Accelerating the Finite-Element Method for Reaction-Diffusion Simulations on GPUs with CUDA
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical System
2.2. Finite Element Method
2.3. Assembly of Stiffness and Damping Matrices
2.4. Resolution of the Matrix Differential Equations
| Algorithm 1 User-level algorithm. |
|
| Algorithm 2 Addition of two vectors. |
|
| Algorithm 3 Naive dot product. |
|
| Algorithm 4 Optimized dot product. |
|
2.5. Comparison GPU and CPU
2.6. Post-Processing
3. Results
3.1. Geometry
3.2. Comparison Performance
3.3. Profiling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brodtkorb, A.; Hagen, T.; Sætra, M. Graphics processing unit (GPU) programming strategies and trends in GPU computing. J. Parallel Distrib. Comput. 2013, 73, 4–13. [Google Scholar] [CrossRef]
- Ghorpade, J.; Parande, J.; Kulkarni, M.; Bawaskar, A. GPGPU processing in CUDA architecture. arXiv 2012, arXiv:1202.4347. [Google Scholar] [CrossRef]
- CUDA Performance Report. Available online: http://developer.download.nvidia.com/compute/cuda/6_5/rel/docs/CUDA_6.5_Performance_Report.pdf (accessed on 7 September 2020).
- Nickolls, J.; Buck, I.; Garland, M.; Skadron, K. Scalable parallel programming with CUDA. Queue 2008, 6, 40–53. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic differentiation in pytorch. In Proceedings of the NIPS 2017 Workshop Autodiff, Long Beach, CA, USA, 9 December 2017. [Google Scholar]
- Rovigatti, L.; Šulc, P.; Reguly, I.; Romano, F. A comparison between parallelization approaches in molecular dynamics simulations on GPUs. J. Comput. Chem. 2015, 36, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Glaser, J.; Nguyen, T.; Anderson, J.; Lui, P.; Spiga, F.; Millan, J.; Morse, D.; Glotzer, S. Strong scaling of general-purpose molecular dynamics simulations on GPUs. Comput. Phys. Commun. 2015, 192, 97–107. [Google Scholar] [CrossRef]
- Le Grand, S.; Götz, A.; Walker, R. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013, 184, 374–380. [Google Scholar] [CrossRef]
- Zienkiewicz, O.; Taylor, R.; Zhu, J. The Finite Element Method: Its Basis and Fundamentals; Elsevier: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Fu, Z.; Lewis, T.; Kirby, R.; Whitaker, R. Architecting the finite element method pipeline for the GPU. J. Comput. Appl. Math. 2014, 257, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Heng, P.A. A hybrid condensed finite element model with GPU acceleration for interactive 3D soft tissue cutting. Comput. Animat. Virtual Worlds 2004, 15, 219–227. [Google Scholar] [CrossRef]
- Goddeke, D.; Buijssen, S.H.; Wobker, H.; Turek, S. GPU acceleration of an unmodified parallel finite element Navier-Stokes solver. In Proceedings of the 2009 International Conference on High Performance Computing & Simulation, Leipzig, Germany, 21–24 June 2009; pp. 12–21. [Google Scholar]
- Komatitsch, D.; Erlebacher, G.; Göddeke, D.; Michéa, D. High-order finite-element seismic wave propagation modeling with MPI on a large GPU cluster. J. Comput. Phys. 2010, 229, 7692–7714. [Google Scholar] [CrossRef]
- Joldes, G.R.; Wittek, A.; Miller, K. Real-time nonlinear finite element computations on GPU–Application to neurosurgical simulation. Comput. Methods Appl. Mech. Eng. 2010, 199, 3305–3314. [Google Scholar] [CrossRef]
- Dziekonski, A.; Sypek, P.; Lamecki, A.; Mrozowski, M. Finite element matrix generation on a GPU. Prog. Electromagn. Res. 2012, 128, 249–265. [Google Scholar] [CrossRef]
- Knepley, M.G.; Terrel, A.R. Finite element integration on GPUs. ACM Trans. Math. Softw. (TOMS) 2013, 39, 1–13. [Google Scholar] [CrossRef]
- Wang, S.; Wang, C.; Cai, Y.; Li, G. A novel parallel finite element procedure for nonlinear dynamic problems using GPU and mixed-precision algorithm. Eng. Comput. 2020, 37. [Google Scholar] [CrossRef]
- Huthwaite, P. Accelerated finite element elastodynamic simulations using the GPU. J. Comput. Phys. 2014, 257, 687–707. [Google Scholar] [CrossRef]
- Johnsen, S.F.; Taylor, Z.A.; Clarkson, M.J.; Hipwell, J.; Modat, M.; Eiben, B.; Han, L.; Hu, Y.; Mertzanidou, T.; Hawkes, D.J.; et al. NiftySim: A GPU-based nonlinear finite element package for simulation of soft tissue biomechanics. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 1077–1095. [Google Scholar] [CrossRef]
- Bauer, P.; Klement, V.; Oberhuber, T.; Žabka, V. Implementation of the Vanka-type multigrid solver for the finite element approximation of the Navier–Stokes equations on GPU. Comput. Phys. Commun. 2016, 200, 50–56. [Google Scholar] [CrossRef]
- Carrascal-Manzanares, C.; Imperiale, A.; Rougeron, G.; Bergeaud, V.; Lacassagne, L. A fast implementation of a spectral finite elements method on CPU and GPU applied to ultrasound propagation. Adv. Parallel Comput. 2018, 32, 339–348. [Google Scholar]
- Comsol, A. COMSOL Multiphysics User’s Guide; COMSOL: Stockholm, Sweden, 2005; Volume 10, p. 333. [Google Scholar]
- Soloveichik, D.; Seelig, G.; Winfree, E. DNA as a universal substrate for chemical kinetics. Proc. Natl. Acad. Sci. USA 2010, 107, 5393–5398. [Google Scholar] [CrossRef]
- Kim, J.; Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 2011, 7, 465. [Google Scholar] [CrossRef]
- Montagne, K.; Plasson, R.; Sakai, Y.; Fujii, T.; Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 2011, 7, 466. [Google Scholar] [CrossRef]
- Fujii, T.; Rondelez, Y. Predator–prey molecular ecosystems. ACS Nano 2013, 7, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Padirac, A.; Fujii, T.; Estévez-Torres, A.; Rondelez, Y. Spatial waves in synthetic biochemical networks. J. Am. Chem. Soc. 2013, 135, 14586–14592. [Google Scholar] [CrossRef]
- Srinivas, N.; Parkin, J.; Seelig, G.; Winfree, E.; Soloveichik, D. Enzyme-free nucleic acid dynamical systems. Science 2017, 358. [Google Scholar] [CrossRef] [PubMed]
- Genot, A.J.; Bath, J.; Turberfield, A.J. Reversible logic circuits made of DNA. J. Am. Chem. Soc. 2011, 133, 20080–20083. [Google Scholar] [CrossRef]
- Genot, A.J.; Bath, J.; Turberfield, A.J. Combinatorial displacement of DNA strands: Application to matrix multiplication and weighted sums. Angew. Chem. Int. Ed. 2013, 52, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Stefanovic, D.; Rudchenko, S. Exercises in molecular computing. Acc. Chem. Res. 2014, 47, 1845–1852. [Google Scholar] [CrossRef]
- Lopez, R.; Wang, R.; Seelig, G. A molecular multi-gene classifier for disease diagnostics. Nat. Chem. 2018, 10, 746–754. [Google Scholar] [CrossRef]
- Cherry, K.M.; Qian, L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 2018, 559, 370–376. [Google Scholar] [CrossRef]
- Woods, D.; Doty, D.; Myhrvold, C.; Hui, J.; Zhou, F.; Yin, P.; Winfree, E. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly. Nature 2019, 567, 366–372. [Google Scholar] [CrossRef]
- Song, T.; Eshra, A.; Shah, S.; Bui, H.; Fu, D.; Yang, M.; Mokhtar, R.; Reif, J. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat. Nanotechnol. 2019, 14, 1075–1081. [Google Scholar] [CrossRef]
- Chirieleison, S.M.; Allen, P.B.; Simpson, Z.B.; Ellington, A.D.; Chen, X. Pattern transformation with DNA circuits. Nat. Chem. 2013, 5, 1000. [Google Scholar] [CrossRef] [PubMed]
- Weitz, M.; Kim, J.; Kapsner, K.; Winfree, E.; Franco, E.; Simmel, F.C. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat. Chem. 2014, 6, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, A.; Zadorin, A.; Rondelez, Y.; Estévez-Torres, A.; Galas, J. Pursuit-and-evasion reaction-diffusion waves in microreactors with tailored geometry. J. Phys. Chem. B 2015, 119, 5349–5355. [Google Scholar] [CrossRef] [PubMed]
- Genot, A.; Baccouche, A.; Sieskind, R.; Aubert-Kato, N.; Bredeche, N.; Bartolo, J.; Taly, V.; Fujii, T.; Rondelez, Y. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 2016, 8, 760. [Google Scholar] [CrossRef] [PubMed]
- Baccouche, A.; Okumura, S.; Sieskind, R.; Henry, E.; Aubert-Kato, N.; Bredeche, N.; Bartolo, J.F.; Taly, V.; Rondelez, Y.; Fujii, T.; et al. Massively parallel and multiparameter titration of biochemical assays with droplet microfluidics. Nat. Protoc. 2017, 12, 1912–1932. [Google Scholar] [CrossRef] [PubMed]
- Kurylo, I.; Gines, G.; Rondelez, Y.; Coffinier, Y.; Vlandas, A. Spatiotemporal control of DNA-based chemical reaction network via electrochemical activation in microfluidics. Sci. Rep. 2018, 8, 6396. [Google Scholar] [CrossRef] [PubMed]
- Amodio, A.; Del Grosso, E.; Troina, A.; Placidi, E.; Ricci, F. Remote Electronic Control of DNA-Based Reactions and Nanostructure Assembly. Nano Lett. 2018, 18, 2918–2923. [Google Scholar] [CrossRef]
- Zadorin, A.S.; Rondelez, Y.; Galas, J.C.; Estevez-Torres, A. Synthesis of programmable reaction-diffusion fronts using DNA catalyzers. Phys. Rev. Lett. 2015, 114, 068301. [Google Scholar] [CrossRef]
- Scalise, D.; Schulman, R. Emulating cellular automata in chemical reaction–diffusion networks. Nat. Comput. 2016, 15, 197–214. [Google Scholar] [CrossRef]
- Zadorin, A.S.; Rondelez, Y.; Gines, G.; Dilhas, V.; Urtel, G.; Zambrano, A.; Galas, J.C.; Estévez-Torres, A. Synthesis and materialization of a reaction–diffusion French flag pattern. Nat. Chem. 2017, 9, 990. [Google Scholar] [CrossRef]
- Abe, K.; Kawamata, I.; Shin-ichiro, M.; Murata, S. Programmable reactions and diffusion using DNA for pattern formation in hydrogel medium. Mol. Syst. Des. Eng. 2019, 4, 639–643. [Google Scholar] [CrossRef]
- Chen, S.; Seelig, G. Programmable patterns in a DNA-based reaction–diffusion system. Soft Matter 2020, 16, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Bardi, I.; Biro, O.; Dyczij-Edlinger, R.; Preis, K.; Richter, K.R. On the treatment of sharp corners in the FEM analysis of high frequency problems. IEEE Trans. Magn. 1994, 30, 3108–3111. [Google Scholar] [CrossRef]
- Molnár, F., Jr.; Izsák, F.; Mészáros, R.; Lagzi, I. Simulation of reaction–diffusion processes in three dimensions using CUDA. Chemom. Intell. Lab. Syst. 2011, 108, 76–85. [Google Scholar] [CrossRef]
- Descombes, S.; Dhillon, D.; Zwicker, M. Optimized CUDA-based PDE Solver for Reaction Diffusion Systems on Arbitrary Surfaces. In International Conference on Parallel Processing and Applied Mathematics; Springer: Berlin/Heidelberg, Germany, 2015; pp. 526–536. [Google Scholar]
- Sanderson, A.R.; Meyer, M.D.; Kirby, R.M.; Johnson, C.R. A framework for exploring numerical solutions of advection–reaction–diffusion equations using a GPU-based approach. Comput. Vis. Sci. 2009, 12, 155–170. [Google Scholar] [CrossRef]
- Sato, D.; Xie, Y.; Weiss, J.N.; Qu, Z.; Garfinkel, A.; Sanderson, A.R. Acceleration of cardiac tissue simulation with graphic processing units. Med. Biol. Eng. Comput. 2009, 47, 1011–1015. [Google Scholar] [CrossRef][Green Version]
- Mena, A.; Ferrero, J.M.; Matas, J.F.R. GPU accelerated solver for nonlinear reaction–diffusion systems. Application to the electrophysiology problem. Comput. Phys. Commun. 2015, 196, 280–289. [Google Scholar] [CrossRef]
- Sjodin, B. What’s the Difference between FEM, FDM, and FVM. Mach. Des. 2016. Available online: https://www.machinedesign.com/3d-printing-cad/fea-and-simulation/article/21832072/whats-the-difference-between-fem-fdm-and-fvm (accessed on 7 September 2020).
- Pera, D.; Málaga, C.; Simeoni, C.; Plaza, R. On the efficient numerical simulation of heterogeneous anisotropic diffusion models for tumor invasion using GPUs. Rend. Mat. E Sue Appl. 2019, 40, 233–255. [Google Scholar]
- Gormantara, A.; Pranowo, P. Parallel simulation of pattern formation in a reaction-diffusion system of FitzHugh-Nagumo using GPU CUDA. In AIP Conference Proceedings; AIP Publishing LLC: College Park, MD, USA, 2020; Volume 2217, p. 030134. [Google Scholar]
- Zaikin, A.; Zhabotinsky, A. Concentration wave propagation in two-dimensional liquid-phase self-oscillating system. Nature 1970, 225, 535–537. [Google Scholar] [CrossRef]
- Turing, A.M. The chemical basis of morphogenesis. Bull. Math. Biol. 1990, 52, 153–197. [Google Scholar] [CrossRef]
- Dalchau, N.; Seelig, G.; Phillips, A. Computational design of reaction-diffusion patterns using DNA-based chemical reaction networks. In International Workshop on DNA-Based Computers; Springer: Berlin/Heidelberg, Germany, 2014; pp. 84–99. [Google Scholar]
- Zenk, J.; Scalise, D.; Wang, K.; Dorsey, P.; Fern, J.; Cruz, A.; Schulman, R. Stable DNA-based reaction–diffusion patterns. RSC Adv. 2017, 7, 18032–18040. [Google Scholar] [CrossRef]
- Smith, S.; Dalchau, N. Beyond activator-inhibitor networks: The generalised Turing mechanism. arXiv 2018, arXiv:1803.07886. [Google Scholar]
- Smith, S.; Dalchau, N. Model reduction enables Turing instability analysis of large reaction–diffusion models. J. R. Soc. Interface 2018, 15, 20170805. [Google Scholar] [CrossRef]
- Joesaar, A.; Yang, S.; Bögels, B.; van der Linden, A.; Pieters, P.; Kumar, B.P.; Dalchau, N.; Phillips, A.; Mann, S.; de Greef, T.F. DNA-based communication in populations of synthetic protocells. Nat. Nanotechnol. 2019, 14, 369–378. [Google Scholar] [CrossRef]
- Urtel, G.; Estevez-Torres, A.; Galas, J.C. DNA-based long-lived reaction–diffusion patterning in a host hydrogel. Soft Matter 2019, 15, 9343–9351. [Google Scholar] [CrossRef]
- Gines, G.; Zadorin, A.; Galas, J.C.; Fujii, T.; Estevez-Torres, A.; Rondelez, Y. Microscopic agents programmed by DNA circuits. Nat. Nanotechnol. 2017, 12, 351–359. [Google Scholar] [CrossRef]
- Dupin, A.; Simmel, F.C. Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits. Nat. Chem. 2019, 11, 32–39. [Google Scholar] [CrossRef]
- Kasahara, Y.; Sato, Y.; Masukawa, M.K.; Okuda, Y.; Takinoue, M. Photolithographic shape control of DNA hydrogels by photo-activated self-assembly of DNA nanostructures. APL Bioeng. 2020, 4, 016109. [Google Scholar] [CrossRef]
- Wolfram Research, Inc. Mathematica, Version 12.1; Wolfram Research, Inc.: Champaign, IL, USA, 2020. [Google Scholar]
- Galerkin, B. Series occurring in various questions concerning the elastic equilibrium of rods and plates. Eng. Bull. (Vestn. Inzhenerov) 1915, 19, 897–908. [Google Scholar]
- Strang, G. On the construction and comparison of difference schemes. SIAM J. Numer. Anal. 1968, 5, 506–517. [Google Scholar] [CrossRef]
- Ahamed, A.; Magoules, F. Conjugate gradient method with graphics processing unit acceleration: CUDA vs. OpenCL. Adv. Eng. Softw. 2017, 111, 32–42. [Google Scholar] [CrossRef]
- Barrett, R.; Berry, M.; Chan, T.; Demmel, J.; Donato, J.; Dongarra, J.; Eijkhout, V.; Pozo, R.; Romine, C.; Van der Vorst, H. Templates for the Solution of Linear Systems: Building Blocks for Iterative Methods; SIAM: Philadelphia, PA, USA, 1994. [Google Scholar]
- Nvidia, C. Cublas Library; NVIDIA Corp.: Santa Clara, CA, USA, 2008; Volume 15, p. 31. [Google Scholar]
- Naumov, M.; Chien, L.; Vandermersch, P.; Kapasi, U. CUSPARSE library: A set of basic linear algebra subroutines for sparse matrices. In Proceedings of the GPU Technology Conference, San Jose, CA, USA, 20–23 September 2010; Volume 2070. [Google Scholar]
- Hestenes, M.; Stiefel, E. Methods of conjugate gradients for solving linear systems. J. Res. Natl. Bur. Stand. 1952, 49, 409–436. [Google Scholar] [CrossRef]
- Hutton, T.; Munafo, R.; Trevorrow, A.; Rokicki, T.; Wills, D. Ready, A Cross-Platform Implementation of Various Reaction-Diffusion Systems. 2015. Available online: https://github.com/GollyGang/ready (accessed on 7 September 2020).
- Du, Q.; Wang, D.; Zhu, L. On mesh geometry and stiffness matrix conditioning for general finite element spaces. SIAM J. Numer. Anal. 2009, 47, 1421–1444. [Google Scholar] [CrossRef]
- Ramage, A.; Wathen, A. On preconditioning for finite element equations on irregular grids. SIAM J. Matrix Anal. Appl. 1994, 15, 909–921. [Google Scholar] [CrossRef]
- Bell, N.; Garland, M. Efficient Sparse Matrix-Vector Multiplication on CUDA; Technical Report, Nvidia Technical Report NVR-2008-004; Nvidia Corporation: Santa Clara, CA, USA, 2008. [Google Scholar]

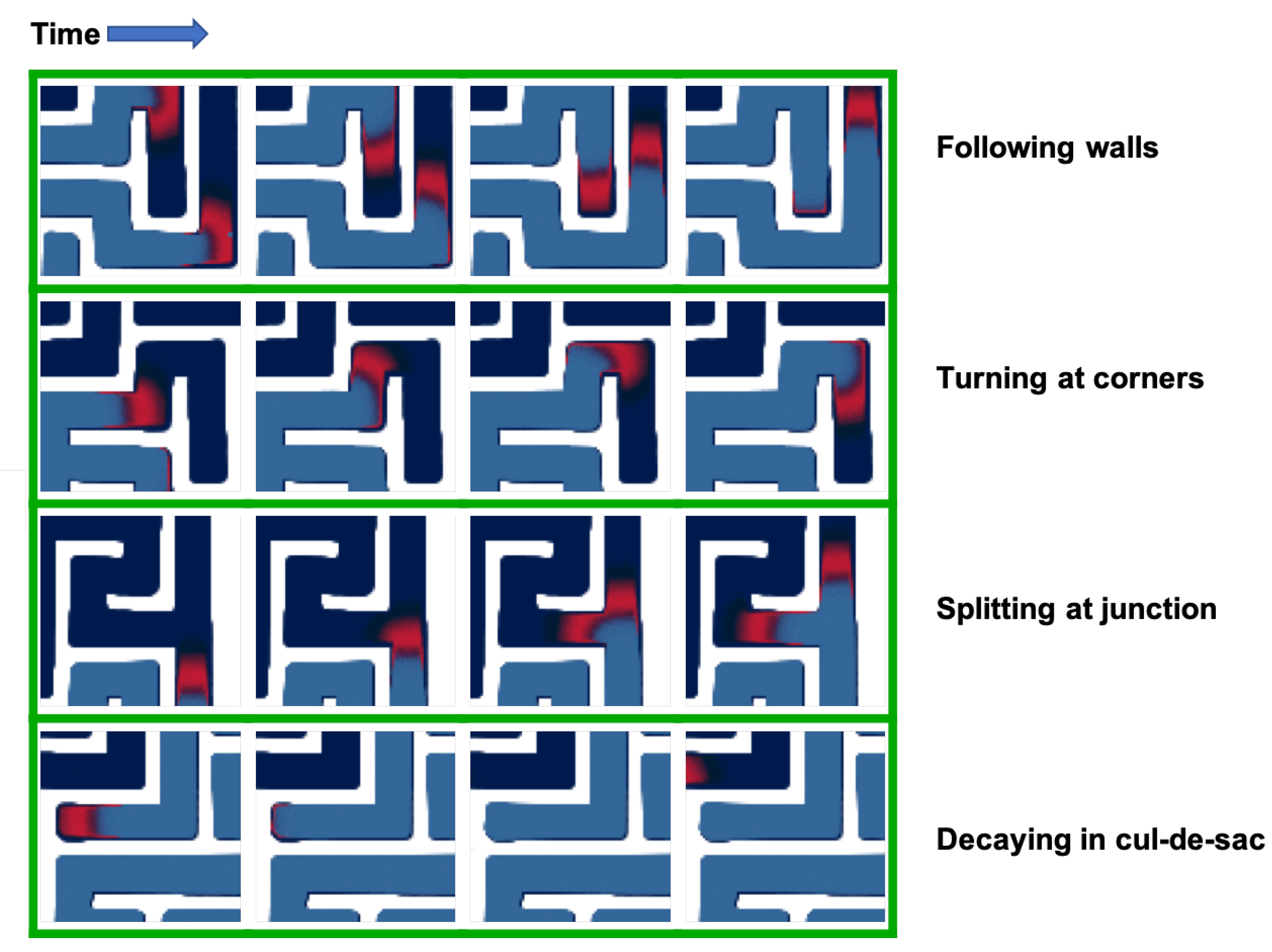
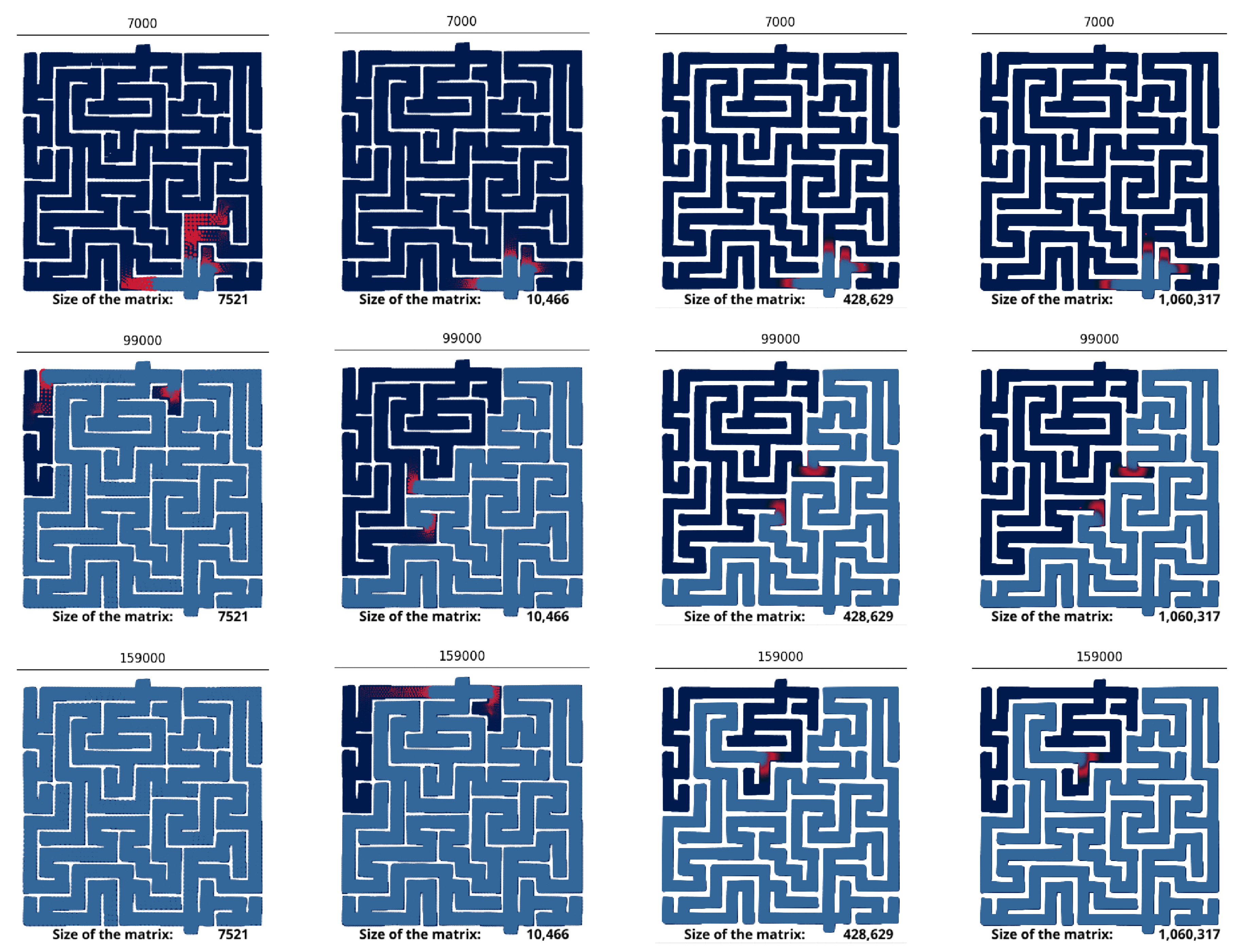
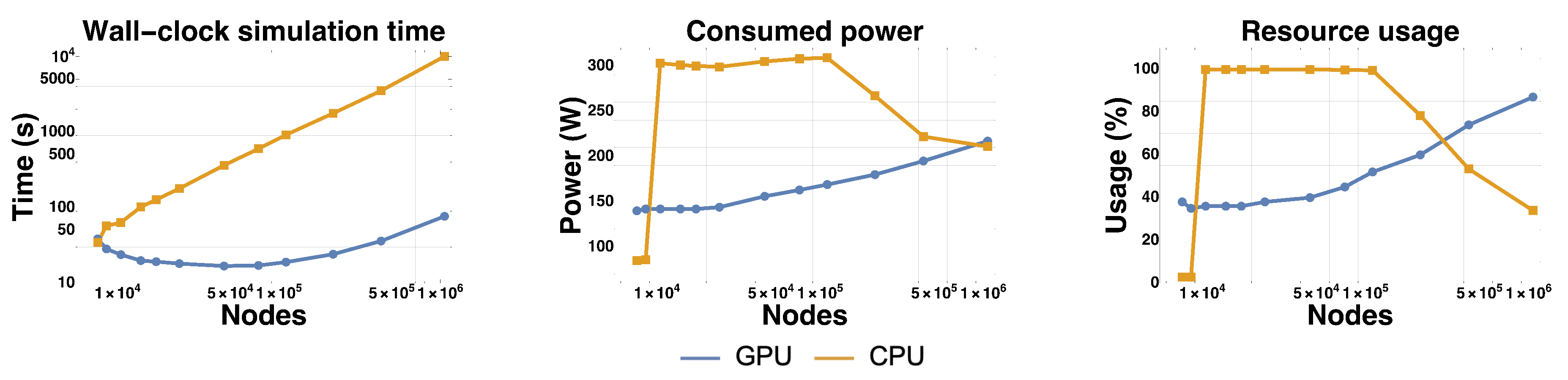
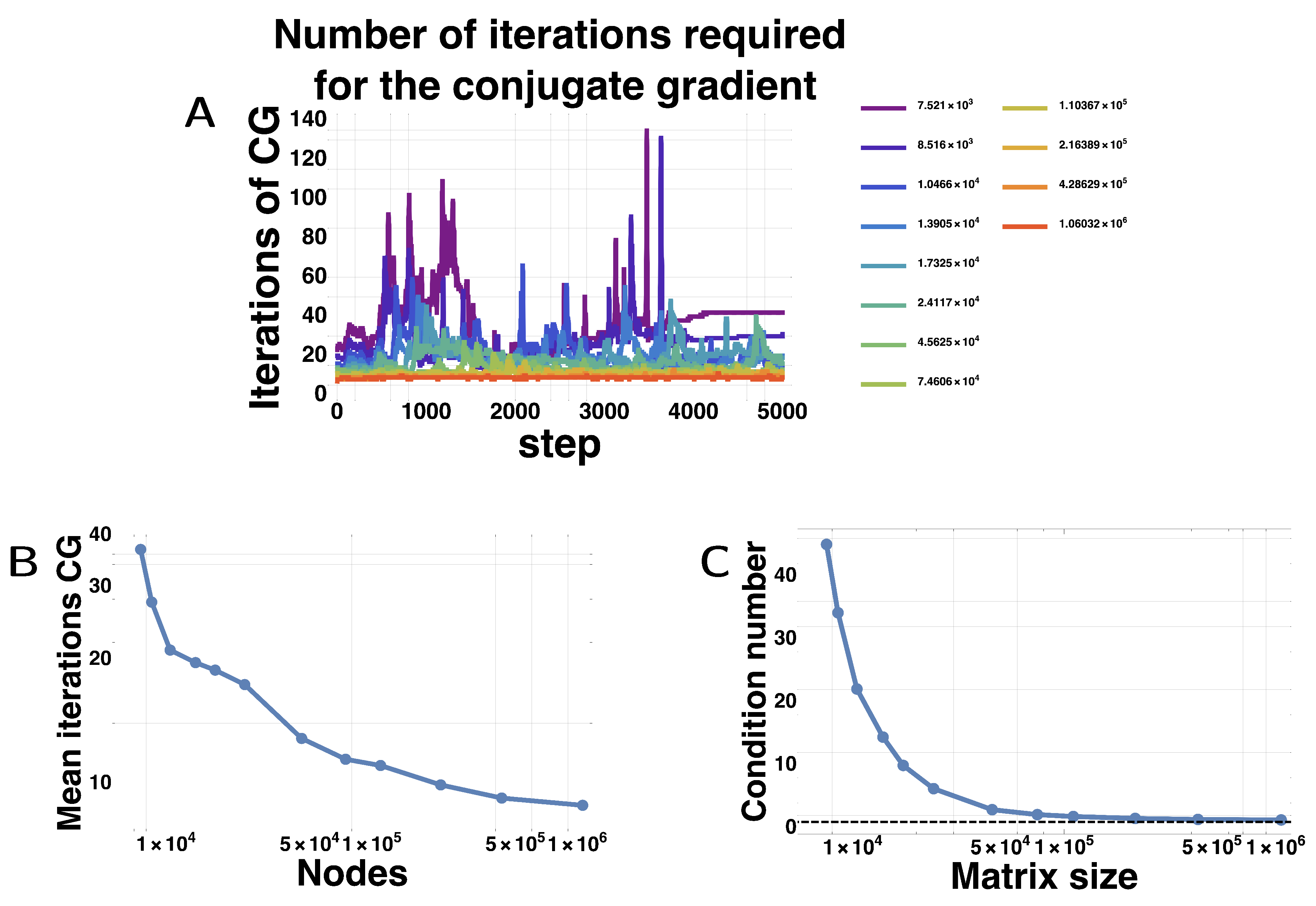
| Paper | Method | Problem | Speedup (CPU vs. GPU) |
|---|---|---|---|
| Sanderson et al., 2009 [52] | FDM | grid mesh, Advection-Reaction-Diffusion | ∼5–10x vs. one CPU core |
| Molnar et al., 2011 [50] | FDM | grid mesh, Turing Patterns, Cahn–Hilliard eq., … | ∼5–40x vs. one CPU thread |
| Pera et al., 2019 [56] | FDM | grid mesh, tumor growth | ∼100–500x vs. 8-core CPU |
| Gormantara et al., 2020 [57] | FDM | grid mesh, FitzHugh-Nahumo model | ∼10x vs. CPU |
| Sato et al., 2009 [53] | FEM+ODE | 3D cardiac simulations | ∼0.6x vs. 32-CPU cluster |
| Mena et al., 2015 [54] | FEM+ODE | 3D cardiac simulations | ∼50x vs. one CPU core |
| Descombes et al., 2015 [51] | FEM | chemotactic reaction-diffusion on arbitrary surface | ∼100–300x vs. 4-core CPU |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sellami, H.; Cazenille, L.; Fujii, T.; Hagiya, M.; Aubert-Kato, N.; Genot, A.J. Accelerating the Finite-Element Method for Reaction-Diffusion Simulations on GPUs with CUDA. Micromachines 2020, 11, 881. https://doi.org/10.3390/mi11090881
Sellami H, Cazenille L, Fujii T, Hagiya M, Aubert-Kato N, Genot AJ. Accelerating the Finite-Element Method for Reaction-Diffusion Simulations on GPUs with CUDA. Micromachines. 2020; 11(9):881. https://doi.org/10.3390/mi11090881
Chicago/Turabian StyleSellami, Hedi, Leo Cazenille, Teruo Fujii, Masami Hagiya, Nathanael Aubert-Kato, and Anthony J. Genot. 2020. "Accelerating the Finite-Element Method for Reaction-Diffusion Simulations on GPUs with CUDA" Micromachines 11, no. 9: 881. https://doi.org/10.3390/mi11090881
APA StyleSellami, H., Cazenille, L., Fujii, T., Hagiya, M., Aubert-Kato, N., & Genot, A. J. (2020). Accelerating the Finite-Element Method for Reaction-Diffusion Simulations on GPUs with CUDA. Micromachines, 11(9), 881. https://doi.org/10.3390/mi11090881





