A Direct Comparison between the Lateral Magnetophoretic Microseparator and AdnaTest for Isolating Prostate Circulating Tumor Cells
Abstract
1. Introduction
2. Materials and Methods
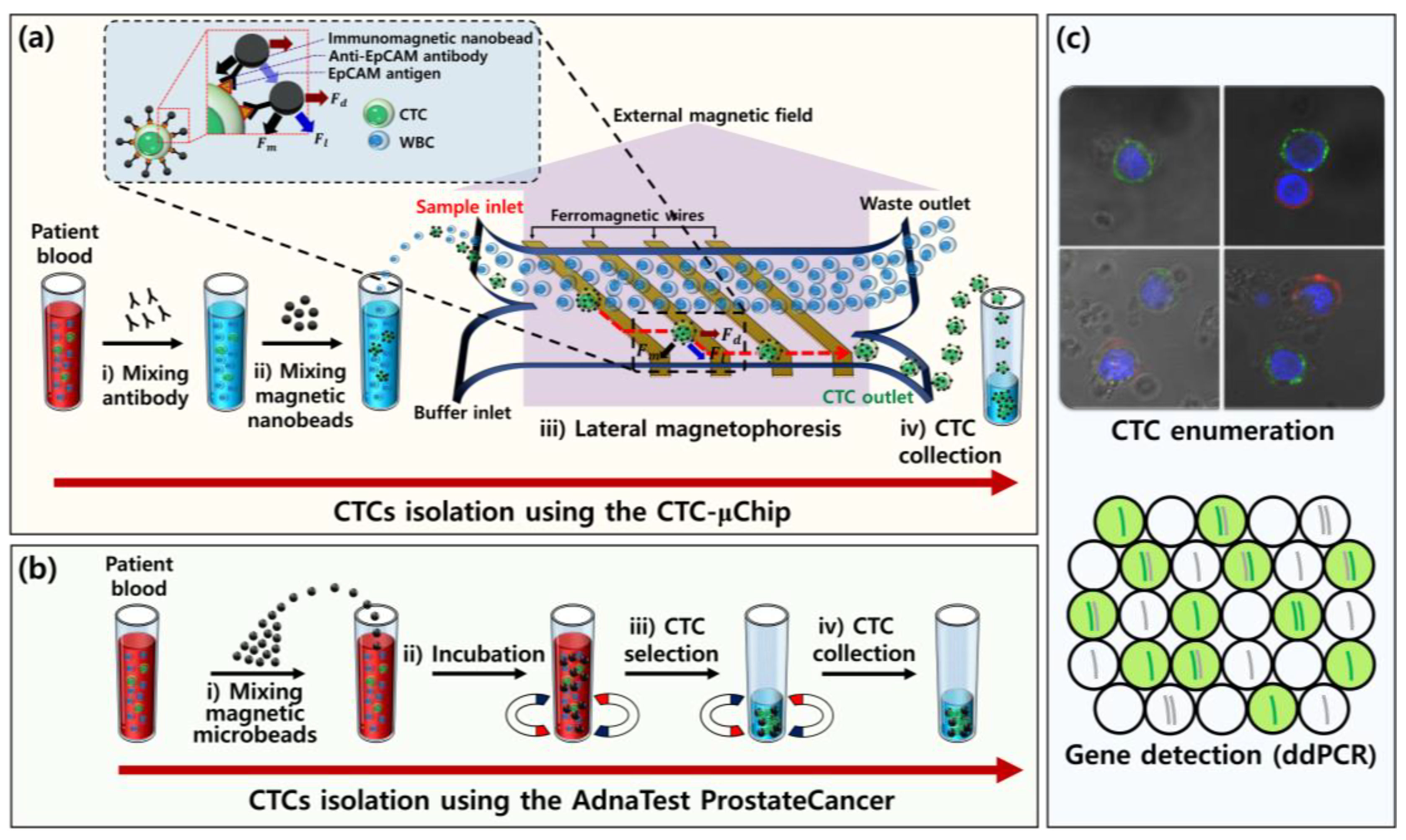
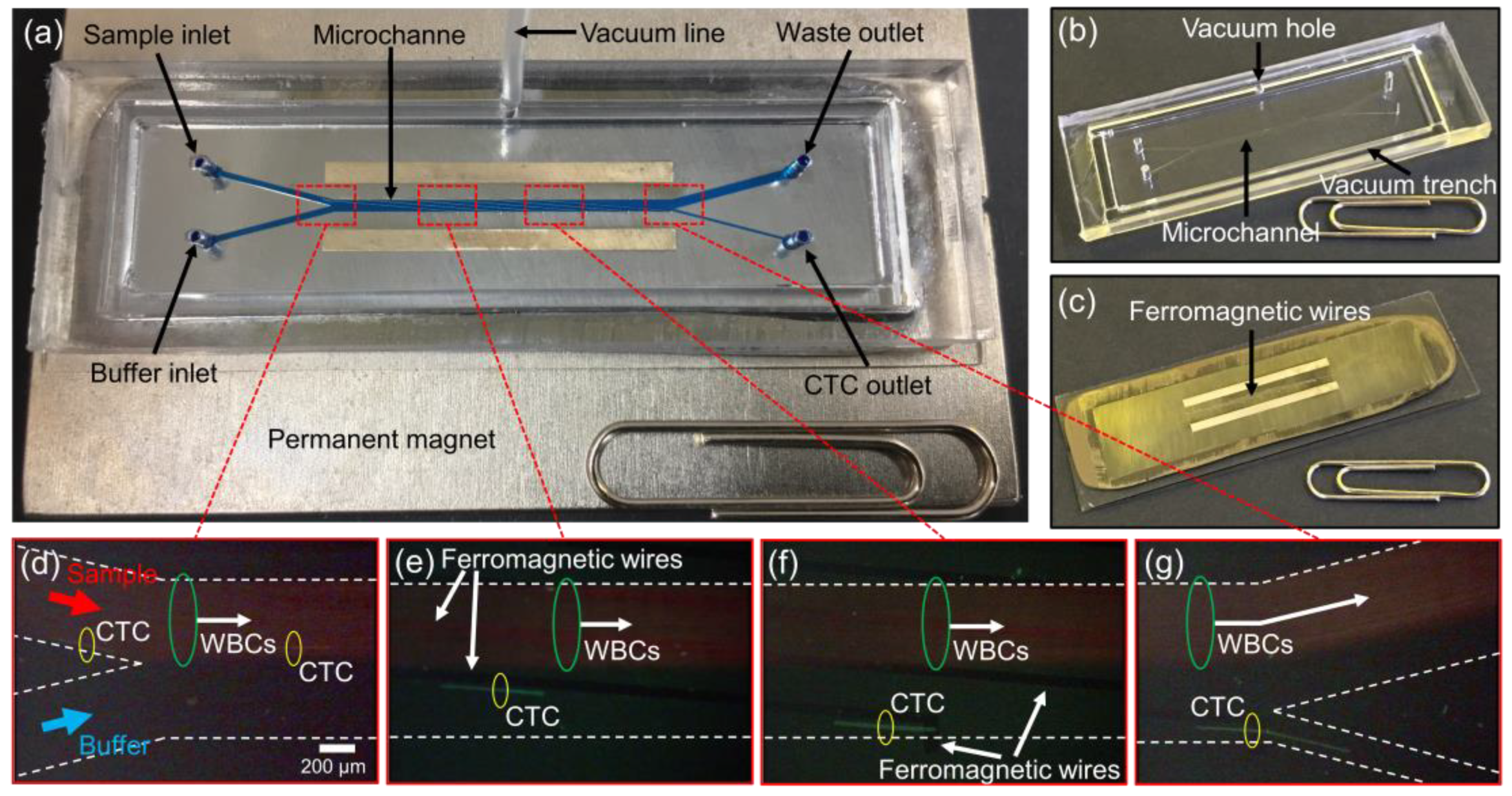
2.1. Experimental Design and Working Principle
2.2. Fabrication of the CTC-µChip
2.3. Preparation of Blood Samples
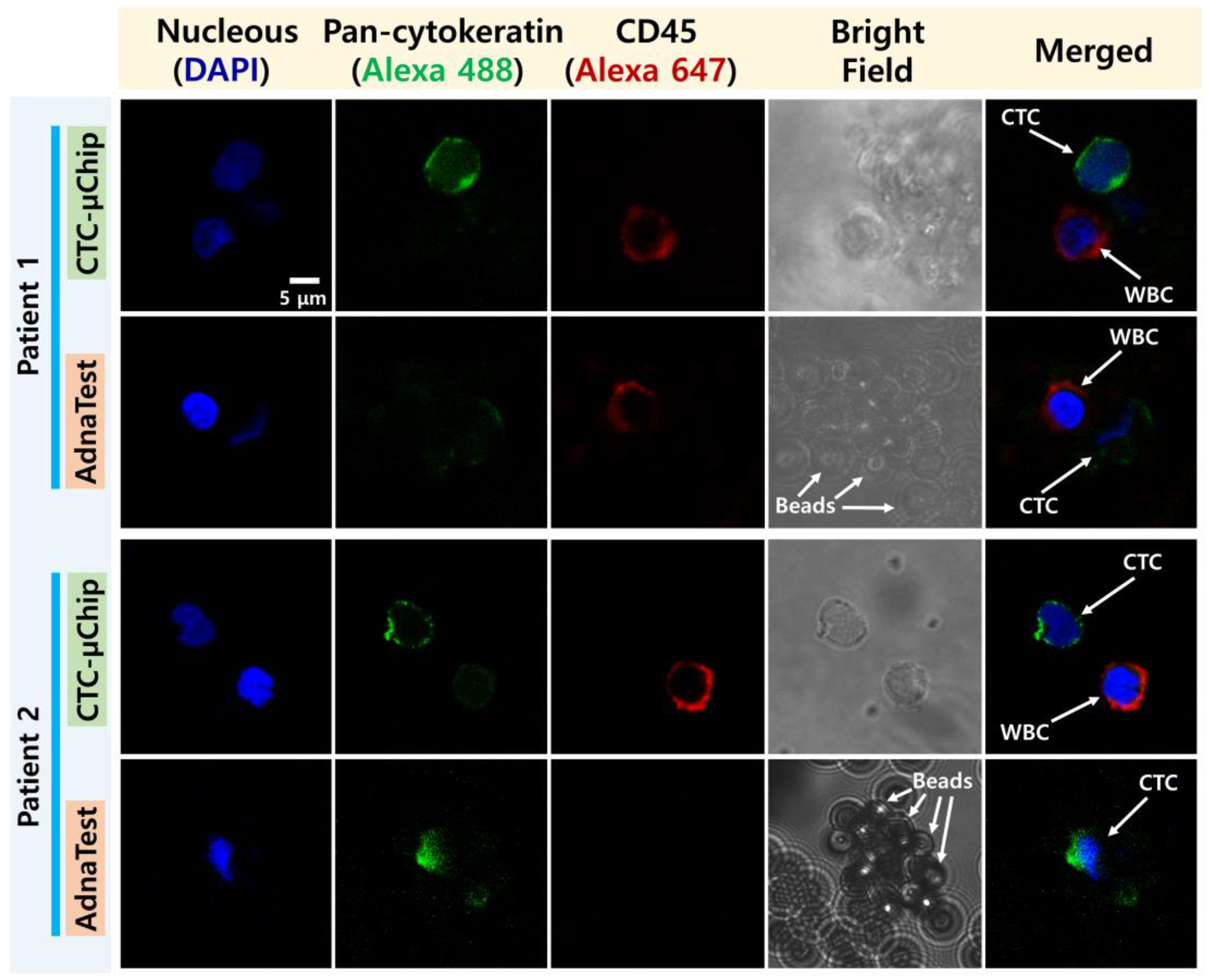
2.4. CTC Enumeration
2.5. Gene Expression Analysis Using RT-ddPCR
3. Results
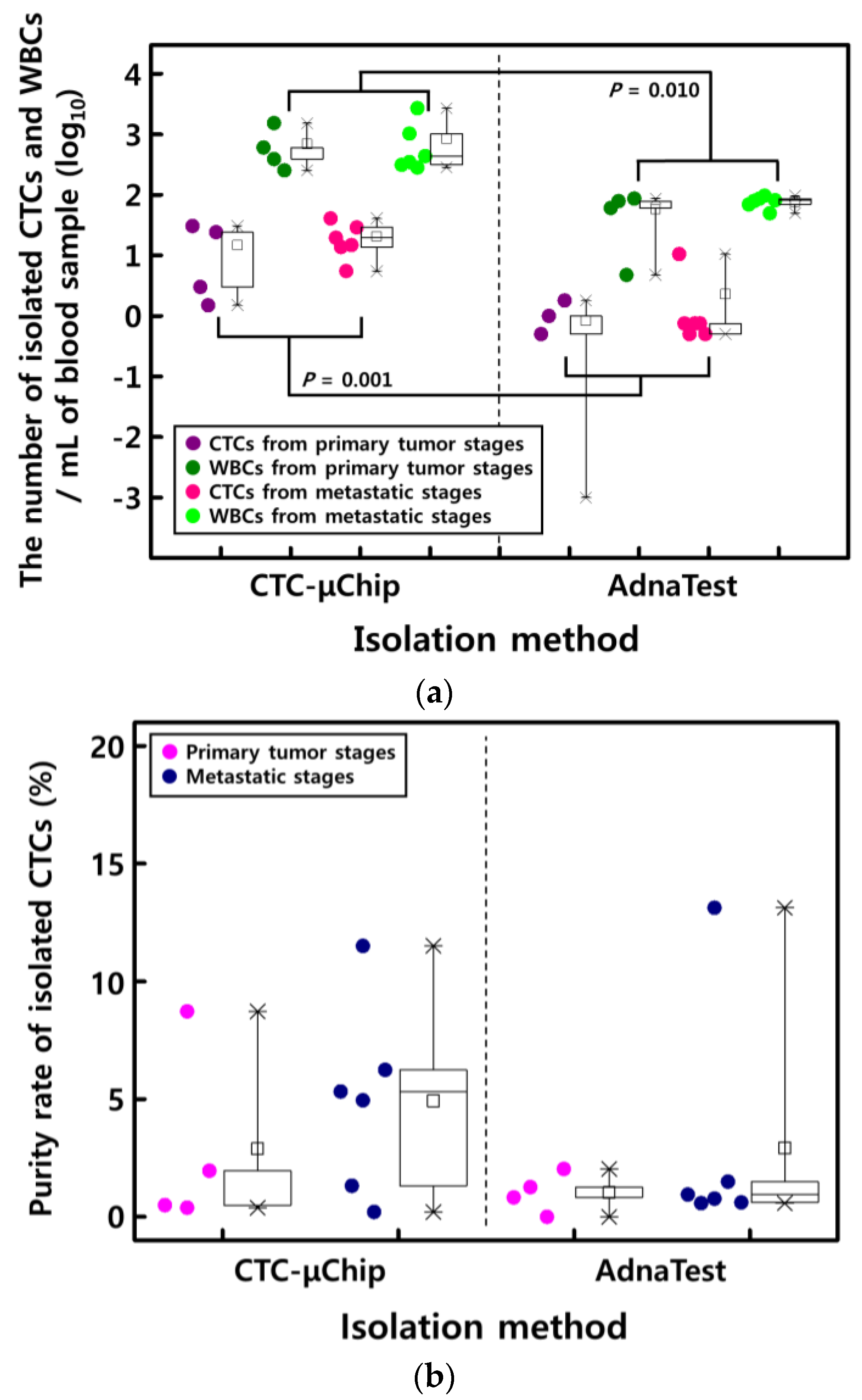
3.1. Comparison of CTC Enumeration
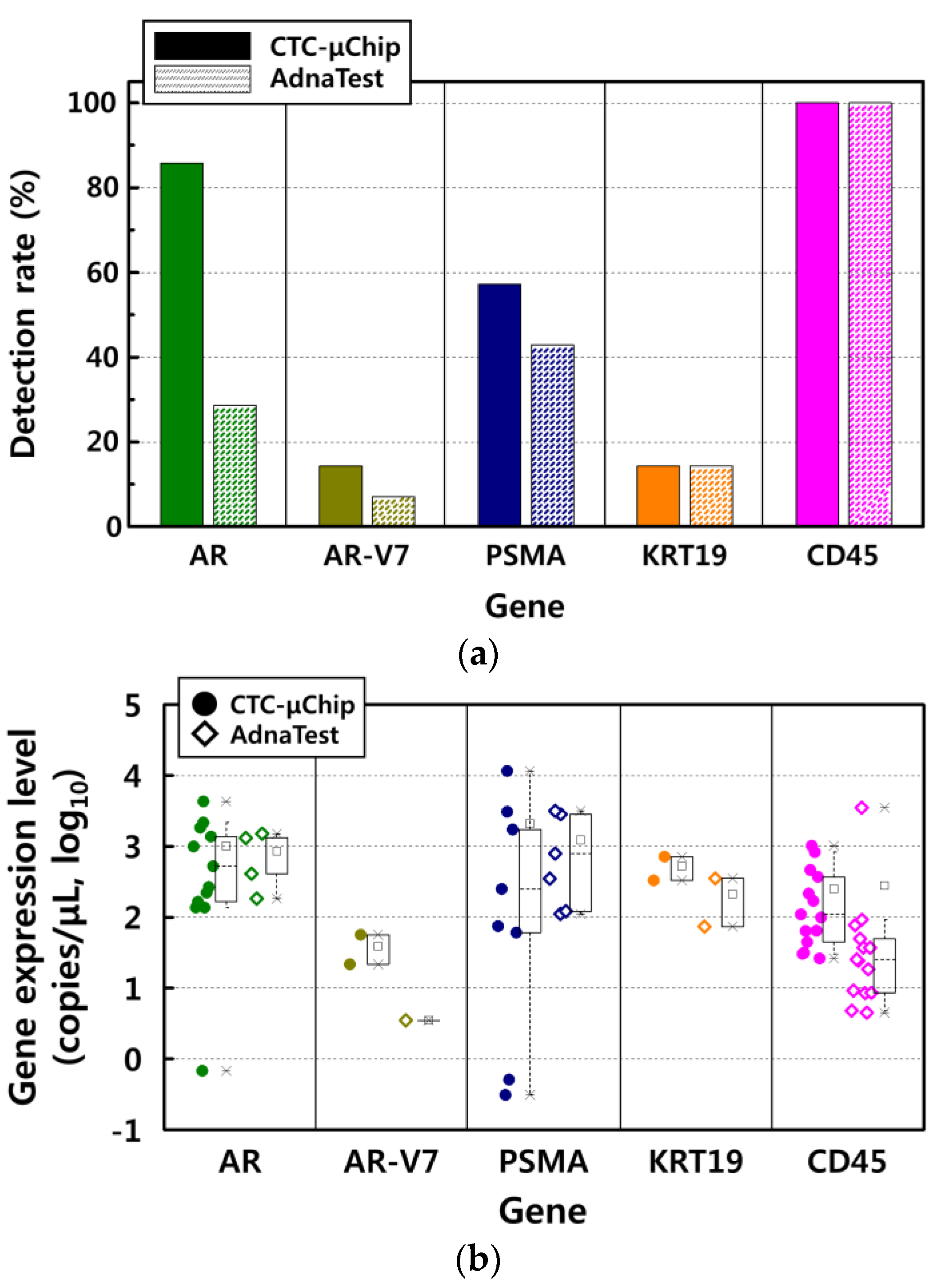
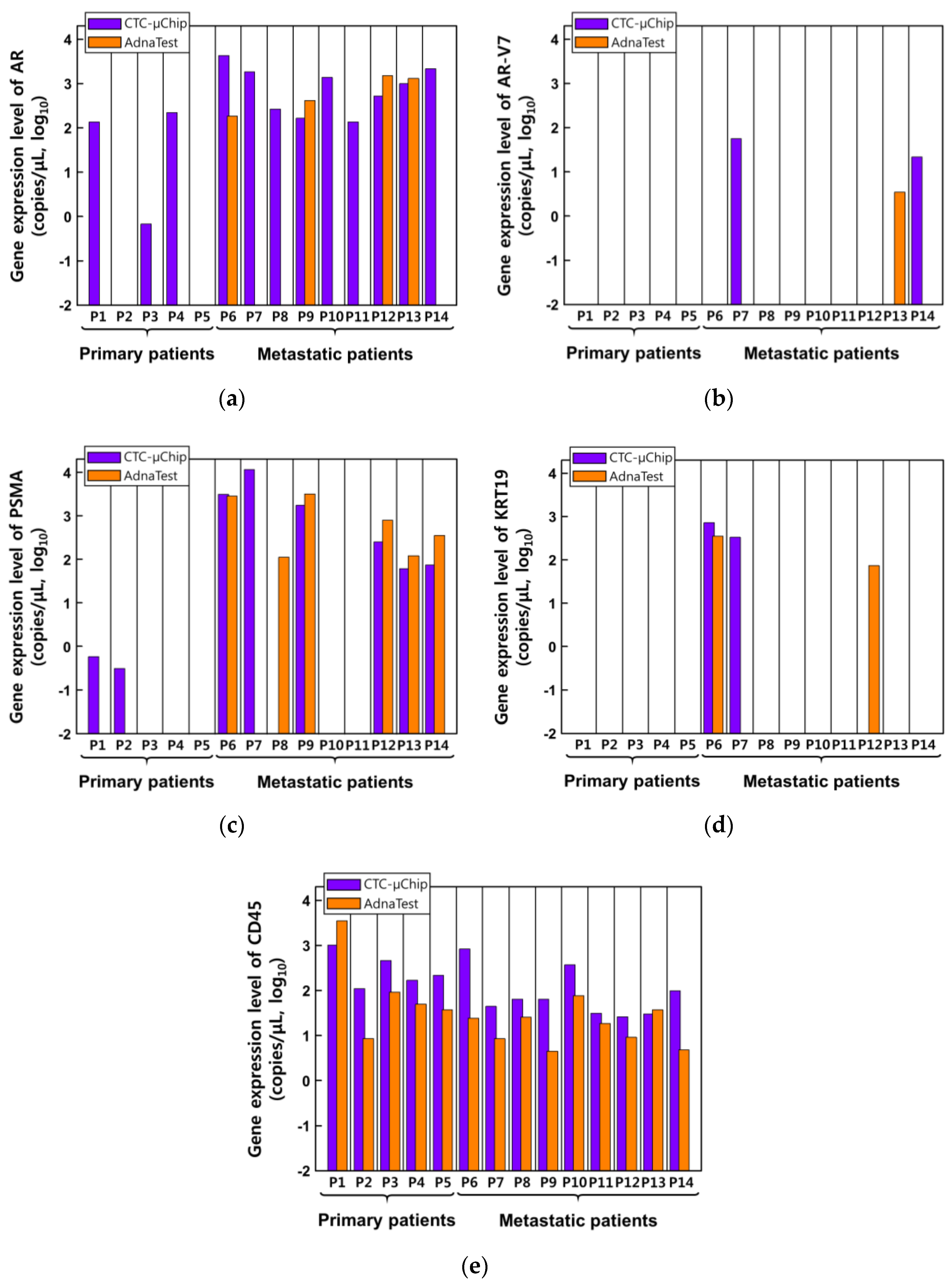
3.2. Comparison of Gene Expression Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cabel, L.; Proudhon, C.; Gortais, H.; Loirat, D.; Coussy, F.; Pierga, J.-Y.; Bidard, F.-C. Circulating tumor cells: Clinical validity and utility. Int. J. Clin. Oncol. 2017, 22, 421–430. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Mavroudis, D. Circulating cancer cells. Ann. Oncol. 2010, 21, vii95–vii100. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Meye, A.; Bilkenroth, U.; Schmidt, U.; Fussel, S.; Robel, K.; Melchior, A.M.; Blumke, K.; Pinkert, D.; Bartel, F.; Linne, C. Isolation and enrichment of urologic tumor cells in blood samples by a semi-automated CD45 depletion autoMACS protocol. Int. J. Oncol. 2002, 21, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Zeni, N.; Mewes, S.; Niestroj, R.; Gasiorowski, L.; Murawa, D.; Nowaczyk, P.; Tomasi, T.; Weber, E.; Dworacki, G.; Morgenthaler, N.G. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int. J. Oncol. 2012, 41, 1241–1250. [Google Scholar]
- Laget, S.; Broncy, L.; Hormigos, K.; Dhingra, D.M.; BenMohamed, F.; Capiod, T.; Osteras, M.; Farinelli, L.; Jackson, S.; Paterlini-Brechot, P. Technical insights into highly sensitive isolation and molecular characterization of fixed and live circulating tumor cells for early detection of tumor invasion. PLoS ONE 2017, 12, e0169427. [Google Scholar] [CrossRef]
- Zieglschmid, V.; Hollmann, C.; Mannel, J.; Albert, W.; Jaeschke-Melli, S.; Eckstein, B.; Hillemann, T.; Greten, T.F.; Gross, E.; Boecher, O. Tumor-associated gene expression in disseminated tumor cells correlates with disease progression and tumor stage in colorectal cancer. Anticancer Res. 2007, 27, 1823–1832. [Google Scholar]
- Cho, H.; Kim, J.; Song, H.; Sohn, K.Y.; Jeon, M.; Han, K.-H. Microfluidic technologies for circulating tumor cell isolation. Analyst 2018, 143, 2936–2970. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Yu, V.; Dhar, M.; Renier, C.; Matsumoto, M.; Heirich, K.; Garon, E.B.; Goldman, J.; Rao, J.; Sledge, G.W. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget 2016, 7, 12748. [Google Scholar] [CrossRef] [PubMed]
- Hvichia, G.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Müller, V.; Loges, S.; Neves, R.; Stoecklein, N. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer 2016, 138, 2894–2904. [Google Scholar] [CrossRef] [PubMed]
- Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; Ionescu-Zanetti, C. Mutational analysis of circulating tumor cells using a novel microfluidic collection device and qPCR assay. Transl. Oncol. 2013, 6, 528. [Google Scholar] [CrossRef]
- Renier, C.; Pao, E.; Che, J.; Liu, H.E.; Lemaire, C.A.; Matsumoto, M.; Triboulet, M.; Srivinas, S.; Jeffrey, S.S.; Rettig, M. Label-free isolation of prostate circulating tumor cells using Vortex microfluidic technology. Npj Precis. Oncol. 2017, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gwak, H.; Kim, J.; Kashefi-Kheyrabadi, L.; Kwak, B.; Hyun, K.-A.; Jung, H.-I. Progress in circulating tumor cell research using microfluidic devices. Micromachines 2018, 9, 353. [Google Scholar] [CrossRef]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular analysis of circulating tumour cells—Biology and biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129. [Google Scholar] [CrossRef]
- Zhang, C.; Guan, Y.; Sun, Y.; Ai, D.; Guo, Q. Tumor heterogeneity and circulating tumor cells. Cancer Lett. 2016, 374, 216–223. [Google Scholar] [CrossRef]
- Müller, V.; Riethdorf, S.; Rack, B.; Janni, W.; Fasching, P.A.; Solomayer, E.; Aktas, B.; Kasimir-Bauer, S.; Pantel, K.; Fehm, T. Prognostic impact of circulating tumor cells assessed with the CellSearch System™ and AdnaTest Breast™ in metastatic breast cancer patients: The DETECT study. Breast Cancer Res. 2012, 14, R118. [Google Scholar] [CrossRef]
- Vojtechova, G.; Benesova, L.; Belsanova, B.; Minarikova, P.; Levy, M.; Lipska, L.; Suchanek, S.; Zavoral, M.; Minarik, M. Monitoring of circulating tumor cells by a combination of immunomagnetic enrichment and RT-PCR in colorectal cancer patients undergoing surgery. Adv. Clin. Exp. Med. 2016, 25, 1273–1279. [Google Scholar] [CrossRef]
- Blassl, C.; Kuhlmann, J.D.; Webers, A.; Wimberger, P.; Fehm, T.; Neubauer, H. Gene expression profiling of single circulating tumor cells in ovarian cancer–Establishment of a multi-marker gene panel. Mol. Oncol. 2016, 10, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.I.; Bier, S.; Hennenlotter, J.; Beger, G.; Pavlenco, L.; van de Flierdt, J.; Hauch, S.; Maas, M.; Walz, S.; Rausch, S. Expression of tumour progression-associated genes in circulating tumour cells of patients at different stages of prostate cancer. BJU Int. 2018, 122, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Andreopoulou, E.; Yang, L.Y.; Rangel, K.; Reuben, J.; Hsu, L.; Krishnamurthy, S.; Valero, V.; Fritsche, H.; Cristofanilli, M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int. J. Cancer 2012, 130, 1590–1597. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, I.; Peeters, D.; Benoy, I.; Elst, H.; Van Laere, S.; Prove, A.; Maes, H.; Huget, P.; Van Dam, P.; Vermeulen, P. Circulating tumour cell detection: A direct comparison between the CellSearch System, the AdnaTest and CK-19/mammaglobin RT–PCR in patients with metastatic breast cancer. Br. J. Cancer 2010, 102, 276–284. [Google Scholar] [CrossRef]
- Brown, L.C.; Lu, C.; Antonarakis, E.S.; Luo, J.; Armstrong, A.J. Androgen receptor variant-driven prostate cancer II: Advances in clinical investigation. Prostate Cancer Prostatic Dis. 2020, 23, 367–380. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Sumanasuriya, S.; Omlin, A.; Armstrong, A.; Attard, G.; Chi, K.N.; Bevan, C.L.; Shibakawa, A.; IJzerman, M.J.; De Laere, B.; Lolkema, M. Consensus statement on circulating biomarkers for advanced prostate cancer. Eur. Urol. Oncol. 2018, 1, 151–159. [Google Scholar] [CrossRef]
- Aktas, B.; Kasimir-Bauer, S.; Müller, V.; Janni, W.; Fehm, T.; Wallwiener, D.; Pantel, K.; Tewes, M.; DETECT Study Group. Comparison of the HER2, estrogen and progesterone receptor expression profile of primary tumor, metastases and circulating tumor cells in metastatic breast cancer patients. BMC Cancer 2016, 16, 522. [Google Scholar] [CrossRef]
- Tewes, M.; Aktas, B.; Welt, A.; Mueller, S.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: An option for monitoring response to breast cancer related therapies. Breast Cancer Res. Treat. 2009, 115, 581. [Google Scholar] [CrossRef]
- Sharp, A.; Welti, J.C.; Lambros, M.B.; Dolling, D.; Rodrigues, D.N.; Pope, L.; Aversa, C.; Figueiredo, I.; Fraser, J.; Ahmad, Z. Clinical utility of circulating tumour cell androgen receptor splice variant-7 status in metastatic castration-resistant prostate cancer. Eur. Urol. 2019, 76, 676–685. [Google Scholar] [CrossRef]
- Gorges, T.M.; Stein, A.; Quidde, J.; Hauch, S.; Röck, K.; Riethdorf, S.; Joosse, S.A.; Pantel, K. Improved detection of circulating tumor cells in metastatic colorectal cancer by the combination of the CellSearch® System and the AdnaTest®. PLoS ONE 2016, 11, e0155126. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, J.; Cho, H.; Han, K.-H. Evaluation of Positive and Negative Methods for Isolation of Circulating Tumor Cells by Lateral Magnetophoresis. Micromachines 2019, 10, 386. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, J.; Jeon, C.-W.; Han, K.-H. A disposable microfluidic device with a reusable magnetophoretic functional substrate for isolation of circulating tumor cells. Lab Chip 2017, 17, 4113–4123. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Han, S.-I.; Park, M.-J.; Jeon, C.-W.; Joo, Y.-D.; Choi, I.-H.; Han, K.-H. Circulating tumor cell microseparator based on lateral magnetophoresis and immunomagnetic nanobeads. Anal. Chem. 2013, 85, 2779–2786. [Google Scholar] [CrossRef]
- Mannweiler, S.; Amersdorfer, P.; Trajanoski, S.; Terrett, J.A.; King, D.; Mehes, G. Heterogeneity of prostate-specific membrane antigen (PSMA) expression in prostate carcinoma with distant metastasis. Pathol. Oncol. Res. 2009, 15, 167–172. [Google Scholar] [CrossRef]
- Sweat, S.D.; Pacelli, A.; Murphy, G.P.; Bostwick, D.G. Prostate-specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology 1998, 52, 637–640. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.; Chung, J.-S.; Han, K.-H. A Direct Comparison between the Lateral Magnetophoretic Microseparator and AdnaTest for Isolating Prostate Circulating Tumor Cells. Micromachines 2020, 11, 870. https://doi.org/10.3390/mi11090870
Cho H, Chung J-S, Han K-H. A Direct Comparison between the Lateral Magnetophoretic Microseparator and AdnaTest for Isolating Prostate Circulating Tumor Cells. Micromachines. 2020; 11(9):870. https://doi.org/10.3390/mi11090870
Chicago/Turabian StyleCho, Hyungseok, Jae-Seung Chung, and Ki-Ho Han. 2020. "A Direct Comparison between the Lateral Magnetophoretic Microseparator and AdnaTest for Isolating Prostate Circulating Tumor Cells" Micromachines 11, no. 9: 870. https://doi.org/10.3390/mi11090870
APA StyleCho, H., Chung, J.-S., & Han, K.-H. (2020). A Direct Comparison between the Lateral Magnetophoretic Microseparator and AdnaTest for Isolating Prostate Circulating Tumor Cells. Micromachines, 11(9), 870. https://doi.org/10.3390/mi11090870






