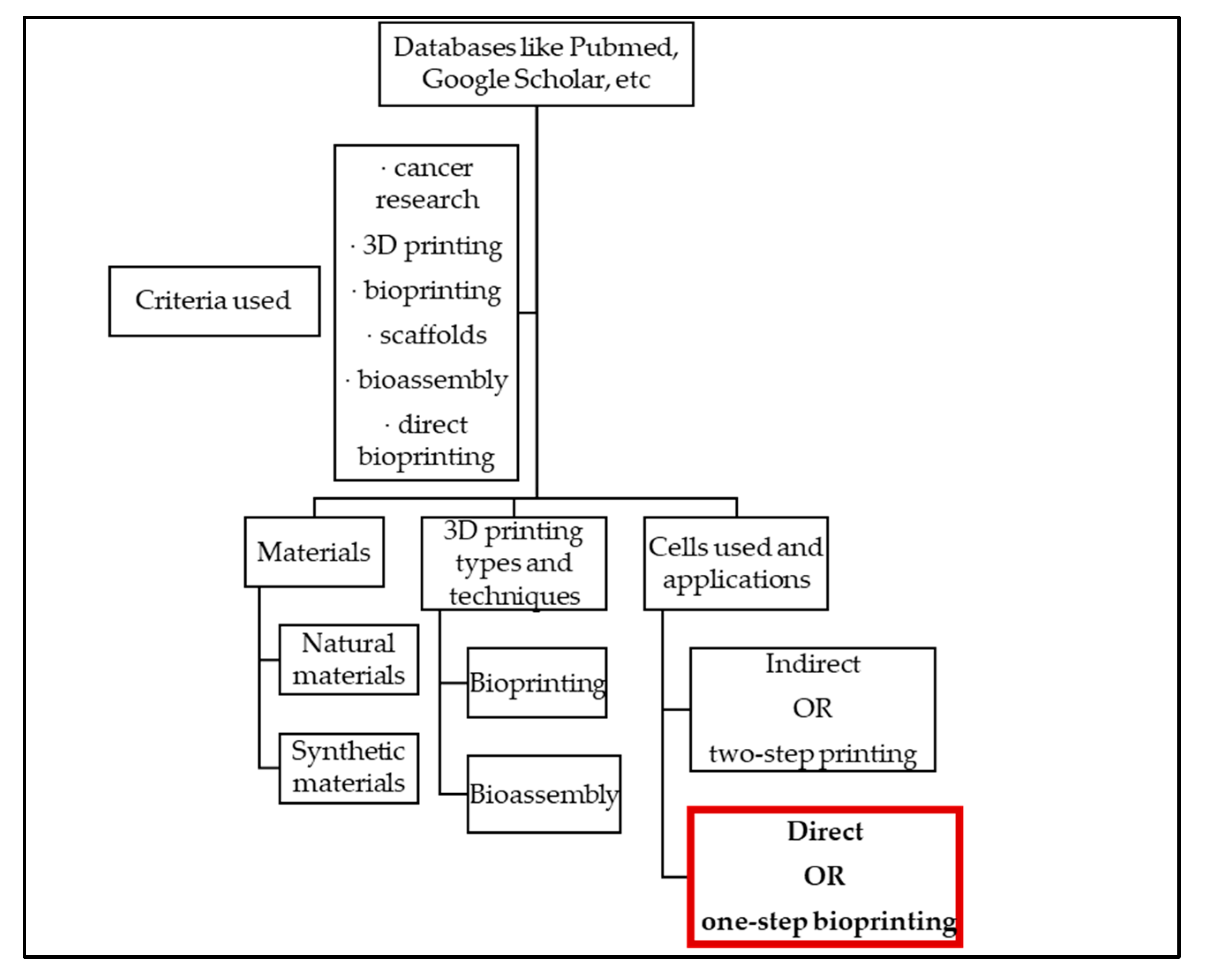
:1. Introduction
2. Materials
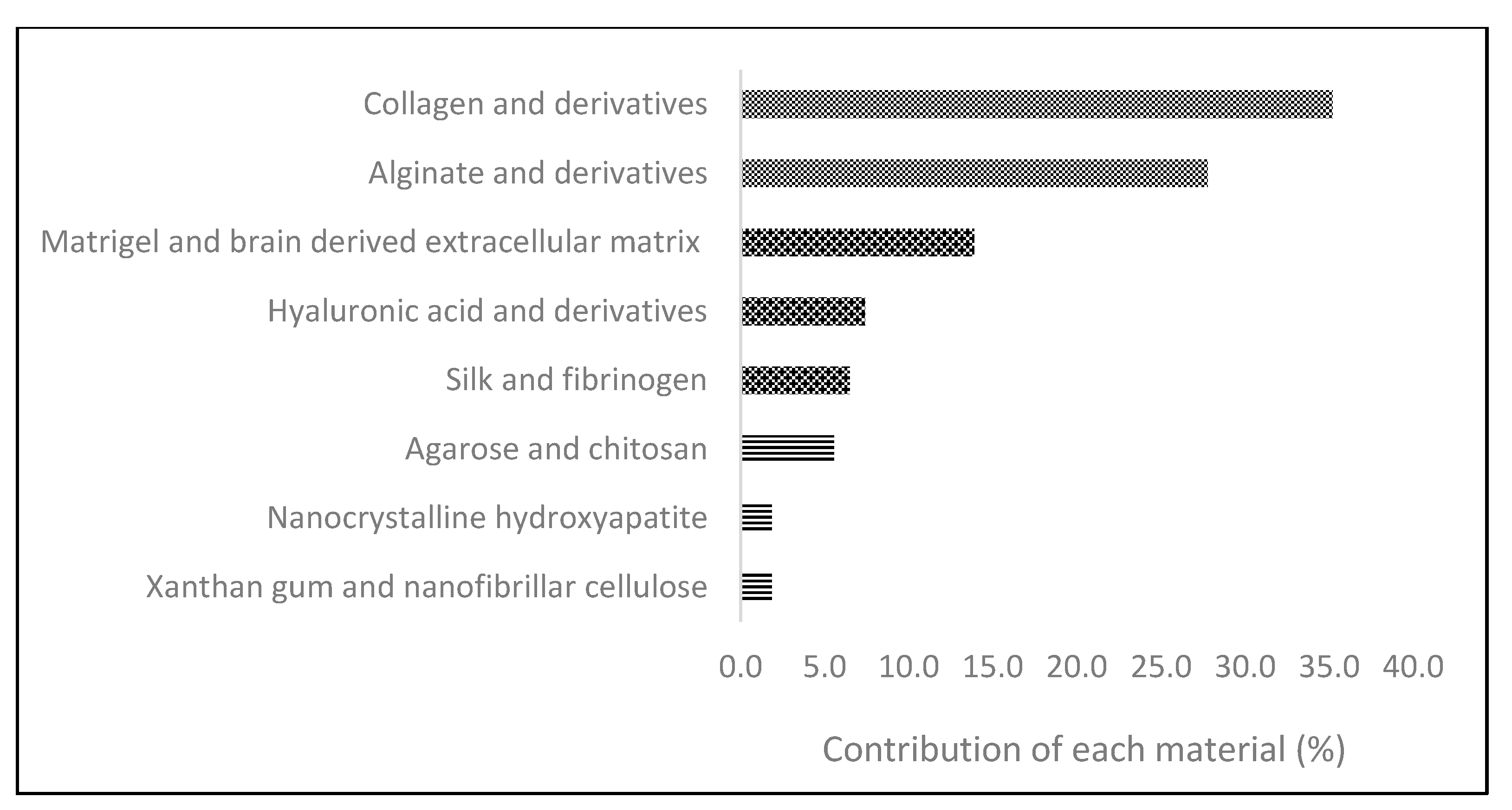
2.1. Natural-Derived Biomaterials
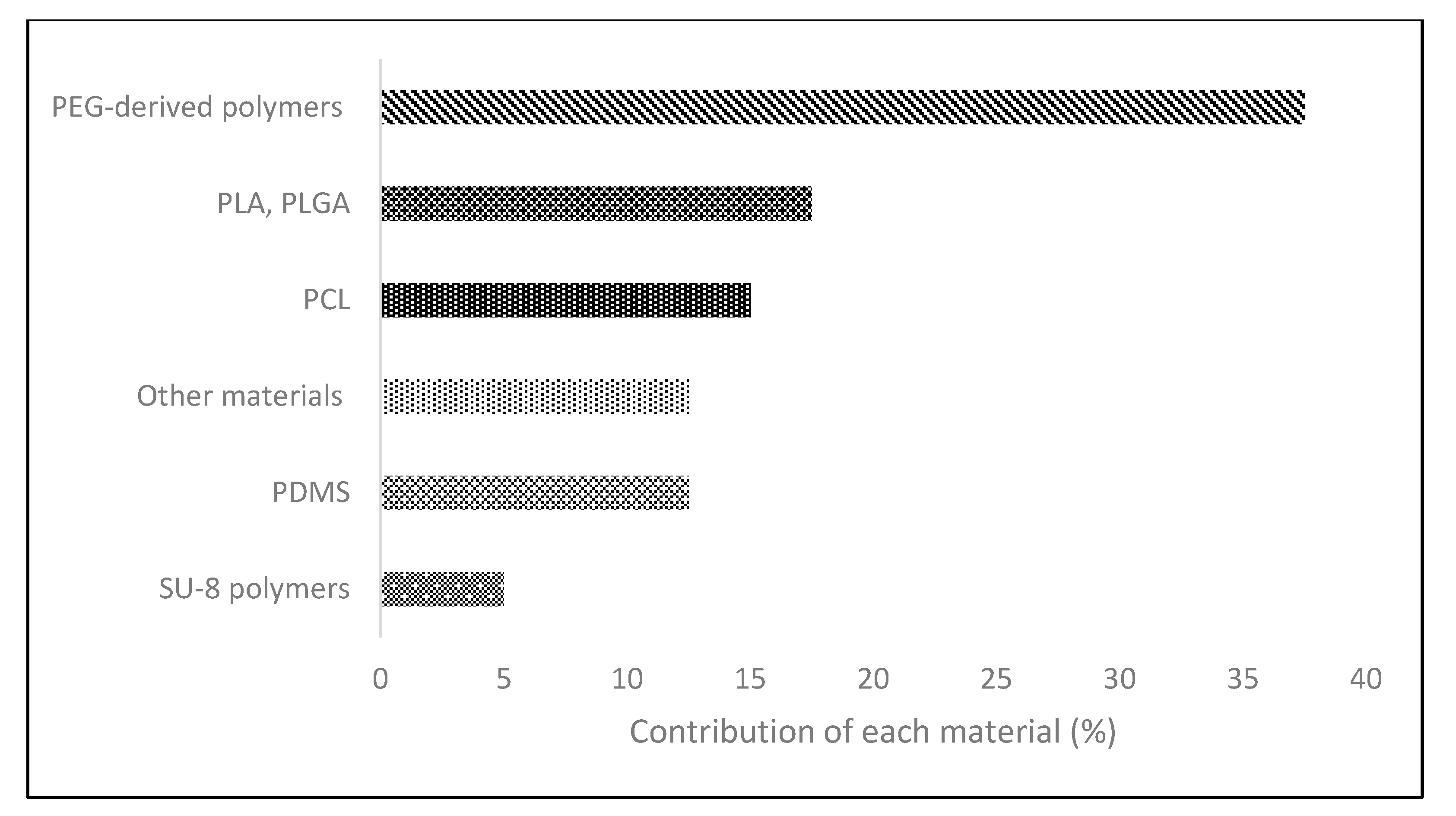
2.2. Synthetic Polymers
3. 3D Printing Techniques
3.1. Bioprinting Methodologies
3.2. Bioassembly Methodologies
4. Cellular Classification for Direct Bioprinting
4.1. Breast Cancer Cells
4.2. Brain-Associated Cancer Cells
4.3. Lung-Associated Cancer Cells
4.4. Liver-Associated Cancer Cells
4.5. Reproductive-Associated Cancer Cells
4.6. Gastric and Colorectal Cancer Cells
4.7. Skin-Associated Cancer Cells
4.8. Urinary Bladder Cancer
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gabriel, S.; Hull, C.W. Apparatus for Production of Three-Dimensional Objects by Stereolithography. U.S. Patent 4575330A, 11 March 1984. [Google Scholar]
- Melchels, F.P.; Feijen, J.; Grijpma, D.W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010, 31, 6121–6130. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Jin, S.; Zhang, F.; Wang, C.; Wang, Y.; Zhou, C.; Cheng, G.J. 3D stereolithography printing of graphene oxide reinforced complex architectures. Nanotechnology 2015, 26, 434003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafezi, F.; Kucukgul, C.; Ozler, S.; Koc, B. Bioprinting: Application of Additive Manufacturing in Medicine. In Additive Manufacturing; CRC Press: Boca Raton, FL, USA, 2015; pp. 197–214. [Google Scholar]
- Mullen, L.; Stamp, R.C.; Brooks, W.K.; Jones, E.; Sutcliffe, C.J. Selective laser melting: A regular unit cell approach for the manufacture of porous, titanium, bone in-growth constructs, suitable for orthopedic applications. J. Biomed. Mater. Res. Part B Appl. Biomater. 2009, 89, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Iwanaga, S.; Henmi, C.; Arai, K.; Nishiyama, Y. Biomatrices and biomaterials for future developments of bioprinting and biofabrication. Biofabrication 2010, 2, 6. [Google Scholar] [CrossRef]
- Thayer, P.; Martinez, H.; Gatenholm, E. History and Trends of 3D Bioprinting. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2020; Volume 2140, pp. 3–18. [Google Scholar]
- Rijal, G.; Li, W. 3D scaffolds in breast cancer research. Biomaterials 2016, 81, 135–156. [Google Scholar] [CrossRef]
- Horvath, L.; Umehara, Y.; Jud, C.; Blank, F.; Petri-Fink, A.; Rothen-Rutishauser, B. Engineering an in vitro air-blood barrier by 3D bioprinting. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; He, R.; Liu, Y. 3D printing scaffolds with hydrogel materials for biomedical applications. Eur. J. Biomed. Res. 2015, 1, 3. [Google Scholar] [CrossRef]
- Tappa, K.; Jammalamadaka, U. Novel biomaterials used in medical 3D printing techniques. J. Funct. Biomater. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Agency for Research on Cancer. 900 World Fact Sheets; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Yabroff, K.R.; Lund, J.; Kepka, D.; Mariotto, A. Economic burden of cancer in the United States: Estimates, projections, and future research. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2006–2014. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kong, D.; Ahmad, A.; Bao, B.; Sarkar, F.H. Pancreatic cancer stem cells: Emerging target for designing novel therapy. Cancer Lett. 2013, 338, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The bioink: A comprehensive review on bioprintable materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Drury, J.L.; Mooney, D.J. Hydrogels for tissue engineering: Scaffold design variables and applications. Biomaterials 2003, 24, 4337–4351. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Atala, A. Biomaterials for Integration with 3-D Bioprinting. Ann. Biomed. Eng. 2015, 43, 730–746. [Google Scholar] [CrossRef]
- Chung, J.H.Y.; Naficy, S.; Yue, Z.; Kapsa, R.; Quigley, A.; Moulton, S.E.; Wallace, G.G. Bio-ink properties and printability for extrusion printing living cells. Biomater. Sci. 2013, 1, 763–773. [Google Scholar] [CrossRef] [Green Version]
- Özbek, S.; Balasubramanian, P.G.; Chiquet-Ehrismann, R.; Tucker, R.P.; Adams, J.C. The evolution of extracellular matrix. Mol. Biol. Cell 2010, 21, 4300–4305. [Google Scholar] [CrossRef] [Green Version]
- Mecham, R.P. Overview of extracellular matrix. Curr. Protoc. Cell Biol. 2012, 57, 10.1.1–10.1.16. [Google Scholar] [CrossRef]
- Ashby, M. Materials Selection in Mechanical Design, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2010; Volume 9780080952, ISBN 9780080952239. [Google Scholar]
- Yue, B. Biology of the Extracellular Matrix: An Overview. J. Glaucoma 2014, 23, S20–S23. [Google Scholar] [CrossRef] [PubMed]
- Chimene, D.; Lennox, K.K.; Kaunas, R.R.; Gaharwar, A.K. Advanced Bioinks for 3D Printing: A Materials Science Perspective. Ann. Biomed. Eng. 2016, 44, 2090–2102. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.Q.; Sun, W.Q.; Connor, J. Sterilization of biomaterials of synthetic and biological origin. In Comprehensive Biomaterials; Elsevier: Amsterdam, The Netherlands, 2011; Volume 4, pp. 127–144. ISBN 9780080552941. [Google Scholar]
- Mewis, J.; Wagner, N.J. Thixotropy. Adv. Colloid Interface Sci. 2009, 147, 214–227. [Google Scholar] [CrossRef]
- Klinge, U.; Klosterhalfen, B.; Müller, M.; Schumpelick, V. Foreign body reaction to meshes used for the repair of abdominal wall hernias. Eur. J. Surg. 1999, 165, 665–673. [Google Scholar] [CrossRef]
- Luttikhuizen, D.T.; Harmsen, M.C.; Van Luyn, M.J.A. Cellular and molecular dynamics in the foreign body reaction. Tissue Eng. 2006, 12, 1955–1970. [Google Scholar] [CrossRef]
- Wells, R.G. The role of matrix stiffness in regulating cell behavior. Hepatology 2008, 47, 1394–1400. [Google Scholar] [CrossRef]
- Kural, M.H.; Billiar, K.L. Regulating tension in three-dimensional culture environments. Exp. Cell Res. 2013, 319, 2447–2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudreau, N.; Werb, Z.; Bissell, M.J. Suppression of apoptosis by basement membrane requires three-dimensional tissue organization and withdrawal from the cell cycle. Proc. Natl. Acad. Sci. USA 1996, 93, 3509–3513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emerman, J.T.; Burwen, S.J.; Pitelka, D.R. Substrate properties influencing ultrastructural differentiation of mammary epithelial cells in culture. Tissue Cell 1979, 11, 109–119. [Google Scholar] [CrossRef]
- Farmer, S.R.; Ben-Ze’ev, A.; Benecke, B.J.; Penman, S. Altered translatability of messenger RNA from suspended anchorage-dependent fibroblasts: Reversal upon cell attachment to a surface. Cell 1978, 15, 627–637. [Google Scholar] [CrossRef]
- Roskelley, C.D.; Desprez, P.Y.; Bissell, M.J. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc. Natl. Acad. Sci. USA 1994, 91, 12378–12382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [Green Version]
- Chaji, S.; Al-Saleh, J.; Gomillion, C.T. Bioprinted three-dimensional cell-laden hydrogels to evaluate adipocyte-breast cancer cell interactions. Gels 2020, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Kievit, F.M.; Florczyk, S.J.; Leung, M.C.; Veiseh, O.; Park, J.O.; Disis, M.L.; Zhang, M. Chitosan-alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials 2010, 31, 5903–5910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermida, M.A.; Kumar, J.D.; Schwarz, D.; Laverty, K.G.; Di Bartolo, A.; Ardron, M.; Bogomolnijs, M.; Clavreul, A.; Brennan, P.M.; Wiegand, U.K.; et al. Three dimensional in vitro models of cancer: Bioprinting multilineage glioblastoma models. Adv. Biol. Regul. 2020, 75. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.G.; Jeong, Y.H.; Kim, Y.; Choi, Y.J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef]
- Wang, X.; Dai, X.; Zhang, X.; Ma, C.; Li, X.; Xu, T.; Lan, Q. 3D bioprinted glioma cell-laden scaffolds enriching glioma stem cells via epithelial–mesenchymal transition. J. Biomed. Mater. Res. Part A 2019, 107, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, L.; Maffini, M.V.; Soto, A.; Sonnenschein, C.; Kaplan, D.L. A complex 3D human tissue culture system based on mammary stromal cells and silk scaffolds for modeling breast morphogenesis and function. Biomaterials 2010, 31, 3920–3929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dippold, D.; Cai, A.; Hardt, M.; Boccaccini, A.R.; Horch, R.E.; Beier, J.P.; Schubert, D.W. Investigation of the batch-to-batch inconsistencies of Collagen in PCL-Collagen nanofibers. Mater. Sci. Eng. C 2019, 95, 217–225. [Google Scholar] [CrossRef]
- Ma, H.; Luo, J.; Sun, Z.; Xia, L.; Shi, M.; Liu, M.; Chang, J.; Wu, C. 3D printing of biomaterials with mussel-inspired nanostructures for tumor therapy and tissue regeneration. Biomaterials 2016, 111, 138–148. [Google Scholar] [CrossRef]
- Gill, B.J.; Gibbons, D.L.; Roudsari, L.C.; Saik, J.E.; Rizvi, Z.H.; Roybal, J.D.; Kurie, J.M.; West, J.L. A synthetic matrix with independently tunable biochemistry and mechanical properties to study epithelial morphogenesis and EMT in a lung adenocarcinoma model. Cancer Res. 2012, 72, 6013–6023. [Google Scholar] [CrossRef] [Green Version]
- Stroock, A.D. Microfluidics. In Optical Biosensors; Elsevier: Amsterdam, The Netherlands, 2008; pp. 659–681. ISBN 9780444531254. [Google Scholar]
- Jeon, J.S.; Bersini, S.; Gilardi, M.; Dubini, G.; Charest, J.L.; Moretti, M.; Kamm, R.D. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl. Acad. Sci. USA 2015, 112, 214–219. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhao, X. A 3D model of ovarian cancer cell lines on peptide nanofiber scaffold to explore the cell-scaffold interaction and chemotherapeutic resistance of anticancer drugs. Int. J. Nanomed. 2011, 6, 303–310. [Google Scholar] [CrossRef] [Green Version]
- Pamfil, D.; Schick, C.; Vasile, C. New Hydrogels Based on Substituted Anhydride Modified Collagen and 2-Hydroxyethyl Methacrylate. Synthesis and Characterization. Ind. Eng. Chem. Res. 2014, 53, 11239–11248. [Google Scholar] [CrossRef]
- Yu, L.; Wei, M. Biomineralization of collagen-based materials for hard tissue repair. Int. J. Mol. Sci. 2021, 22, 1–17. [Google Scholar]
- Aldana, A.A.; Abraham, G.A. Current advances in electrospun gelatin-based scaffolds for tissue engineering applications. Int. J. Pharm. 2017, 523, 441–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J., II; Park, K.M. Advances in gelatin-based hydrogels for wound management. J. Mater. Chem. B 2021, 9, 1503–1520. [Google Scholar] [CrossRef]
- Farokhi, M.; Jonidi Shariatzadeh, F.; Solouk, A.; Mirzadeh, H. Alginate Based Scaffolds for Cartilage Tissue Engineering: A Review. Int. J. Polym. Mater. Polym. Biomater. 2020, 69, 230–247. [Google Scholar] [CrossRef]
- Valente, J.F.A.; Valente, T.A.M.; Alves, P.; Ferreira, P.; Silva, A.; Correia, I.J. Alginate based scaffolds for bone tissue engineering. Mater. Sci. Eng. C 2012, 32, 2596–2603. [Google Scholar] [CrossRef]
- Birgersdotter, A.; Sandberg, R.; Ernberg, I. Gene expression perturbation in vitro—A growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol. 2005, 15, 405–412. [Google Scholar] [CrossRef]
- Kubinova, S. Extracellular matrix based biomaterials for central nervous system tissue repair: The benefits and drawbacks. Neural Regen. Res. 2017, 12, 1430. [Google Scholar] [CrossRef]
- Collins, M.N.; Birkinshaw, C. Hyaluronic acid based scaffolds for tissue engineering—A review. Carbohydr. Polym. 2013, 92, 1262–1279. [Google Scholar] [CrossRef]
- Wang, T.W.; Spector, M. Development of hyaluronic acid-based scaffolds for brain tissue engineering. Acta Biomater. 2009, 5, 2371–2384. [Google Scholar] [CrossRef]
- Tandon, S.; Kandasubramanian, B.; Ibrahim, S.M. Silk-Based Composite Scaffolds for Tissue Engineering Applications. Ind. Eng. Chem. Res. 2020, 59, 17593–17611. [Google Scholar] [CrossRef]
- Santi, S.; Mancini, I.; Dirè, S.; Callone, E.; Speranza, G.; Pugno, N.; Migliaresi, C.; Motta, A. A Bio-inspired Multifunctionalized Silk Fibroin. ACS Biomater. Sci. Eng. 2021, 7, 507–516. [Google Scholar] [CrossRef]
- Rajangam, T.; An, S.S.A. Fibrinogen and fibrin based micro and nano scaffolds incorporated with drugs, proteins, cells and genes for therapeutic biomedical applications. Int. J. Nanomed. 2013, 8, 3641–3662. [Google Scholar]
- Zarrintaj, P.; Manouchehri, S.; Ahmadi, Z.; Saeb, M.R.; Urbanska, A.M.; Kaplan, D.L.; Mozafari, M. Agarose-based biomaterials for tissue engineering. Carbohydr. Polym. 2018, 187, 66–84. [Google Scholar] [CrossRef] [PubMed]
- Salati, M.A.; Khazai, J.; Tahmuri, A.M.; Samadi, A.; Taghizadeh, A.; Taghizadeh, M.; Zarrintaj, P.; Ramsey, J.D.; Habibzadeh, S.; Seidi, F.; et al. Agarose-Based biomaterials: Opportunities and challenges in cartilage tissue engineering. Polymers 2020, 12, 1150. [Google Scholar] [CrossRef] [PubMed]
- Gnavi, S.; Barwig, C.; Freier, T.; Haastert-Talini, K.; Grothe, C.; Geuna, S. The use of chitosan-based scaffolds to enhance regeneration in the nervous system. In International Review of Neurobiology; Academic Press Inc.: Cambridge, MA, USA, 2013; Volume 109, pp. 1–62. [Google Scholar]
- Sivashankari, P.R.; Prabaharan, M. Prospects of chitosan-based scaffolds for growth factor release in tissue engineering. Int. J. Biol. Macromol. 2016, 93, 1382–1389. [Google Scholar] [CrossRef]
- Family, R.; Solati-Hashjin, M.; Nik, S.N.; Nemati, A. Surface modification for titanium implants by hydroxyapatite nanocomposite. Casp. J. Intern. Med. 2012, 3, 460–465. [Google Scholar]
- Mortensen, A.; Aguilar, F.; Crebelli, R.; Di Domenico, A.; Frutos, M.J.; Galtier, P.; Gott, D.; Gundert-Remy, U.; Lambré, C.; Leblanc, J.C.; et al. Re-evaluation of xanthan gum (E 415) as a food additive. EFSA J. 2017, 15, 4909. [Google Scholar] [CrossRef] [Green Version]
- Salmoria, G.V.; Klauss, P.; Paggi, R.A.; Kanis, L.A.; Lago, A. Structure and mechanical properties of cellulose based scaffolds fabricated by selective laser sintering. Polym. Test. 2009, 28, 648–652. [Google Scholar] [CrossRef]
- Müller, F.A.; Müller, L.; Hofmann, I.; Greil, P.; Wenzel, M.M.; Staudenmaier, R. Cellulose-based scaffold materials for cartilage tissue engineering. Biomaterials 2006, 27, 3955–3963. [Google Scholar] [CrossRef]
- Sheikholeslam, M.; Wright, M.E.E.; Jeschke, M.G.; Amini-Nik, S. Biomaterials for Skin Substitutes. Adv. Healthc. Mater. 2018, 7. [Google Scholar] [CrossRef]
- Han, D.K.; Park, K.D.; Hubbell, J.A.; Kim, Y.H. Surface characteristics and biocompatibility of lactide-based poly(ethylene glycol) scaffolds for tissue engineering. J. Biomater. Sci. Polym. Ed. 1998, 9, 667–680. [Google Scholar] [CrossRef]
- Ghalia, M.A.; Dahman, Y. Biodegradable poly(lactic acid)-based scaffolds: Synthesis and biomedical applications. J. Polym. Res. 2017, 24. [Google Scholar] [CrossRef]
- Niaounakis, M. Biopolymers: Processing and Products, 1st ed.; Andrew, W., Ed.; Elsevier Science: New York, NY, USA, 2014; ISBN 9780323279383. [Google Scholar]
- Zhang, Y.Z.; Venugopal, J.; Huang, Z.M.; Lim, C.T.; Ramakrishna, S. Characterization of the surface biocompatibility of the electrospun PCL-Collagen nanofibers using fibroblasts. Biomacromolecules 2005, 6, 2583–2589. [Google Scholar] [CrossRef]
- Osathanon, T.; Chuenjitkuntaworn, B.; Nowwarote, N.; Supaphol, P.; Sastravaha, P.; Subbalekha, K.; Pavasant, P. The responses of human adipose-derived mesenchymal stem cells on polycaprolactone-based scaffolds: An in vitro study. Tissue Eng. Regen. Med. 2014, 11, 239–246. [Google Scholar] [CrossRef]
- McDonald, J.C.; Whitesides, G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002, 35, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Conradie, E.H.; Moore, D.F. SU-8 thick photoresist processing as a functional material for MEMS applications. J. Micromech. Microeng. 2002, 12, 368–374. [Google Scholar] [CrossRef]
- Charbe, N.; McCarron, P.A.; Tambuwala, M.M. Three-dimensional bio-printing: A new frontier in oncology research. World J. Clin. Oncol. 2017, 8, 21–36. [Google Scholar] [CrossRef]
- Mao, H.; Yang, L.; Zhu, H.; Wu, L.; Ji, P.; Yang, J.; Gu, Z. Recent advances and challenges in materials for 3D bioprinting. Prog. Nat. Sci. Mater. Int. 2020, 30, 618–634. [Google Scholar] [CrossRef]
- Heinrich, M.A.; Bansal, R.; Lammers, T.; Zhang, Y.S.; Michel Schiffelers, R.; Prakash, J. 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics. Adv. Mater. 2019, 31. [Google Scholar] [CrossRef] [PubMed]
- Vinson, B.T.; Phamduy, T.B.; Shipman, J.; Riggs, B.; Strong, A.L.; Sklare, S.C.; Murfee, W.L.; Burow, M.E.; Bunnell, B.A.; Huang, Y.; et al. Laser direct-write based fabrication of a spatially-defined, biomimetic construct as a potential model for breast cancer cell invasion into adipose tissue. Biofabrication 2017, 9. [Google Scholar] [CrossRef]
- Chen, J.; Liu, C.Y.; Wang, X.; Sweet, E.; Liu, N.; Gong, X.; Lin, L. 3D printed microfluidic devices for circulating tumor cells (CTCs) isolation. Biosens. Bioelectron. 2019, 150, 111900. [Google Scholar] [CrossRef]
- Ried, K.; Eng, P.; Sali, A. Screening for circulating tumour cells allows early detection of cancer and monitoring of treatment effectiveness: An observational study. Asian Pac. J. Cancer Prev. 2017, 18, 2275–2285. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Martinez, A.D.; Sole-Gras, M.; Dykes, S.S.; Wakefield, Z.R.; Bauer, K.; Majbour, D.; Bundy, A.; Pampo, C.; Burow, M.E.; Siemann, D.W.; et al. Bioprinting on Live Tissue for Investigating Cancer Cell Dynamics. Tissue Eng. Part A 2020, 27, 1–43. [Google Scholar] [CrossRef]
- Polonio-Alcalá, E.; Rabionet, M.; Gallardo, X.; Angelats, D.; Ciurana, J.; Ruiz-Martínez, S.; Puig, T. PLA electrospun scaffolds for three-dimensional triple-negative breast cancer cell culture. Polymers 2019, 11, 916. [Google Scholar] [CrossRef] [Green Version]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef]
- Redwood, B.; Schffer, F.; Garret, B. The 3D Printing Handbook: Technologies, Design and Applications, 1st ed.; Wiley: Hoboken, NJ, USA, 2017; ISBN 9082748509. [Google Scholar]
- Chhabra, D. Comparison and analysis of different 3d printing techniques. Int. J. Latest Trends Eng. Technol. 2017, 8. [Google Scholar] [CrossRef]
- Dahlin, R.L.; Kasper, F.K.; Mikos, A.G. Polymeric Nanofibers in Tissue Engineering. Tissue Eng. Part B Rev. 2011, 17, 349–364. [Google Scholar] [CrossRef] [Green Version]
- Carter, P.; Bhattarai, N. Bioscaffolds: Fabrication and Performance. In Engineered Biomimicry; Elsevier: Amsterdam, The Netherlands, 2013; pp. 161–188. ISBN 9780124159952. [Google Scholar]
- Gaudillere, C.; Serra, J.M. Freeze-casting: Fabrication of highly porous and hierarchical ceramic supports for energy applications. Bol. Soc. Esp. Cerám. Vidr. 2016, 55, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Groll, J.; Boland, T.; Blunk, T.; Burdick, J.A.; Cho, D.-W.; Dalton, P.D.; Derby, B.; Forgacs, G.; Li, Q.; Mironov, V.A.; et al. Biofabrication: Reappraising the definition of an evolving field. Biofabrication 2016, 8, 013001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Evans, D. Tissue Engineering: The Future of Stem Cells. Top. Tissue Eng. 2005, 2, 1–22. [Google Scholar]
- Cui, H.; Nowicki, M.; Fisher, J.P.; Zhang, L.G. 3D Bioprinting for Organ Regeneration. Adv. Healthc. Mater. 2017, 6, 1601118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browne, S.; Pandit, A. Multi-modal delivery of therapeutics using biomaterial scaffolds. J. Mater. Chem. B 2014, 2, 6692–6707. [Google Scholar] [CrossRef]
- Langer, E.M.; Allen-Petersen, B.L.; King, S.M.; Kendsersky, N.D.; Turnidge, M.A.; Kuziel, G.M.; Riggers, R.; Samatham, R.; Amery, T.S.; Jacques, S.L.; et al. Modeling Tumor Phenotypes In Vitro with Three-Dimensional Bioprinting. Cell Rep. 2019, 26, 608–623.e6. [Google Scholar] [CrossRef] [Green Version]
- Grolman, J.M.; Zhang, D.; Smith, A.M.; Moore, J.S.; Kilian, K.A. Rapid 3D Extrusion of Synthetic Tumor Microenvironments. Adv. Mater. 2015, 27, 5512–5517. [Google Scholar] [CrossRef] [Green Version]
- Verissimo, A.R.; Nakayama, K. Scaffold-Free Biofabrication. In 3D Printing and Biofabrication; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–20. [Google Scholar]
- Ozbolat, I.T. Scaffold-Based or Scaffold-Free Bioprinting: Competing or Complementing Approaches? J. Nanotechnol. Eng. Med. 2015, 6. [Google Scholar] [CrossRef]
- Rosso, F.; Giordano, A.; Barbarisi, M.; Barbarisi, A. From Cell-ECM Interactions to Tissue Engineering. J. Cell. Physiol. 2004, 199, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.S.; Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Bartosh, T.J.; Ylostalo, J.H. Preparation of anti-inflammatory mesenchymal stem/precursor cells (MSCs) through sphere formation using hanging-drop culture technique. Curr. Protoc. Stem Cell Biol. 2014, 1. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Takayama, S.; Park, J. Formation and manipulation of cell spheroids using a density adjusted PEG/DEX aqueous two phase system. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Yip, D.; Cho, C.H. A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem. Biophys. Res. Commun. 2013, 433, 327–332. [Google Scholar] [CrossRef]
- Van Pel, D.M.; Harada, K.; Song, D.; Naus, C.C.; Sin, W.C. Modelling glioma invasion using 3D bioprinting and scaffold-free 3D culture. J. Cell Commun. Signal. 2018, 12, 723–730. [Google Scholar] [CrossRef]
- Campbell, A.; Philipovskiy, A.; Heydarian, R.; Varela-Ramirez, A.; Gutierrez, D.A.; Solis, L.H.; Furth, M.E.; Boland, T. 2D and 3D thermally bioprinted human MCF-7 breast cancer cells: A promising model for drug discovery. J. Clin. Oncol. 2019, 37, 2605. [Google Scholar] [CrossRef]
- Reid, J.A.; Palmer, X.L.; Mollica, P.A.; Northam, N.; Sachs, P.C.; Bruno, R.D. A 3D bioprinter platform for mechanistic analysis of tumoroids and chimeric mammary organoids. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, D.M.; Dias, A.D.; Chrisey, D.B.; Corr, D.T. Single-step laser-based fabrication and patterning of cell-encapsulated alginate microbeads. Biofabrication 2013, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, K.; Huang, G.; Liu, J.; Zhang, X.; Ma, Y.; Lu, T.; Xu, F. Bioprinting-Based High-Throughput Fabrication of Three-Dimensional MCF-7 Human Breast Cancer Cellular Spheroids. Engineering 2015, 1, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Peela, N.; Sam, F.S.; Christenson, W.; Truong, D.; Watson, A.W.; Mouneimne, G.; Ros, R.; Nikkhah, M. A three dimensional micropatterned tumor model for breast cancer cell migration studies. Biomaterials 2016, 81, 72–83. [Google Scholar] [CrossRef]
- Chen, L.; Xiao, Z.; Meng, Y.; Zhao, Y.; Han, J.; Su, G.; Chen, B.; Dai, J. The enhancement of cancer stem cell properties of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and anti-cancer drugs. Biomaterials 2012, 33, 1437–1444. [Google Scholar] [CrossRef]
- Bersini, S.; Jeon, J.S.; Dubini, G.; Arrigoni, C.; Chung, S.; Charest, J.L.; Moretti, M.; Kamm, R.D. A microfluidic 3D invitro model for specificity of breast cancer metastasis to bone. Biomaterials 2014, 35, 2454–2461. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, W.; Kuss, M.; Mirza, S.; Qi, D.; Krasnoslobodtsev, A.; Zeng, J.; Band, H.; Band, V.; Duan, B. 3D Bioprinting of Breast Cancer Models for Drug Resistance Study. ACS Biomater. Sci. Eng. 2018, 4, 4401–4411. [Google Scholar] [CrossRef]
- Swaminathan, S.; Hamid, Q.; Sun, W.; Clyne, A.M. Bioprinting of 3D breast epithelial spheroids for human cancer models. Biofabrication 2019, 11, 025003. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Munguia-Lopez, J.G.; Flores-Torres, S.; Grant, J.; Vijayakumar, S.; de Leon-Rodriguez, A.; Kinsella, J.M. Directing the Self-Assembly of Tumour Spheroids by Bioprinting Cellular Heterogeneous Models within Alginate/Gelatin Hydrogels. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zhu, W.; Nowicki, M.; Miao, S.; Cui, H.; Holmes, B.; Glazer, R.I.; Zhang, L.G. 3D Bioprinting a Cell-Laden Bone Matrix for Breast Cancer Metastasis Study. ACS Appl. Mater. Interfaces 2016, 8, 30017–30026. [Google Scholar] [CrossRef]
- Schmid, R.; Schmidt, S.K.; Hazur, J.; Detsch, R.; Maurer, E.; Boccaccini, A.R.; Hauptstein, J.; Teßmar, J.; Blunk, T.; Schrüfer, S.; et al. Comparison of hydrogels for the development of well-defined 3d cancer models of breast cancer and melanoma. Cancers 2020, 12, 2320. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Vecchione-Koval, T.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncology 2017, 19, v1–v88. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, X.; Dai, X.; Zhang, X.; Zhang, J.; Xu, T.; Lan, Q. Coaxial extrusion bioprinted shell-core hydrogel microfibers mimic glioma microenvironment and enhance the drug resistance of cancer cells. Colloids Surf. B Biointerfaces 2018, 171, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, F.; He, Z.; Ma, Y.; Uchiyama, K.; Lin, J.M. A novel approach for precisely controlled multiple cell patterning in microfluidic chips by inkjet printing and the detection of drug metabolism and diffusion. Analyst 2016, 141, 2940–2947. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Ma, C.; Lan, Q.; Xu, T. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.F.D.; Marquez, A.B.; O’seanain, C.; Fischer, H.; Blaeser, A.; Vogt, M.; Corallo, D.; Aveic, S. Exploring cancer cell behavior in vitro in three-dimensional multicellular bioprintable collagen-based hydrogels. Cancers 2019, 11, 180. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Roman, N.; Chalmers, A.J. Patient-specific 3D-printed glioblastomas. Nat. Biomed. Eng. 2019, 3, 498–499. [Google Scholar] [CrossRef]
- Rao, S.S.; Dejesus, J.; Short, A.R.; Otero, J.J.; Sarkar, A.; Winter, J.O. Glioblastoma behaviors in three-dimensional collagen-hyaluronan composite hydrogels. ACS Appl. Mater. Interfaces 2013, 5, 9276–9284. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, L.; Ouyang, J.; Li, X.; Zhang, X.; Lan, Q.; Xu, T. Coaxial 3D bioprinting of self-assembled multicellular heterogeneous tumor fibers. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wald, N.J.; Nanchahal, K.; Thompson, S.G.; Cuckle, H.S. Does breathing other people’s tobacco smoke cause lung cancer? Br. Med. J. (Clin. Res. Ed). 1986, 293, 1217–1222. [Google Scholar] [CrossRef] [Green Version]
- Kjuus, H.; Skjaerven, R.; Langard, S.; Lien, J.T.; Aamodt, T. A case-referent study of lung cancer, occupational exposures and smoking. II Role of asbestos exposure. Scand. J. Work. Environ. Health 1986, 12, 203–209. [Google Scholar] [CrossRef]
- Yang, P.; Wentzlaff, K.A.; Katzmann, J.A.; Marks, R.S.; Allen, M.S.; Lesnick, T.G.; Lindor, N.M.; Myers, J.L.; Wiegert, E.; Midthun, D.E.; et al. Alpha1-antitrypsin deficiency allele carriers among lung cancer patients. Cancer Epidemiol. Biomark. Prev. 1999, 8, 461–465. [Google Scholar]
- Doll, R.; Peto, R. Mortality in relation to smoking: 20 years’ observations on male British doctors. Br. Med. J. 1976, 2, 1525–1536. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, X.; Dai, X.; Wang, X.; Li, X.; Diao, J.; Xu, T. Tumor-like lung cancer model based on 3D bioprinting. 3 Biotech 2018, 8, 501. [Google Scholar] [CrossRef] [PubMed]
- Roudsari, L.C.; Jeffs, S.E.; Witt, A.S.; Gill, B.J.; West, J.L. A 3D Poly(ethylene glycol)-based Tumor Angiogenesis Model to Study the Influence of Vascular Cells on Lung Tumor Cell Behavior. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Mao, S.; He, J.; Zhao, Y.; Liu, T.; Xie, F.; Yang, H.; Mao, Y.; Pang, Y.; Sun, W. Bioprinting of patient-derived in vitro intrahepatic cholangiocarcinoma tumor model: Establishment, evaluation and anti-cancer drug testing. Biofabrication 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level. JAMA Oncol. 2017, 3, 1683. [Google Scholar] [CrossRef]
- Ng, A.W.T.; Poon, S.L.; Huang, M.N.; Lim, J.Q.; Boot, A.; Yu, W.; Suzuki, Y.; Thangaraju, S.; Ng, C.C.Y.; Tan, P.; et al. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci. Transl. Med. 2017, 9, eaan6446. [Google Scholar] [CrossRef] [Green Version]
- Schulze, K.; Imbeaud, S.; Letouzé, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef]
- Xu, X.X.; Liu, C.; Liu, Y.; Li, N.; Guo, X.; Wang, S.J.; Sun, G.W.; Wang, W.; Ma, X.J. Encapsulated human hepatocellular carcinoma cells by alginate gel beads as an in vitro metastasis model. Exp. Cell Res. 2013, 319, 2135–2144. [Google Scholar] [CrossRef]
- Rahman, N. Realizing the promise of cancer predisposition genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Knudson, A.G. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [Green Version]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Alsop, K.; Fereday, S.; Meldrum, C.; DeFazio, A.; Emmanuel, C.; George, J.; Dobrovic, A.; Birrer, M.J.; Webb, P.M.; Stewart, C.; et al. BRCA Mutation Frequency and Patterns of Treatment Response in BRCA Mutation–Positive Women With Ovarian Cancer: A Report From the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012, 30, 2654–2663. [Google Scholar] [CrossRef] [Green Version]
- Ketabi, Z.; Bartuma, K.; Bernstein, I.; Malander, S.; Grönberg, H.; Björck, E.; Holck, S.; Nilbert, M. Ovarian cancer linked to lynch syndrome typically presents as early-onset, non-serous epithelial tumors. Gynecol. Oncol. 2011, 121, 462–465. [Google Scholar] [CrossRef]
- Moorman, P.G.; Havrilesky, L.J.; Gierisch, J.M.; Coeytaux, R.R.; Lowery, W.J.; Peragallo Urrutia, R.; Dinan, M.; McBroom, A.J.; Hasselblad, V.; Sanders, G.D.; et al. Oral Contraceptives and Risk of Ovarian Cancer and Breast Cancer Among High-Risk Women: A Systematic Review and Meta-Analysis. J. Clin. Oncol. 2013, 31, 4188–4198. [Google Scholar] [CrossRef]
- Bassuk, S.S.; Manson, J.E. Oral contraceptives and menopausal hormone therapy: Relative and attributable risks of cardiovascular disease, cancer, and other health outcomes. Ann. Epidemiol. 2015, 25, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Rice, M.S.; Hankinson, S.E.; Tworoger, S.S. Tubal ligation, hysterectomy, unilateral oophorectomy, and risk of ovarian cancer in the Nurses’ Health Studies. Fertil. Steril. 2014, 102. [Google Scholar] [CrossRef] [Green Version]
- Gaitskell, K.; Green, J.; Pirie, K.; Reeves, G. Tubal ligation and ovarian cancer risk in a large cohort: Substantial variation by histological type. Int. J. Cancer 2016, 138, 1076–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannioto, R.A.; Moysich, K.B. Epithelial ovarian cancer and recreational physical activity: A review of the epidemiological literature and implications for exercise prescription. Gynecol. Oncol. 2015, 137, 559–573. [Google Scholar] [CrossRef] [Green Version]
- Wentzensen, N.; Poole, E.M.; Trabert, B.; White, E.; Arslan, A.A.; Patel, A.V.; Setiawan, V.W.; Visvanathan, K.; Weiderpass, E.; Adami, H.-O.; et al. Ovarian Cancer Risk Factors by Histologic Subtype: An Analysis From the Ovarian Cancer Cohort Consortium. J. Clin. Oncol. 2016, 34, 2888–2898. [Google Scholar] [CrossRef]
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef]
- Small, W.; Bacon, M.A.; Bajaj, A.; Chuang, L.T.; Fisher, B.J.; Harkenrider, M.M.; Jhingran, A.; Kitchener, H.C.; Mileshkin, L.R.; Viswanathan, A.N.; et al. Cervical cancer: A global health crisis. Cancer 2017, 123, 2404–2412. [Google Scholar] [CrossRef] [Green Version]
- Olorunfemi, G.; Ndlovu, N.; Masukume, G.; Chikandiwa, A.; Pisa, P.T.; Singh, E. Temporal trends in the epidemiology of cervical cancer in South Africa (1994–2012). Int. J. Cancer 2018, 143, 2238–2249. [Google Scholar] [CrossRef]
- Halkitis, P.N.; Valera, P.; LoSchiavo, C.E.; Goldstone, S.E.; Kanztanou, M.; Maiolatesi, A.J.; Ompad, D.C.; Greene, R.E.; Kapadia, F. Human Papillomavirus Vaccination and Infection in Young Sexual Minority Men: The P18 Cohort Study. AIDS Patient Care STDS 2019, 33, 149–156. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Kim, H.; Barron, J.A.; Krizman, D.B.; Chrisey, D.B.; Jackman, S.; Auyeung, R.Y.C.; Spargo, B.J. Laser Printing of Pluripotent Embryonal Carcinoma Cells. Tissue Eng. 2004, 10, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yao, R.; Ouyang, L.; Ding, H.; Zhang, T.; Zhang, K.; Cheng, S.; Sun, W. Three-dimensional printing of Hela cells for cervical tumor model in vitro. Biofabrication 2014, 6. [Google Scholar] [CrossRef]
- Xu, F.; Celli, J.; Rizvi, I.; Moon, S.; Hasan, T.; Demirci, U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol. J. 2011, 6, 204–212. [Google Scholar] [CrossRef]
- Xu, G.; Yin, F.; Wu, H.; Hu, X.; Zheng, L.; Zhao, J. In vitro ovarian cancer model based on three-dimensional agarose hydrogel. J. Tissue Eng. 2014, 5, 204173141352043. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Gastroenterol. Rev. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Fang, W.-L.; Wang, R.-F.; Liu, C.-A.; Yang, M.-H.; Lo, S.-S.; Wu, C.-W.; Li, A.F.-Y.; Shyr, Y.-M.; Huang, K.-H. Clinicopathological Variation of Lauren Classification in Gastric Cancer. Pathol. Oncol. Res. 2016, 22, 197–202. [Google Scholar] [CrossRef]
- Liang, Y.-X. Characteristics and prognosis of gastric cancer in patients aged ≥70 years. World J. Gastroenterol. 2013, 19, 6568. [Google Scholar] [CrossRef]
- Petrelli, F.; Berenato, R.; Turati, L.; Mennitto, A.; Steccanella, F.; Caporale, M.; Dallera, P.; de Braud, F.; Pezzica, E.; Di Bartolomeo, M.; et al. Prognostic value of diffuse versus intestinal histotype in patients with gastric cancer: A systematic review and meta-analysis. J. Gastrointest. Oncol. 2017, 8, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Crew, K.D.; Neugut, A.I. Epidemiology of gastric cancer. World J. Gastroenterol. 2006, 12, 354. [Google Scholar] [CrossRef]
- Han, J.P.; Hong, S.J.; Kim, H.K. Long-term outcomes of early gastric cancer diagnosed as mixed adenocarcinoma after endoscopic submucosal dissection. J. Gastroenterol. Hepatol. 2015, 30, 316–320. [Google Scholar] [CrossRef]
- Bae, J.-M.; Kim, E.H. Epstein-Barr Virus and Gastric Cancer Risk: A Meta-analysis With Meta-regression of Case-control Studies. J. Prev. Med. Public Health 2016, 49, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Wroblewski, L.E.; Peek, R.M.; Wilson, K.T. Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef] [Green Version]
- Henrikson, N.B.; Webber, E.M.; Goddard, K.A.; Scrol, A.; Piper, M.; Williams, M.S.; Zallen, D.T.; Calonge, N.; Ganiats, T.G.; Janssens, A.C.J.W.; et al. Family history and the natural history of colorectal cancer: Systematic review. Genet. Med. 2015, 17, 702–712. [Google Scholar] [CrossRef] [Green Version]
- Jess, T.; Rungoe, C.; Peyrin-Biroulet, L. Risk of Colorectal Cancer in Patients With Ulcerative Colitis: A Meta-analysis of Population-Based Cohort Studies. Clin. Gastroenterol. Hepatol. 2012, 10, 639–645. [Google Scholar] [CrossRef]
- Cottet, V.; Jooste, V.; Fournel, I.; Bouvier, A.M.; Faivre, J.; Bonithon-Kopp, C. Long-term risk of colorectal cancer after adenoma removal: A population-based cohort study. Gut 2012, 61, 1180–1186. [Google Scholar] [CrossRef] [Green Version]
- Shakibaei, M.; Kraehe, P.; Popper, B.; Shayan, P.; Goel, A.; Buhrmann, C. Curcumin potentiates antitumor activity of 5-fluorouracil in a 3D alginate tumor microenvironment of colorectal cancer. BMC Cancer 2015, 15, 250. [Google Scholar] [CrossRef] [Green Version]
- Amorim, S.; Soares Da Costa, D.; Pashkuleva, I.; Reis, C.A.; Reis, R.L.; Pires, R.A. 3D hydrogel mimics of the tumor microenvironment: The interplay among hyaluronic acid, stem cells and cancer cells. Biomater. Sci. 2021, 9, 252–260. [Google Scholar] [CrossRef]
- De Vries, E.; Willem Coebergh, J. Cutaneous malignant melanoma in Europe. Eur. J. Cancer 2004, 40, 2355–2366. [Google Scholar] [CrossRef]
- Dennis, L.K. Analysis of the Melanoma Epidemic, Both Apparent and Real. Arch. Dermatol. 1999, 135, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Perera, E.; Gnaneswaran, N.; Staines, C.; Win, A.K.; Sinclair, R. Incidence and prevalence of non-melanoma skin cancer in Australia: A systematic review. Australas. J. Dermatol. 2015, 56, 258–267. [Google Scholar] [CrossRef]
- National Cancer Institute SEER. Stats Fact Sheets: Melanoma of the Skin. Available online: https://seer.cancer.gov/statfacts/html/melan.html (accessed on 14 May 2021).
- Katalinic, A.; Kunze, U.; Schafer, T. Epidemiology of cutaneous melanoma and non-melanoma skin cancer in Schleswig-Holstein, Germany: Incidence, clinical subtypes, tumour stages and localization (epidemiology of skin cancer). Br. J. Dermatol. 2003, 149, 1200–1206. [Google Scholar] [CrossRef]
- Leiter, U.; Garbe, C. Epidemiology of Melanoma and Nonmelanoma Skin Cancer—The Role of Sunlight. In Sunlight, Vitamin D and Skin Cancer; Springer: New York, NY, USA, 2008; Volume 624, pp. 89–103. ISBN 9780387775739. [Google Scholar]
- Rubin, A.I.; Chen, E.H.; Ratner, D. Basal-Cell Carcinoma. N. Engl. J. Med. 2005, 353, 2262–2269. [Google Scholar] [CrossRef]
- Wehner, M.R.; Shive, M.L.; Chren, M.-M.; Han, J.; Qureshi, A.A.; Linos, E. Indoor tanning and non-melanoma skin cancer: Systematic review and meta-analysis. BMJ 2012, 345, e5909. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.K.; Schmid, R.; Arkudas, A.; Kengelbach-Weigand, A.; Bosserhoff, A.K. Tumor Cells Develop Defined Cellular Phenotypes After 3D-Bioprinting in Different Bioinks. Cells 2019, 8, 1295. [Google Scholar] [CrossRef] [Green Version]
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef]
- Hendricksen, K.; Aziz, A.; Bes, P.; Chun, F.K.H.; Dobruch, J.; Kluth, L.A.; Gontero, P.; Necchi, A.; Noon, A.P.; van Rhijn, B.W.G.; et al. Discrepancy Between European Association of Urology Guidelines and Daily Practice in the Management of Non–muscle-invasive Bladder Cancer: Results of a European Survey. Eur. Urol. Focus 2019, 5, 681–688. [Google Scholar] [CrossRef] [Green Version]
- Shariat, S.F.; Milowsky, M.; Droller, M.J. Bladder cancer in the elderly. Urol. Oncol. Semin. Orig. Investig. 2009, 27, 653–667. [Google Scholar] [CrossRef] [Green Version]
- Dobruch, J.; Daneshmand, S.; Fisch, M.; Lotan, Y.; Noon, A.P.; Resnick, M.J.; Shariat, S.F.; Zlotta, A.R.; Boorjian, S.A. Gender and Bladder Cancer: A Collaborative Review of Etiology, Biology, and Outcomes. Eur. Urol. 2016, 69, 300–310. [Google Scholar] [CrossRef]
- Ishida, K.; Hsieh, M.H. Understanding Urogenital Schistosomiasis-Related Bladder Cancer: An Update. Front. Med. 2018, 5, 223. [Google Scholar] [CrossRef] [Green Version]
- Moschini, M.; Zaffuto, E.; Karakiewicz, P.I.; Andrea, D.D.; Foerster, B.; Abufaraj, M.; Soria, F.; Mattei, A.; Montorsi, F.; Briganti, A.; et al. External Beam Radiotherapy Increases the Risk of Bladder Cancer When Compared with Radical Prostatectomy in Patients Affected by Prostate Cancer: A Population-based Analysis. Eur. Urol. 2019, 75, 319–328. [Google Scholar] [CrossRef]
- Warschkow, R.; Güller, U.; Cerny, T.; Schmied, B.M.; Plasswilm, L.; Putora, P.M. Secondary malignancies after rectal cancer resection with and without radiation therapy: A propensity-adjusted, population-based SEER analysis. Radiother. Oncol. 2017, 123, 139–146. [Google Scholar] [CrossRef]
- Kim, M.J.; Chi, B.H.; Yoo, J.J.; Ju, Y.M.; Whang, Y.M.; Chang, I.H. Structure establishment of three-dimensional (3D) cell culture printing model for bladder cancer. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Das Gupta, T.; Brasfield, R. Metastatic melanoma.A clinicopathological study. Cancer 1964, 17, 1323–1339. [Google Scholar] [CrossRef]
- Atala, A.; Bauer, S.B.; Soker, S.; Yoo, J.J.; Retik, A.B. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet 2006, 367, 1241–1246. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.M.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Cheng, Y.; Wang, X.; Wang, J.; Shi, X.; Li, X.; Tan, W.; Tan, Z. 3D printed in vitro tumor tissue model of colorectal cancer. Theranostics 2020, 10, 12127–12143. [Google Scholar] [CrossRef] [PubMed]
- Rios De La Rosa, J.M.; Wubetu, J.; Tirelli, N.; Tirella, A. Colorectal tumor 3D in vitro models: Advantages of biofabrication for the recapitulation of early stages of tumour development. Biomed. Phys. Eng. Express 2018, 4, 45010. [Google Scholar] [CrossRef]
- Gastaldi, D.; Parisi, G.; Lucchini, R.; Contro, R.; Bignozzi, S.; Ginestra, P.S.; Filardo, G.; Kon, E.; Vena, P. A Predictive Model for the Elastic Properties of a Collagen-Hydroxyapatite Porous Scaffold for Multi-Layer Osteochondral Substitutes. Int. J. Appl. Mech. 2015, 07, 1550063. [Google Scholar] [CrossRef] [Green Version]
- Ginestra, P.S.; Rovetta, R.; Fiorentino, A.; Ceretti, E. Bioprinting process optimization: Evaluation of parameters influence on the extrusion of inorganic polymers. Procedia CIRP 2020, 89, 104–109. [Google Scholar] [CrossRef]
- Ginestra, P.; Pandini, S.; Ceretti, E. Hybrid multi-layered scaffolds produced via grain extrusion and electrospinning for 3D cell culture tests. Rapid Prototyp. J. 2019, 26, 593–602. [Google Scholar] [CrossRef]
- Angelats Lobo, D.; Ginestra, P. Cell bioprinting: The 3D-bioplotterTM case. Materials 2019, 12, 4005. [Google Scholar] [CrossRef] [Green Version]
| Material | Source | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Collagen | Natural, peptide | · Good for cell adhesion · Biocompatible · Low toxicity · Low immunogenicity | · Problems on mechanical strength · Problems on sterilization · Unstable in aqueous conditions | [48,49] |
| Gelatin | Natural, peptide | · Good cell adhesion and infiltration · Stable at high temperatures · Biodegradable · Non immunogenic | · Low stability · Controversial bioactivity | [50,51] |
| Alginate | Natural, polysaccharide | · Mimic functions of extracellular matrix · Biocompatible and cytocompatible · Biodegradable and bioabsorbable | · Problems on sterilization · Low cell adherence · Poor mechanical properties | [52,53] |
| Matrigel | Natural, derived from animal sarcoma | · Mimic more the in vivo microenvironment | · Batch-to-batch variability · Complexity of the composition | [54] |
| BdECM 1 | Natural, derived from brain | · Easy to obtain · Tissue specificity | · Potentially immunogenic | [55] |
| Hyaluronic acid | Natural, polysaccharide | · Non immunogenic · Biocompatible · Osseocompatible | · Fragile · Low biodegradability | [56,57] |
| Silk | Natural, peptide | · Great strength and elasticity · Biocompatible · Thermostable · Assists on cell migration and vascularization | · Induction of degradation · Possible immunogenicity | [58,59] |
| Fibrinogen | Natural, peptide | · Biocompatible · Cell-adhesive and binding properties · Non immunogenic | · Poor mechanical strength · High degradation | [60] |
| Agarose | Natural, polysaccharide | · Great biocompatibility · Non immunogenic · Reversible gelation | · Low cell adhesion · Non-degradable | [61,62] |
| Chitosan | Natural, polysaccharide | · Promotes cell adhesion · Anti-inflammatory · Non-toxic | · Low mechanical strength · Low solubility · Fast degradation in vivo | [63,64] |
| Hydroxyapatite-based | Natural, mineral | · Similar chemical and crystallographic structures to human bone · Biocompatible | · Fragile · Low tensile strength | [65] |
| Xantham gum | Natural, polysaccharide | · Non-toxic · Safe to use | N.A. | [66] |
| Cellulose-based | Natural, polysaccharide | · Stable structure · Good mechanical properties · Biocompatible and cytocompatible | · Inside the human body, it behaves as non-degradable | [67,68] |
| PEG 2 | Synthetic | · Biocompatible · Elastic · Bio adhesive · Non immunogenic | · Insoluble networks · Bioinert origin | [69,70] |
| PLA 3 | Synthetic | · Biocompatible and cytocompatible · Good mechanical strength and degradation rate | · Fragile · Hydrophobic | [71] |
| PLGA 4 | Synthetic | · Great cell adhesion and proliferation · Good mechanical properties | · Possible biocompatibility issues | [72] |
| PCL 5 | Synthetic | · Non-toxic · Cytocompatible · Good mechanical properties · Controls cell proliferation and angiogenesis | · Hydrophobicity · Low bioactivity | [73,74] |
| PDMS 6 | Synthetic | · Inert · Non-toxic | · Hydrophobic · Elasticity restrictions | [75] |
| SU-8 polymers | Synthetic | · Chemical stability · Good mechanical and optical properties | · Restrict adhesion selection | [76] |
| Main Materials | Cost | Speed | Problems | References | |
|---|---|---|---|---|---|
| Laser printing | Mainly metal powders | Expensive | Fast | Requires post-processing techniques | [86] |
| Lithography | Resin and photocurable polymers | Expensive | Fast | Possible cytotoxicities | [87] |
| Inkjet printing | Mainly ceramic powders and thermoplastics | Cheap | Fast | Low mechanical strength | [87] |
| Droplet-based printing | Mainly photocurable polymers | Cheap | Fast | Low mechanical strength | [86] |
| Electrospinning | Mainly thermoplastics | Cheap | Fast | Limited control on pores size | [88] |
| Gas foaming | Polymers | Cheap | - | Limited reproducibility | [89] |
| Freeze-casting | Mainly metal powders | Cheap | - | Limitations on gas diffusion | [90] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| Sodium alginate beads | MDA-MB-231 1 | Laser-direct writing | [107] |
| PED-DMA 2 and gelatin type A | MCF-7 3 | Extrusion printing (valve-based) | [108] |
| Alginate-collagen microbeads | MDA-MB-231, MCF-7 and adipose cells | Laser-direct writing | [80] |
| PEG 4 coating–TMSPM 5 photoinitiator and ME-GEL 6 | MDA-MB-231, MCF-7 and MCF-10A 7 | Photolithography (photomask) | [109] |
| Collagen | MCF-7 | Extrusion printing | [110] |
| Microfluidic device (PDMS 8), collagen type I, and Matrigel | MDA-MB-231, hBM-MSCs 9 and HUVEC 10 | Extrusion printing | [111] |
| PEG–DEX 11 system (not scaffolds) | MDA-MB-231, MCF-7, HepG2 12, HCT-116 13, ES D3 cells 14, NIH-3T3 15 | Hanging drop (spheroids) | [102] |
| Sodium alginate | Dil-positive cells (NT, CTSL KD 16), MDA-4T1 17 | Inkjet printing | [83] |
| Cell suspension in PBS 18 | MCF-7 | Thermal inkjet printing | [105] |
| Neutralized rat tail collagen type I | MCF-7, MDA-MB-468 19 and MCF-12A 20 | Customized Felix 3.0 extrusion printer | [106] |
| MeHA 21, HA 22, ME-GEL, gelatin | 21 PT 23 and ADMSCs 24 | Extrusion printing | [112] |
| Matrigel; sodium alginate-gelatin (hydrogel I) and alginate-collagen (hydrogel II) | MDA-MB-231, MCF-7, MCF-10A, MCF-10A-NeuN, breast epithelial cells | Coaxial extrusion printing | [113] |
| Alginate and gelatin | MCF-7, HCC1143 25, SKBR3 26, MDA-MB-231, HUVEC and fibroblasts | Extrusion printing | [95] |
| Alginate and gelatin | MDA-MB-231 and IMR-90 27 | Extrusion printing | [114] |
| ME-GEL, nHa, and Irgacure 2959 photoinitiator | MDA-MB-231 and hBM-MSCS | Laser printing | [115] |
| Peptide-conjugated alginate fibers | MDA-MB-231 and RAW26.7 28 | Extrusion printing | [96] |
| Alginate-gelatin (3:2) | MCF-7 and ADSCs 29 | Extrusion printing | [36] |
| Alginate; ADA-GEL 30; HA-SH 31 and PEGDA 32 | MDA-MB-231, MCF-7, Mel Im 33 and MV3 34 | Extrusion printing | [116] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| Collagen or BdECM 1 | U-118 2 and HUVEC 3 | Extrusion printing | [39] |
| Alginate | GSC23 4 and U-118 | Coaxial extrusion printing | [118] |
| Gelatin-alginate-fibrinogen (GAF hydrogel) | SU3 5 and U-87 6 | Extrusion printing | [120] |
| Alginate, a microfluidic device of PDMS 7 and SU-8 2050 epoxy | U-251 8 and HepG2 9 | Inkjet printing | [119] |
| Direct bioprinting of cells | U-118 GFP 10 labeled, GBM4 11, CD1 12, C57BL 13, and Ipsc 14-derived hnp cells | Extrusion printing | [104] |
| Alginate, gelatin, and fibrinogen (GAF hydrogel) | U-118 | Extrusion printing | [40] |
| Gelatin methacryloyl and gelatin | GL261 15, GAMs 16 and RAW 264.7 17 | Extrusion printing | [79] |
| Agarose and collagen type I | SH-SY5Y 18, UC-MSCs 19, and HUVEC | Droplet printing | [121] |
| Alginate modified with RGDS 20, HA 21, and collagen type I | U-87MG, GSCs 22, GASCs 23, microglia, WI-38 24 and MM6 cells 25 | Fab@Home or Renishaw PLC multi-nozzle extrusion printers | [38] |
| BdECM and silicone | Glioblastoma cells and HUVEC | Extrusion printing | [122] |
| Collagen type I, III or IV, and thiol-HA 26 | OSU2 cells 27 and astrocytes | Extrusion printing | [123] |
| Alginate and gelatin (shell) and fibrinogen (core) | GSC23 and hMSCs 28 | Coaxial extrusion printing | [124] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| PEG 1-RGDS 2, PEG-PQ 3 scaffolds | 344SQ 4, 393P 5 and 344P 6 | Droplet printing (white light polymerization) | [44] |
| PEG-SVA 7, PEG-RGDS, PEG-PQ-PEG 8, and microfluidic device of PDMS 9 | 344SQ, HVP 10 and HUVEC 11 | Extrusion printing | [130] |
| Gelatin-alginate hydrogel | A549 12 and A95D 13 | Livprint Norm extrusion printer | [129] |
| Gelatin-sodium alginate-Matrigel hydrogel | A549 and Primary ICC 14 cells | SUNP ALPHA-CPT1 Multinozzle extrusion printer | [131] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| Alginate, a microfluidic device of PDMS 1 and SU-8 2050 epoxy | HepG2 2 and U-251 3 | Inkjet printing | [119] |
| PEG 4–DEX 5 system (not scaffolds) | HepG2 and MDA-MB-231 6, MCF-7 7, HCT-116 8, ES D3 cells 9, NIH-3T3 10 | Hanging drop (spheroids) | [102] |
| Alginate beads | MHCC97L 11 and HCCLM3 12 | Extrusion printing | [138] |
| Rat tail collagen type I | HepG2 and 3T3-J2 13 | Hanging drop (spheroids) | [103] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| Gelatin-alginate-fibrinogen (1:2:1) hydrogel | HeLa 1 cells | Extrusion printing | [155] |
| Matrigel | OVCAR-5 2 and MRC-5 3 | Extrusion printing (two extruders) | [156] |
| Agarose | SkOV3 4 | Extrusion printing | [157] |
| RADA16-I hydrogel | A2780 5, A2780/DDP 6 and SkOV3 | Extrusion printing | [47] |
| Matrigel | P19 7 cells | Laser direct writing (MAPLE direct writing) | [154] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| Alginate | HCT-116 1 and HCT-116R 2 | Extrusion printing | [169] |
| Alginate-hyaluronic acid hydrogel | MKN45 3 and bmMSCs 4 | Extrusion printing (spheres) | [170] |
| Bioink Composition | Cells Used | 3D Bioprinting Technology | Reference |
|---|---|---|---|
| Matrigel:cells (11:1) | MV3dc 1 | Pneumatic extrusion printing | [179] |
| Alginate; ADA-GEL 2; HA-SH 3 and PEGDA 4 | Mel Im 5 and MV3 6, MDA-MB-231 7, and MCF-7 8 | Extrusion printing | [116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobo, D.A.; Ginestra, P.; Ceretti, E.; Miquel, T.P.; Ciurana, J. Cancer Cell Direct Bioprinting: A Focused Review. Micromachines 2021, 12, 764. https://doi.org/10.3390/mi12070764
Lobo DA, Ginestra P, Ceretti E, Miquel TP, Ciurana J. Cancer Cell Direct Bioprinting: A Focused Review. Micromachines. 2021; 12(7):764. https://doi.org/10.3390/mi12070764
Chicago/Turabian StyleLobo, David Angelats, Paola Ginestra, Elisabetta Ceretti, Teresa Puig Miquel, and Joaquim Ciurana. 2021. "Cancer Cell Direct Bioprinting: A Focused Review" Micromachines 12, no. 7: 764. https://doi.org/10.3390/mi12070764
APA StyleLobo, D. A., Ginestra, P., Ceretti, E., Miquel, T. P., & Ciurana, J. (2021). Cancer Cell Direct Bioprinting: A Focused Review. Micromachines, 12(7), 764. https://doi.org/10.3390/mi12070764








