Accumulation of Tetrahymena pyriformis on Interfaces
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Cells
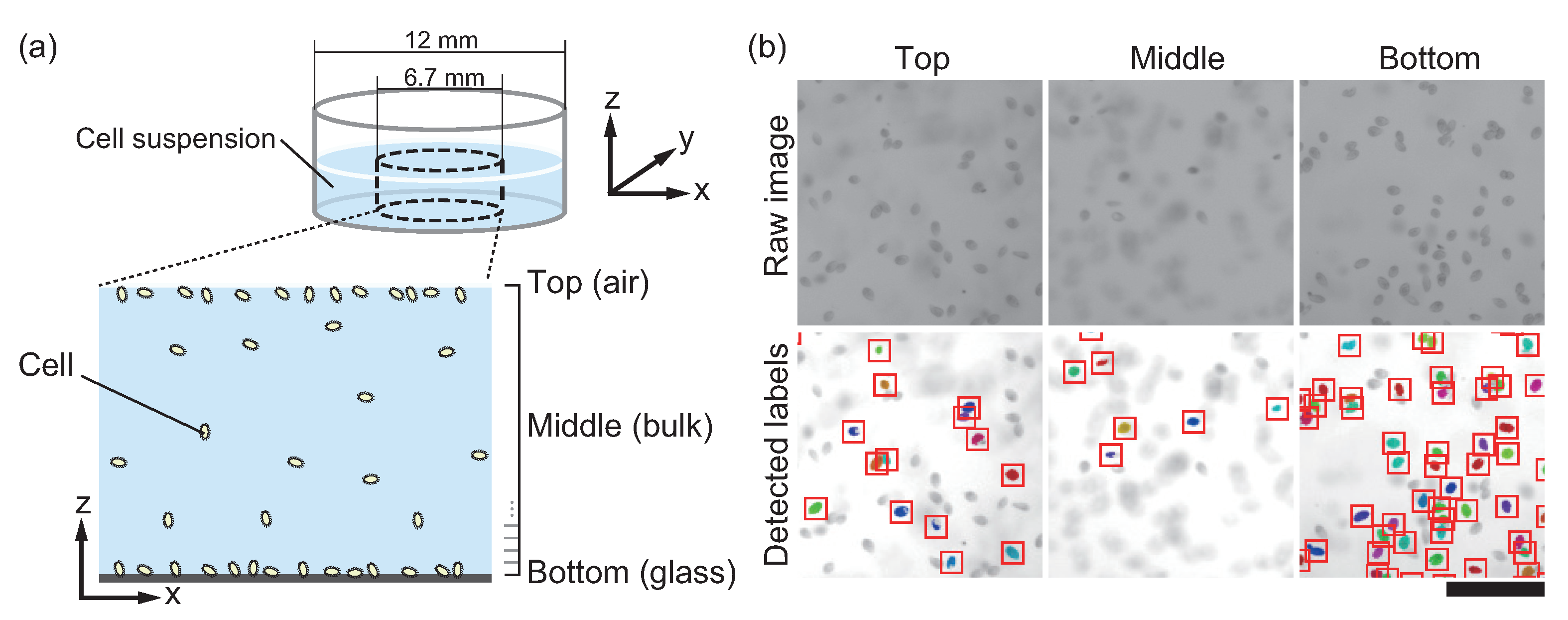
2.2. Observation of Cells
2.3. Counting of Cells
3. Results
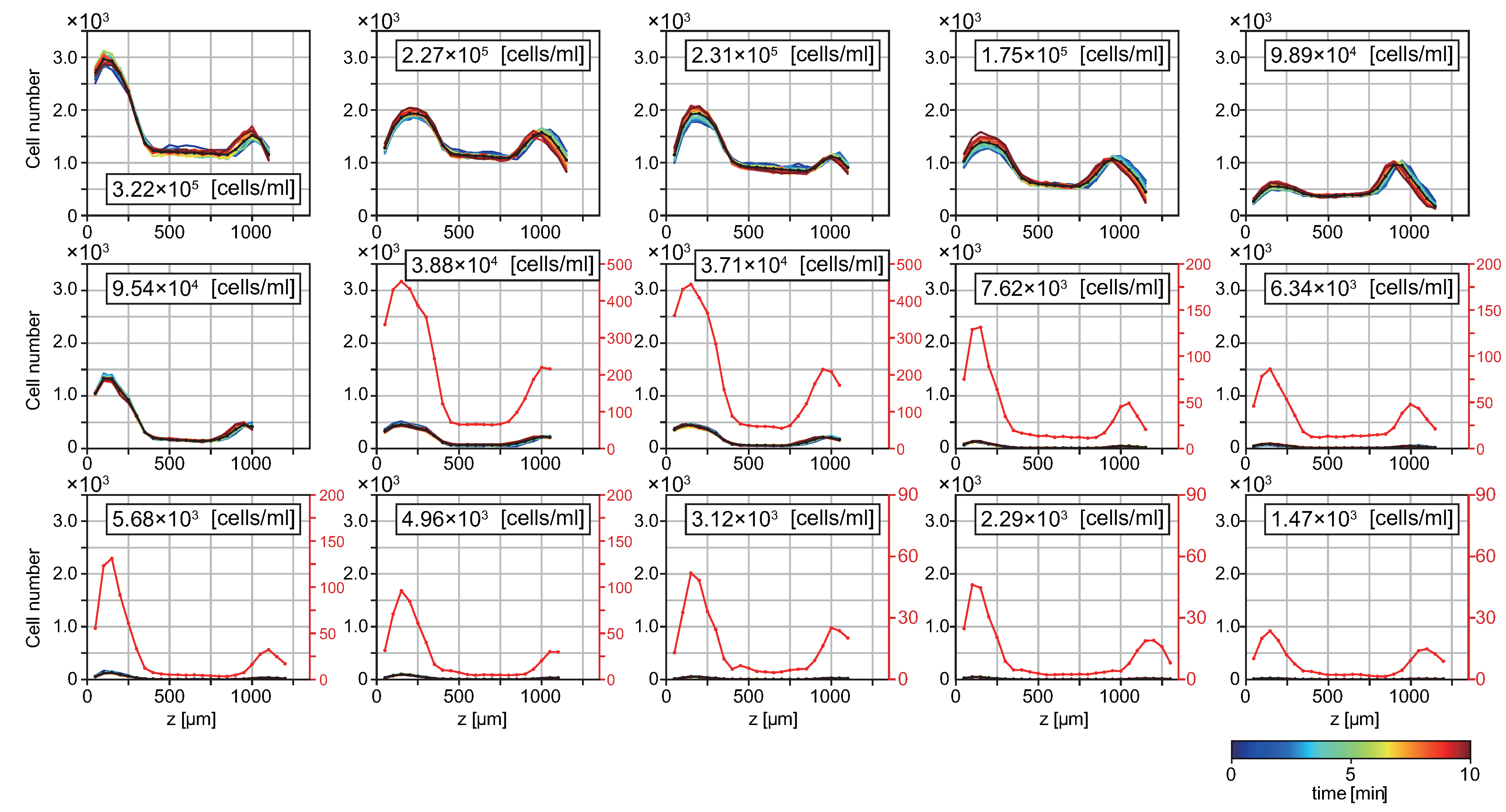
3.1. T. pyriformis Distribute Close to Interfaces Rather than in Bulk Water
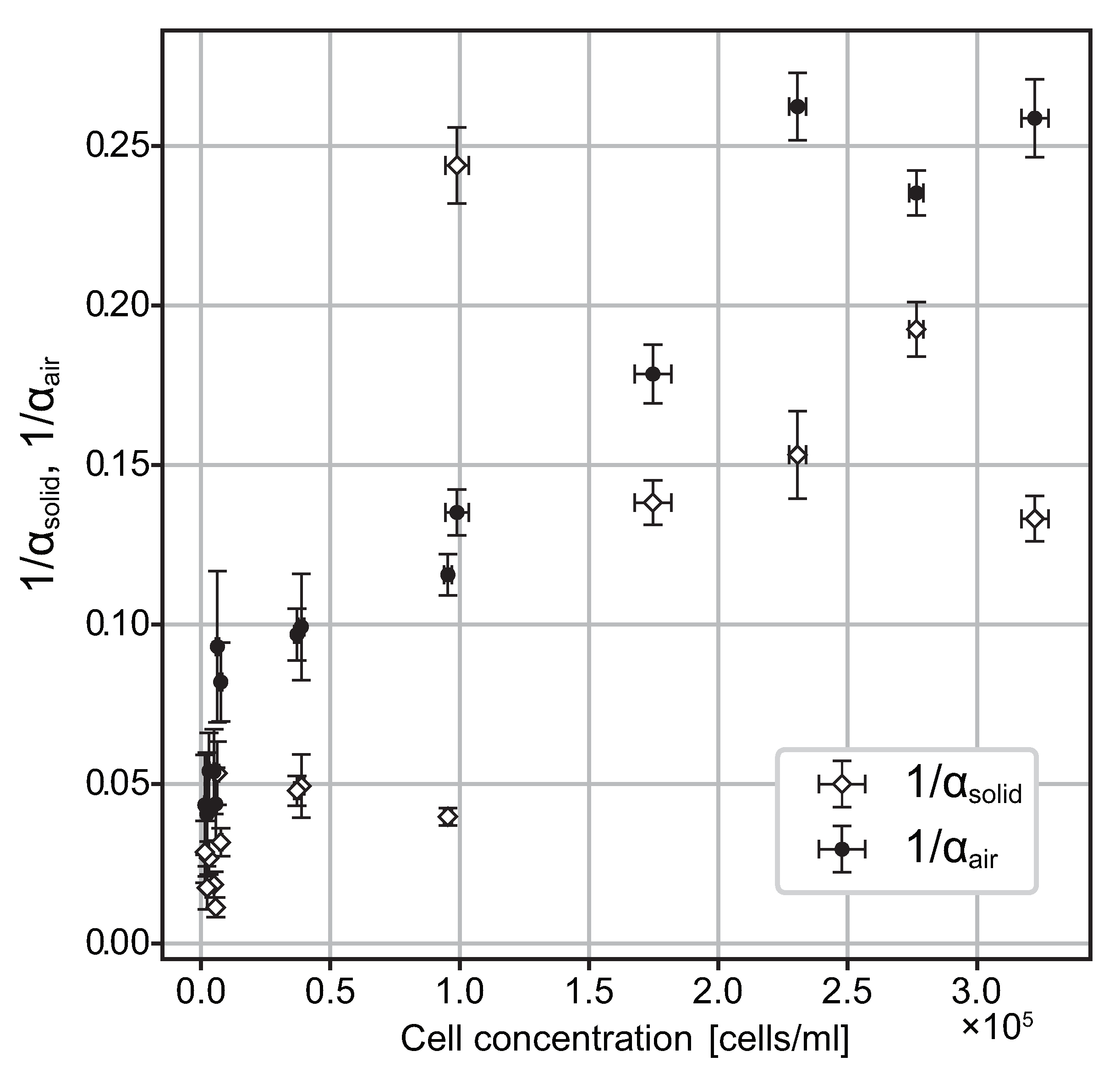
3.2. Probability of Accumulation Depends on Cell Density of Suspension
4. Discussion
4.1. Accumulation Coefficients
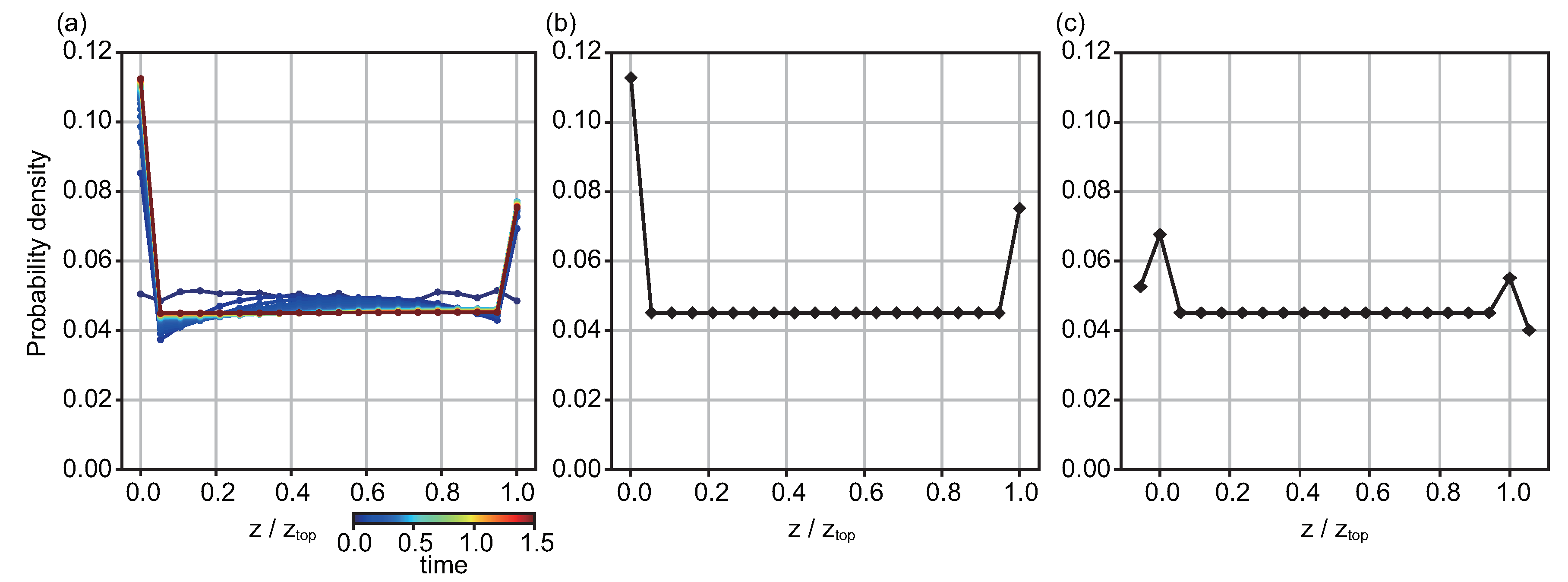
4.2. Diffusion Model
4.3. Additional Discussion for the Model for Representing Experiment
4.4. Comparison with the Suspension of Squirmer Model
4.5. Biological Aspects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Adl, S.M.; Simpson, A.G.; Farmer, M.A.; Andersen, R.A.; Anderson, O.R.; Barta, J.R.; Bowser, S.S.; Brugerolle, G.; Fensome, R.A.; Fredericq, S.; et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005, 52, 399–451. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.; Lane, C.E.; Lukeš, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–514. [Google Scholar] [CrossRef] [Green Version]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [Green Version]
- Corliss, J.O. Why the World Needs Protists! 1. J. Eukaryot. Microbiol. 2004, 51, 8–22. [Google Scholar] [CrossRef]
- Anderson, O.R. Protozoan ecology. eLS 2001. [Google Scholar] [CrossRef]
- Lynn, D.H. The Ciliated Protozoa: Characterization, Classification and Guide to the Literature; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Lynn, D.H. Handbook of the Protists; Springer: Cham, Switzerland, 2017; Chapter Ciliophora; pp. 679–729. [Google Scholar]
- Hill, D.G. The Biochemistry and Physiology of Tetrahymena; Academic Press: New York, NY, USA, 1972. [Google Scholar]
- Sauvant, N.; Pepin, D.; Piccinni, E. Tetrahymena pyriformis: A tool for toxicological studies. A review. Chemosphere 1999, 38, 1631–1669. [Google Scholar] [CrossRef]
- Hellung-Larsen, P.; Leick, V.; Tommerup, N.; Kronborg, D. Chemotaxis in tetrahymena. Eur. J. Protistol. 1990, 25, 229–233. [Google Scholar] [CrossRef]
- And, V.L.; Hellung-Larsen, P. Chemosensory behaviour of Tetrahymena. BioEssays 1992, 14, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Koppelhus, U.; Hellung-Larsen, P.; Leick, V. An improved quantitative assay for chemokinesis in Tetrahymena. Biol. Bull. 1994, 187, 8–15. [Google Scholar] [CrossRef]
- Kunita, I.; Yamaguchi, T.; Tero, A.; Akiyama, M.; Kuroda, S.; Nakagaki, T. A ciliate memorizes the geometry of a swimming arena. J. R. Soc. Interface 2016, 13, 20160155. [Google Scholar] [CrossRef] [Green Version]
- Schenz, D.; Nishigami, Y.; Sato, K.; Nakagaki, T. Uni-cellular integration of complex spatial information in slime moulds and ciliates. Curr. Opin. Genet. Dev. 2019, 57, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Ferracci, J.; Ueno, H.; Numayama-Tsuruta, K.; Imai, Y.; Yamaguchi, T.; Ishikawa, T. Entrapment of ciliates at the water-air interface. PLoS ONE 2013, 8, e75238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manabe, J.; Omori, T.; Ishikawa, T. Shape matters: Entrapment of a model ciliate at interfaces. J. Fluid Mech. 2020, 892, A15. [Google Scholar] [CrossRef]
- Ohmura, T.; Nishigami, Y.; Taniguchi, A.; Nonaka, S.; Manabe, J.; Ishikawa, T.; Ichikawa, M. Simple mechanosense and response of cilia motion reveal the intrinsic habits of ciliates. Proc. Natl. Acad. Sci. USA 2018, 115, 3231–3236. [Google Scholar] [CrossRef] [Green Version]
- Ohmura, T.; Nishigami, Y.; Taniguchi, A.; Nonaka, S.; Ishikawa, T.; Ichikawa, M. Near-wall rheotaxis of the ciliate Tetrahymena induced by the kinesthetic sensing of cilia. Sci. Adv. 2021, 7, eabi5878. [Google Scholar] [CrossRef] [PubMed]
- Lighthill, M.J. On the squirming motion of nearly spherical deformable bodies through liquids at very small Reynolds numbers. Commun. Pure Appl. Math. 1952, 5, 109–118. [Google Scholar] [CrossRef]
- Blake, J.R. A spherical envelope approach to ciliary propulsion. J. Fluid Mech. 1971, 46, 199–208. [Google Scholar] [CrossRef]
- Ishikawa, T.; Simmonds, M.; Pedley, T.J. Hydrodynamic interaction of two swimming model micro-organisms. J. Fluid Mech. 2006, 568, 119–160. [Google Scholar] [CrossRef]
- Downton, M.T.; Stark, H. Simulation of a model microswimmer. J. Phys. Condens. Matter 2009, 21, 204101. [Google Scholar] [CrossRef]
- Lauga, E.; Powers, T.R. The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 2009, 72, 096601. [Google Scholar] [CrossRef]
- Llopis, I.; Pagonabarraga, I. Hydrodynamic interactions in squirmer motion: Swimming with a neighbour and close to a wall. J. -Non-Newton. Fluid Mech. 2010, 165, 946–952. [Google Scholar] [CrossRef]
- Spagnolie, S.E.; Lauga, E. Hydrodynamics of self-propulsion near a boundary: Predictions and accuracy of far-field approximations. J. Fluid Mech. 2012, 700, 105–147. [Google Scholar] [CrossRef] [Green Version]
- Ishimoto, K.; Gaffney, E.A. Squirmer dynamics near a boundary. Phys. Rev. E 2013, 88, 062702. [Google Scholar] [CrossRef] [Green Version]
- Li, G.J.; Ardekani, A.M. Hydrodynamic interaction of microswimmers near a wall. Phys. Rev. E 2014, 90, 013010. [Google Scholar] [CrossRef] [Green Version]
- Schaar, K.; Zöttl, A.; Stark, H. Detention times of microswimmers close to surfaces: Influence of hydrodynamic interactions and noise. Phys. Rev. Lett. 2015, 115, 038101. [Google Scholar] [CrossRef] [Green Version]
- Oyama, N.; Molina, J.J.; Yamamoto, R. Purely hydrodynamic origin for swarming of swimming particles. Phys. Rev. E 2016, 93, 043114. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.V.; Kishore, V.; Santomauro, G.; Yasa, O.; Bill, J.; Sitti, M. Mechanical Coupling of Puller and Pusher Active Microswimmers Influences Motility. Langmuir 2020, 36, 5435–5443. [Google Scholar] [CrossRef] [PubMed]
- Wensink, H.H.; Dunkel, J.; Heidenreich, S.; Drescher, K.; Goldstein, R.E.; Löwen, H.; Yeomans, J.M. Meso-scale turbulence in living fluids. Proc. Natl. Acad. Sci. USA 2012, 109, 14308–14313. [Google Scholar] [CrossRef] [Green Version]
- Dunkel, J.; Heidenreich, S.; Drescher, K.; Wensink, H.H.; Bär, M.; Goldstein, R.E. Fluid dynamics of bacterial turbulence. Phys. Rev. Lett. 2013, 110, 228102. [Google Scholar] [CrossRef] [Green Version]
- Berke, A.P.; Turner, L.; Berg, H.C.; Lauga, E. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys. Rev. Lett. 2008, 101, 038102. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Bensson, J.; Nisimova, L.; Munger, D.; Mahautmr, P.; Tang, J.X.; Maxey, M.R.; Brun, Y.V. Accumulation of swimming bacteria near a solid surface. Phys. Rev. E 2011, 84, 041932. [Google Scholar] [CrossRef] [Green Version]
- Pedley, T.; Kessler, J. Bioconvection. Sci. Prog. 1992, 76, 105–123. [Google Scholar]
- Hill, N.; Pedley, T. Bioconvection. Fluid Dyn. Res. 2005, 37, 1. [Google Scholar] [CrossRef]
- Mogami, Y.; Yamane, A.; Gino, A.; Baba, S.A. Bioconvective pattern formation of Tetrahymena under altered gravity. J. Exp. Biol. 2004, 207, 3349–3359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef] [PubMed]
- Nishigami, Y.; Ohmura, T.; Taniguchi, A.; Nonaka, S.; Manabe, J.; Ishikawa, T.; Ichikawa, M. Influence of cellular shape on sliding behavior of ciliates. Commun. Integr. Biol. 2018, 11, e1506666. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Quang, T.; Nguyen, T.H.; Guichard, F.; Nicolau, A.; Szatmari, G.; LePalec, G.; Dusser, M.; Lafossee, J.; Bonnet, J.L.; Bohatier, J. Two-dimensional gravitactic bioconvection in a protozoan (Tetrahymena pyriformis) culture. Zool. Sci. 2009, 26, 54–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doerder, F.P.; Brunk, C. Natural populations and inbred strains of Tetrahymena. Methods Cell Biol. 2012, 109, 277–300. [Google Scholar]
- Singh, A.V.; Ansari, M.H.D.; Laux, P.; Luch, A. Micro-nanorobots: Important considerations when developing novel drug delivery platforms. Expert Opin. Drug Deliv. 2019, 16, 1259–1275. [Google Scholar] [CrossRef]
- Bozuyuk, U.; Yasa, O.; Yasa, I.C.; Ceylan, H.; Kizilel, S.; Sitti, M. Light-Triggered Drug Release from 3D-Printed Magnetic Chitosan Microswimmers. ACS Nano 2018, 12, 9617–9625. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okuyama, K.; Nishigami, Y.; Ohmura, T.; Ichikawa, M. Accumulation of Tetrahymena pyriformis on Interfaces. Micromachines 2021, 12, 1339. https://doi.org/10.3390/mi12111339
Okuyama K, Nishigami Y, Ohmura T, Ichikawa M. Accumulation of Tetrahymena pyriformis on Interfaces. Micromachines. 2021; 12(11):1339. https://doi.org/10.3390/mi12111339
Chicago/Turabian StyleOkuyama, Kohei, Yukinori Nishigami, Takuya Ohmura, and Masatoshi Ichikawa. 2021. "Accumulation of Tetrahymena pyriformis on Interfaces" Micromachines 12, no. 11: 1339. https://doi.org/10.3390/mi12111339
APA StyleOkuyama, K., Nishigami, Y., Ohmura, T., & Ichikawa, M. (2021). Accumulation of Tetrahymena pyriformis on Interfaces. Micromachines, 12(11), 1339. https://doi.org/10.3390/mi12111339





