Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices
Abstract
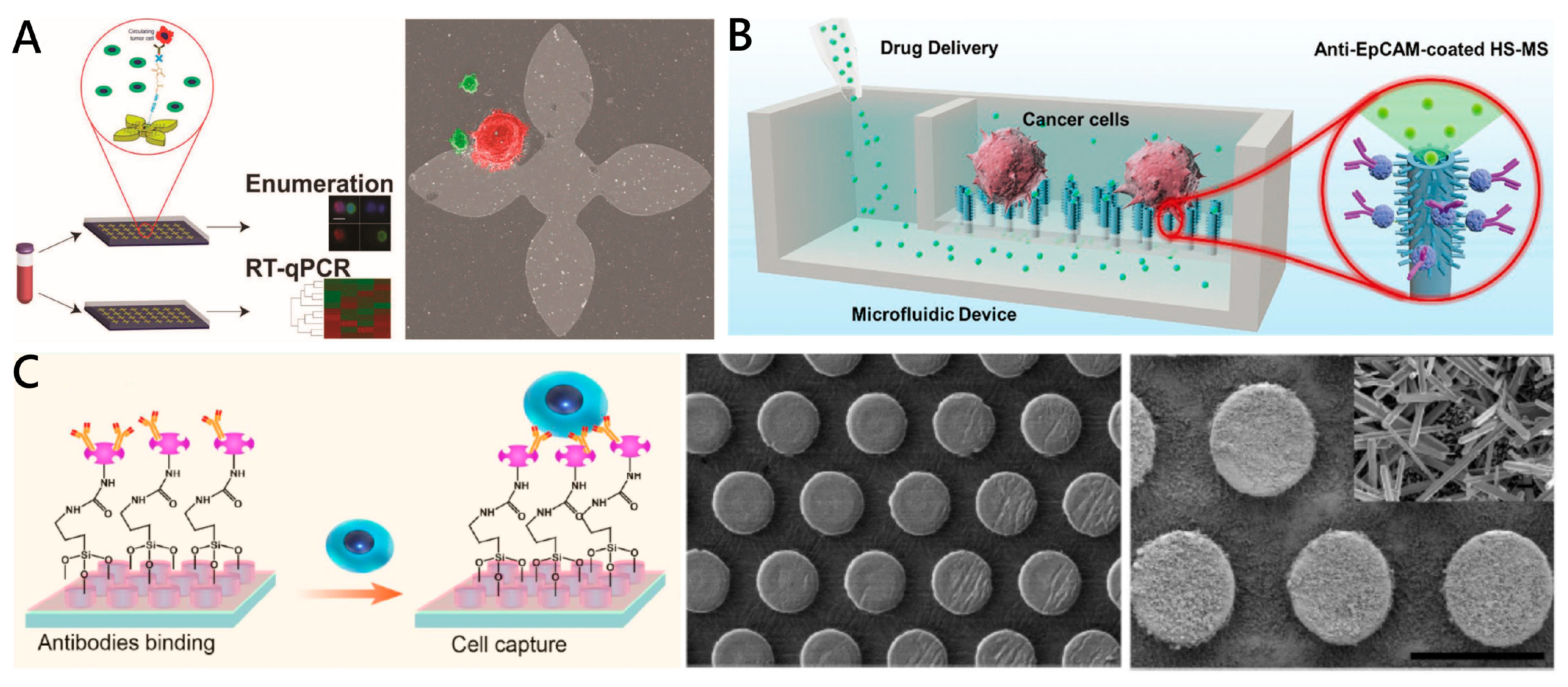
1. Introduction
2. Nanotechnology-Assisted Circulating Tumor Cell (CTC) Isolation Using Microfluidic Devices
2.1. Physical and Biochemical Properties of CTCs
| Properties | CTCs | Normal Cells (e.g., Leukocytes) | Ref. |
|---|---|---|---|
| Size | Average diameter ranging from 15 to 25 μm | Erythrocytes have an average diameter of 7–8.5 μm and leukocytes are 5–13 μm in diameter. | [35,36] |
| Density | CTCs are mainly deposited in the monocyte enrichment layer. | Erythrocytes and granulocytes have a density > 1.077 g/mL, monocytes and lymphocytes have a density < 1.077 g/mL. | [37,38] |
| Morphology | Most CTCs from breast, colorectal and prostate cancer have the roundness of 1.5 ± 0.6, 1.5 ± 1.4 and 1.5 ± 0.8, respectively | The roundness of WBCs is 1.8 ± 1.2. | [39] |
| Deformability | The stiffness ranges from 200 to 2000 Pa. | The stiffness of neutrophils ranges from 156 ± 87 Pa. | [40,41] |
| Dielectric properties | CTCs have more negative charges. | Different cell types have significant differences in the cell membrane capacitance. | [42,43,44] |
| Biomarkers | EpCAM, DAPI CK19, HER2, EP, PR and MUC1 (used for breast cancer diagnosis); PSA, AR, EGR and PTEN (used for prostate cancer diagnosis); CK, ASGPR1, N-cadherin, Vimentin, Gpc3, AFP and albumin (used for hepatocellular carcinoma diagnosis) | CD45, DAPI | [45,46,47] |
2.2. Nanotechnology-Assisted CTC Capture
2.2.1. Nanoparticles in Suspensions for CTC Capture
Magnetic Nanoparticles
Nanoparticles of Gold and Other Materials

2.2.2. Nanostructures on Substrates for CTC Capture
Nanopillars, Nanowires, Nanofibers
| Microchip-Based Nanosubstrate | Materials | Optimal Throughput | Capture Efficiency % | Purity % | Viability % | Cancer Cell Type | Highlights | Ref. |
|---|---|---|---|---|---|---|---|---|
| Magnetic nanoparticles | Fe3O4 MNPs + anti-EpCAM antibody | 10 mL/h | 90% (COLO-205 cells) 86% (SKBR3 cells) | N/A | N/A | COLO-205 and SKBR3 cells | Effective capture achieved by arrayed magnets with alternate polarities | [63] |
| 540 μL/h | 80~90% | ≈91% | >95% | HCT-116 cells | Wavy-herringbone structure for effectively and selectively capturing and releasing CTCs | [68] | ||
| Gold and other materials nanoparticles | Au NPs + Aptamer-SYL3C | 1 mL/h | 74~84% | 99.99% (WBC removal rate) | 96% (SW480 cells) | SW480, LNCap and KATO III cells | Multivalent aptamer–antigen binding; rotated triangular micropillars based on the DLD principle | [53] |
| SiO2 NPs + CD71 antibody | N/A | 94 ± 3% (HL-60 cells) 86 ± 3% (Ramos cells) 89 ± 9% (Jurkat cells) 81 ± 6% (MDA-231 cells) 74 ± 14% (PC-3 cells) | 41~65% (COG-LL-332 cells) | N/A | HL-60, Ramos, Jurkat, MDA-231, PC-3 cells | High capture purity; a wide range of cell separations | [73] | |
| Nanopillars, Nanowires | SiNPs + anti-EpCAM antibody | 1 mL/h | 95% | N/A | N/A | MCF-7 cells | High efficiency achieved with embedded chevron-shaped micropatterns | [77] |
| SiNWs + CKAAKN peptides and anti-EpCAM antibody | 1 mL/h | ≈96% | ≈26% | ≈94% | BxPC3, Panc-1 and AsPC-1 cells | Novel biological method of enzymatic release | [54] | |
| SiNWs + anti-EpCAM antibody | 1 mL/h | 88~94% | ≈200 WBCs | N/A | NSCLC cells | Click reaction; Disulfide cleavage-mediated CTC capture | [79] | |
| Nanofibers | MnO2 + anti-EpCAM antibody | 0.1 mL/h | 80% | N/A | ≈90% | MCF-7 cells | CTC release by oxalic acid dissolution of MnO2 | [81] |
| PLGA + Hyaluronic acid | 1 mL/h | >80% | N/A | N/A | HeLa, A549 and MCF-7 cells | Specific capture of CD44-positive tumor cells | [55] | |
| Other nanoroughened structures | GO nanosheets + anti-EpCAM antibody | 1 mL/h | 73% | N/A | N/A | PC-3 cells | Higher sensitivity; Gentle capture | [82,83] |
| ZnO/Al2O3 HS-MSA + anti-EpCAM antibody | N/A | ≈85% | ≈2.6% (per mL artificial CTCs blood samples) | >95% | MCF-7 cells | 3D micro/nanostructures; in situ extracellular drug delivery of the captured CTCs | [84,85] | |
| ZnO nanograss + anti-EpCAM antibody | N/A | >80% | N/A | 90%~95% | MCF-7 and MDA-MB231 cells | Self-sterilizing and regeneratable | [86] |
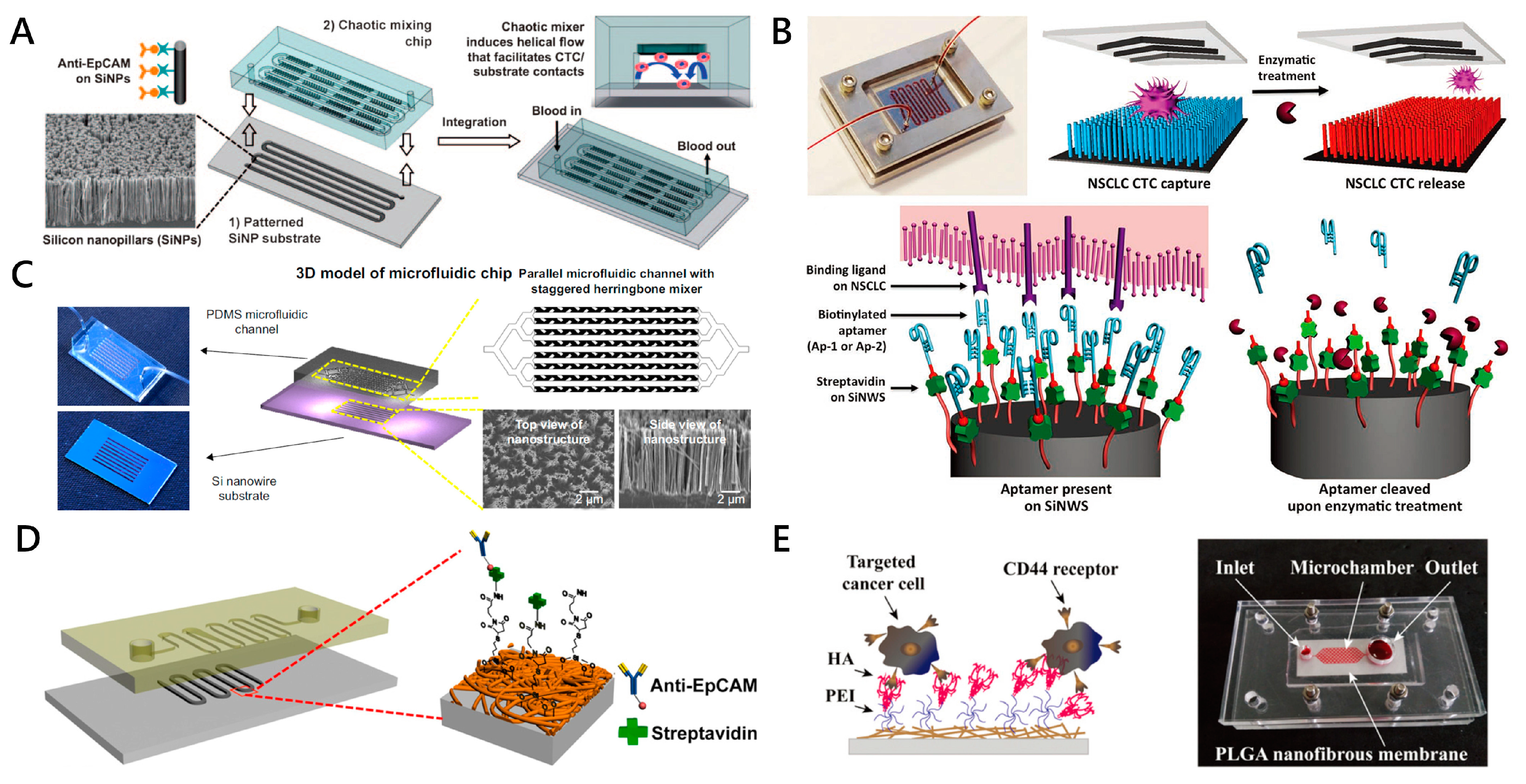
Other Nanoroughened Structures
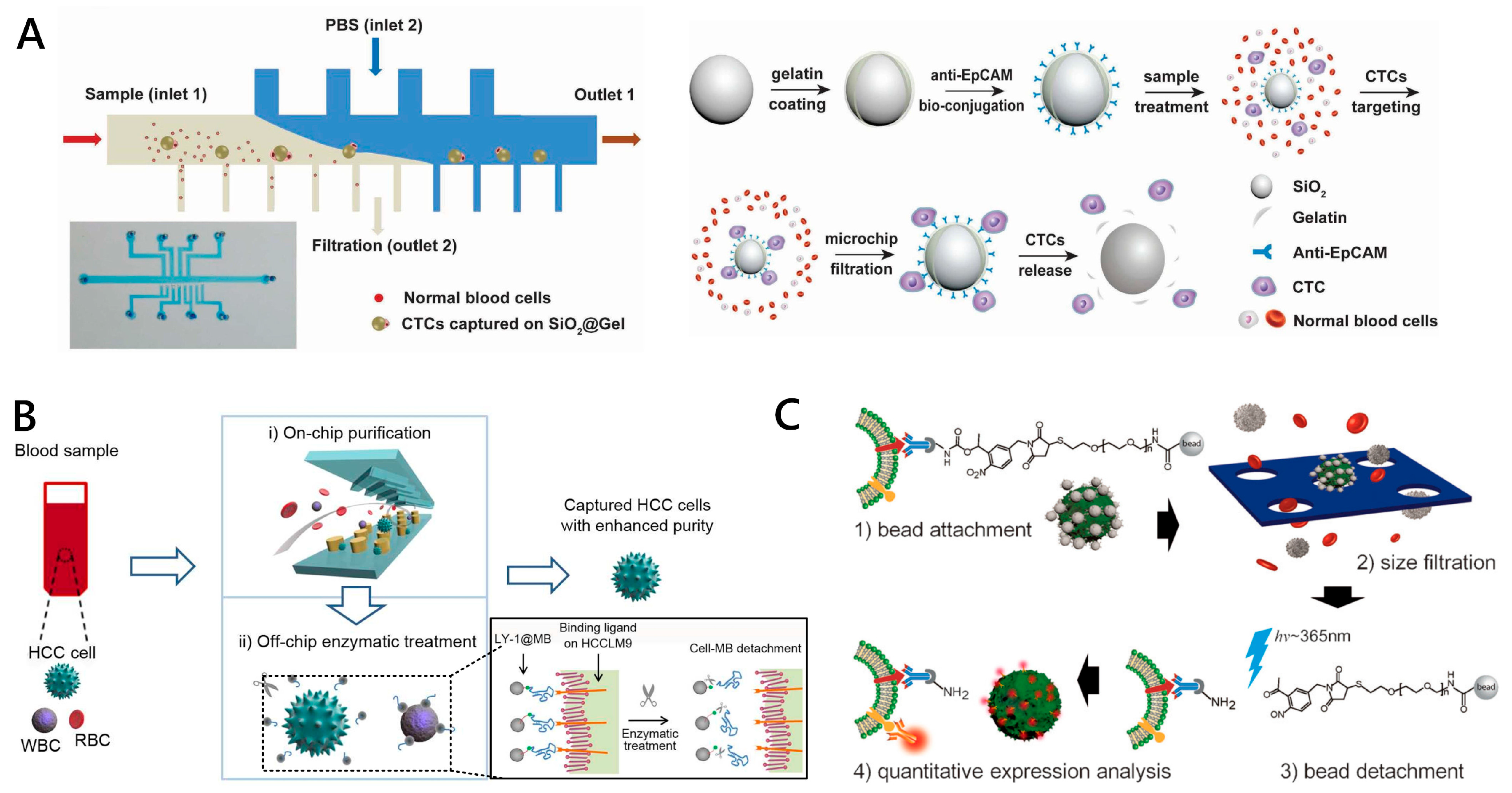
2.3. Approaches to CTC Release from Nanostructures
2.3.1. Flowing Fluid-Mediated CTC Release
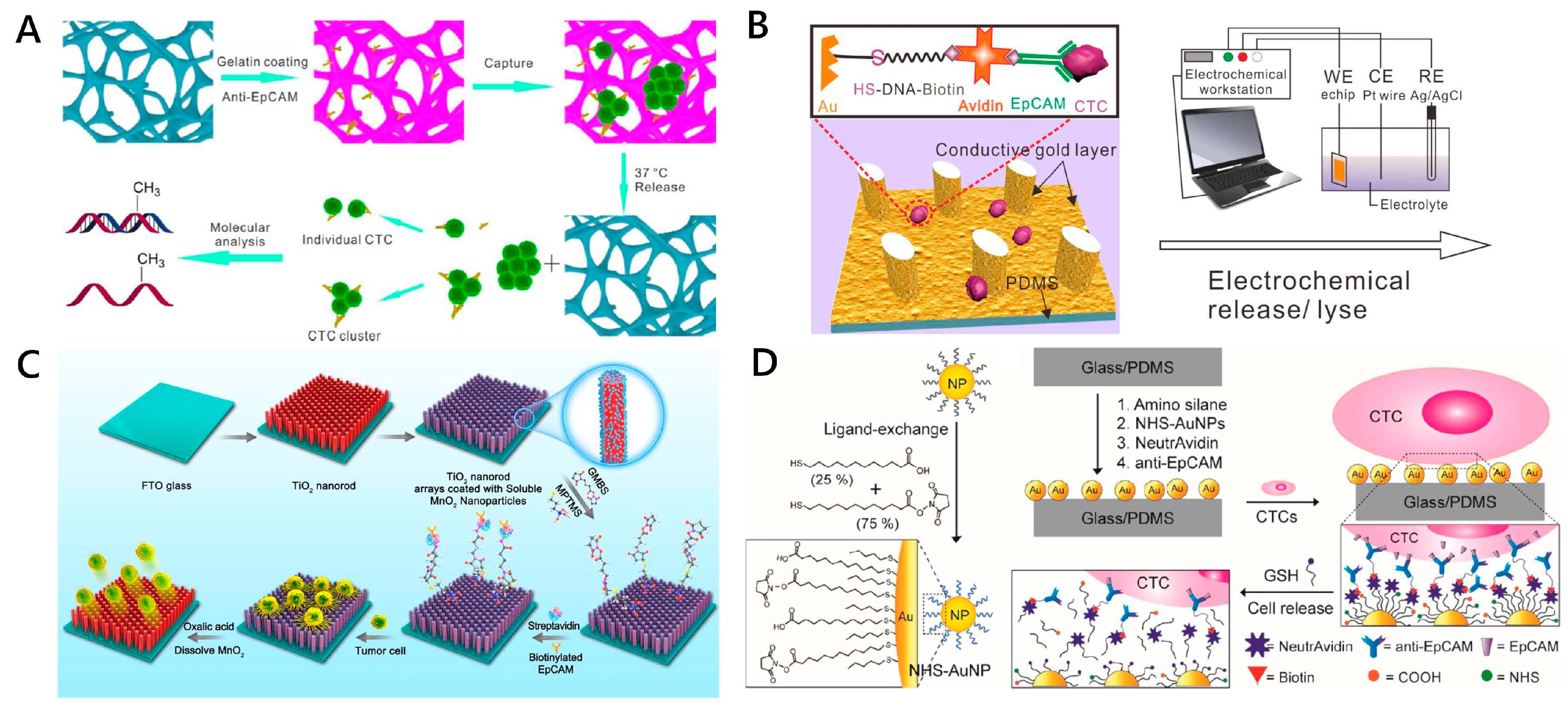
2.3.2. Enzymatic Degradation-Based CTC Release
2.3.3. Light-Controlled CTC Release
2.3.4. Thermal-Controlled CTC Release
2.3.5. Electrochemical and Chemical Reagent-Triggered CTC Release
2.3.6. Ligand Competition for CTC Release
| Mechanism | Material | Reference | Treatment Conditions | Capture Efficiency | Release Efficiency | Cell Viability | Cell Types |
|---|---|---|---|---|---|---|---|
| Enzymatic Degradation | SiO2@Gelbead + PDMS microfluidic filter | [95] | Solution of MMP-9 enzyme | 75% | 97.2% ± 2.1% | >97% | HCT116 cells |
| PDMS herringbone structure + aptamer-conjugated MBs were trapped on the substrate of Ni pillar arrays | [56] | Exonuclease at 37 °C for 15 min | ~91% | ~68.4% | NA | HCCLM9 cells | |
| Light-Controlled | Detachable beads conjugated to antiEpCAM antibodies + 8 μm microporemembrane | [57] | Irradiation from an LED light source (λ = 365 nm) | 89.1% | 98.4% | 97% | SKBR3 cells |
| 7-Aminocoumarin reacted with biotin to bridge the antiEpCAM antibogshdy and streptavidin (SA) modified MBs | [96] | UV or near-infrared (NIR) light irradiation | ~91% | 73% ± 4% and 52% ± 6% | 90% and 97% | HeLa, MCF-7 and SKBR3 cells | |
| Thermal-Controlled | Silicon nanopillars coated with PNIPAAm + antibody | [98] | Alternating temperatures between 37 °C and 20 °C | 97.3% ± 0.4% | 98.8% ± 0.5% | ~95% | MCF-7 cells |
| 3D PDMS scaffold chip with gelatin coating + antibody | [58] | Incubated for 10 min at 37 °C | 88% | 86% | 90% | MCF-7 cells and Hela cells | |
| Aptamer-functionalized tapered chamber + temperature control chip | [100] | Heated at 48 °C for 2 min | ~92% | ~80% | Normal proliferation | CCRF-CEM cells | |
| Electrochemical and Chemical Reagent-Triggered | Thiol-containing ligand assembled gold surfaces of PDMS micropillar-array | [59] | At voltage of −1.2 V for 10 min | 85~100% | >93% | >95% | MCF-7 cells and HepG2 cells |
| 3D PEDOT-based nanorod arrays coated with PLL-g-PEG-biotin | [102] | 20 cycles of cyclic voltammetry sweeping from −0.8 to 0.5 V | NA | >90% | ~90% | MCF-7 cells and Hela cells | |
| TiO2 nanorod arrays coated with transparent MnO2 nanoparticles | [104] | Oxalic acid solution | >60% | 89.9% | >90% | SW480 and MCF-7 cells | |
| Anti-EpCAM functionalized zinc oxide nanowires on glass substrate | [105] | 1% sodium citrate in water (pH 6.5) | 90% ± 1% | 88% ± 4% | 92% ± 1% | MCF-7 cell | |
| Ligand Competition | Anti-EpCAM- deposit alginate coating on fluorescent-magnetic nanospheres | [106] | EDTA treatment | >85% | ~65% | ~70% | SKBR3 cells |
| Thiol-modified AuNPs + anti-EpCAM assembled on a PDMS HB chip | [65] | GSH | Average of 68% ± 29.2% (MDA-MB-231) and 72% ± 26.4% (PC3 cells) | 92% (PC3) and 91% (MDA-MB-231) | 78% (MDA-MB-231) and 87% (PC3) | PC3 cells and MDA-MB-231 cells | |
| PBA-grafted PEDOT NanoVelcro chip | [107] | 0.5 M sorbitol solution | ~70% | >95% | >96% | LNCaP and PC3 cells |
3. Nanotechnology-Assisted CTC Analysis in Microfluidic Chips
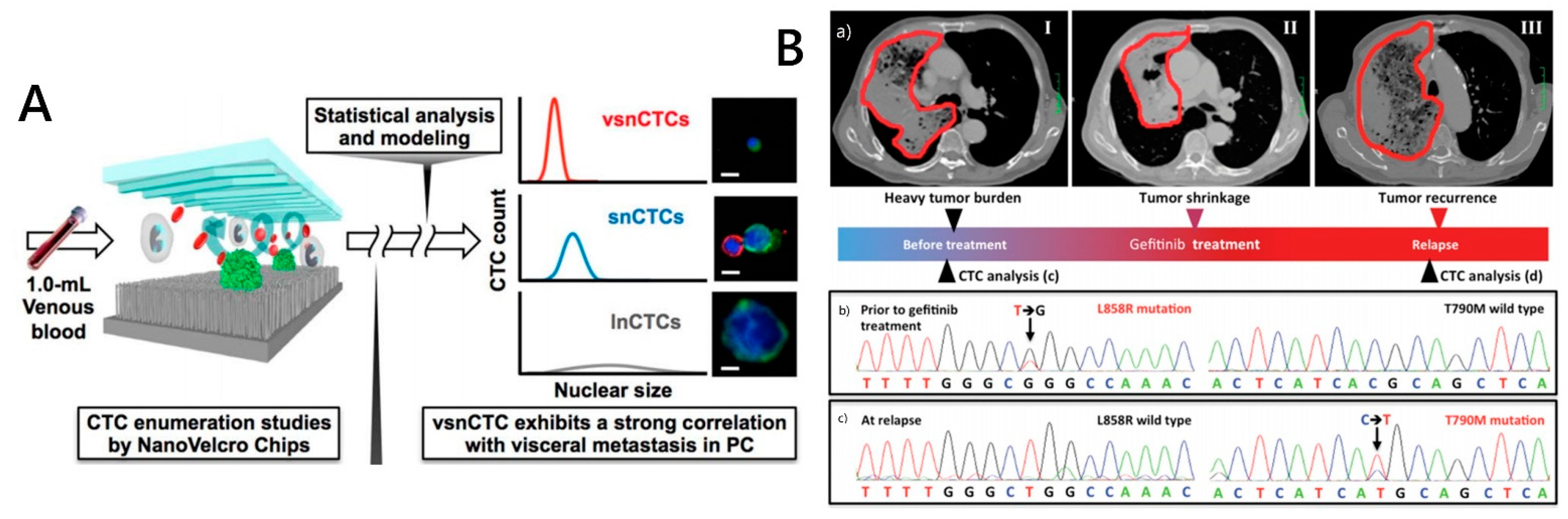
3.1. CTC Morphologic Analysis
3.2. CTC Genomic Analysis
3.3. CTC Transcriptomic Profiling
3.4. CTC Protein Analysis

3.5. CTC Functional Profiling
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Oskarsson, T.; Acharyya, S.; Nguyen, D.X.; Zhang, X.H.F.; Norton, L.; Massague, J. Tumor self-seeding by circulating cancer cells. Cell 2009, 139, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Aceto, N.; Bersani, F.; Madden, M.W.; Donaldson, M.C.; Desai, R.; Zhu, H.; Comaills, V.; Zheng, Z.; et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [Google Scholar] [CrossRef]
- Plaks, V.; Koopman, C.D.; Werb, Z. Circulating Tumor Cells. Science 2013, 341, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Jin, F.; Zhou, M.; Jiang, Y. Recent advances in single cell manipulation and biochemical analysis on microfluidics. Analyst 2019, 144, 766–781. [Google Scholar] [CrossRef]
- Krebs, M.G.; Hou, J.M.; Sloane, R.; Lancashire, L.; Priest, L.; Nonaka, D.; Ward, T.H.; Backen, A.; Clack, G.; Hughes, A.; et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J. Thorac. Oncol. 2012, 7, 306–315. [Google Scholar] [CrossRef]
- Dotan, E.; Cohen, S.J.; Alpaugh, K.R.; Meropol, N.J. Circulating tumor cells: Evolving evidence and future challenges. Oncologist 2009, 14, 1070–1082. [Google Scholar] [CrossRef]
- Stone, H.A.; Stroock, A.D.; Ajdari, A. Engineering flows in small devices: Microfluidics toward a lab-on-a-chip. Ann. Rev. Fluid Mech. 2004, 36, 381–411. [Google Scholar] [CrossRef]
- Dressler, O.J.; Casadevall i Solvas, X.; deMello, A.J. Chemical and biological dynamics using droplet-based microfluidics. Annu. Rev. Anal. Chem. 2017, 10, 1–24. [Google Scholar] [CrossRef]
- Zheng, W.; Jiang, X. Synthesizing living tissues with microfluidics. Acc. Chem. Res. 2018, 51, 3166–3173. [Google Scholar] [CrossRef]
- Hao, S.J.; Wan, Y.; Xia, Y.Q.; Zou, X.; Zheng, S.Y. Size-based separation methods of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Cheung, L.S.L.; Zheng, X.; Stopa, A.; Baygents, J.C.; Guzman, R.; Schroeder, J.A.; Heimark, R.L.; Zohar, Y. Detachment of captured cancer cells under flow acceleration in a bio-functionalized microchannel. Lab Chip 2009, 9, 1721–1731. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.W.; Warkiani, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.; Lim, W.T.; Han, J.; Bhagat, A.A.; Lim, C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013, 3, 1259. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ozcelik, A.; Rufo, J.; Wang, Z.; Fang, R.; Huang, T.J. Acoustofluidic separation of cells and particles. Microsyst. Nanoeng. 2019, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Chen, Z.; Wu, M.; Zhang, Y. Nanomaterial-based microfluidic chips for the capture and detection of circulating tumor cells. Nanotheranostics 2017, 1, 389–402. [Google Scholar] [CrossRef]
- Dong, J.; Chen, J.F.; Smalley, M.; Zhao, M.; Ke, Z.; Zhu, Y.; Tseng, H.R. Nanostructured substrates for detection and characterization of circulating rare cells: From materials research to clinical applications. Adv. Mater. 2019, 32, 1903663. [Google Scholar] [CrossRef]
- Yu, L.; Ng, S.R.; Xu, Y.; Dong, H.; Wang, Y.J.; Li, C.M. Advances of lab-on-a-chip in isolation, detection and post-processing of circulating tumour cells. Lab Chip 2013, 13, 3163–3182. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.; Sun, Y. Microfluidic approaches for cancer cell detection, characterization, and separation. Lab Chip 2012, 12, 1753–1767. [Google Scholar] [CrossRef]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef]
- Shields, C.W., IV; Reyes, C.D.; López, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef]
- Kim, T.H.; Lim, M.; Park, J.; Oh, J.M.; Kim, H.; Jeong, H.; Lee, S.J.; Park, H.C.; Jung, S.; Kim, B.C.; et al. FAST: Size-Selective, Clog-Free Isolation of Rare Cancer Cells from Whole Blood at a Liquid–Liquid Interface. Anal. Chem. 2017, 89, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ye, X.; Ma, Z.; Xie, S.; Wang, W. High-throughput and clogging-free microfluidic filtration platform for on-chip cell separation from undiluted whole blood. Biomicrofluidics 2016, 10, 014118. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.S.; Ho, B.D.; Beech, J.P.; Tegenfeldt, J.O. Open channel deterministic lateral displacement for particle and cell sorting. Lab Chip 2017, 17, 3592–3600. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Ma, C.; Zhao, L.; Wang, Y.; Wang, J.C.; Xu, J.; Li, T.; Pang, L.; Wang, J. High-throughput rare cell separation from blood samples using steric hindrance and inertial microfluidics. Lab Chip 2014, 14, 2525–2538. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Guan, G.; Luan, K.B.; Lee, W.C.; Bhagat, A.A.; Chaudhuri, P.K.; Tan, D.S.; Lim, W.T.; Lee, S.C.; Chen, P.C.; et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip 2014, 14, 128–137. [Google Scholar] [CrossRef]
- Ren, L.; Yang, S.; Zhang, P.; Qu, Z.; Mao, Z.; Huang, P.H.; Chen, Y.; Wu, M.; Wang, L.; Li, P.; et al. Standing Surface Acoustic Wave (SSAW)-Based Fluorescence-Activated Cell Sorter. Small 2018, 14, e1801996. [Google Scholar] [CrossRef]
- Moon, H.S.; Kwon, K.; Kim, S.I.; Han, H.; Sohn, J.; Lee, S.; Jung, H.I. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 2011, 11, 1118–1125. [Google Scholar] [CrossRef]
- Went, P.T.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam protein expression in human carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; Der Spoel, P.V.; Elstrodt, F.; Schutte, M.; Martens, J.W.M.; Gratama, J.W.; Sleijfer, S.; Foekens, J.A. Anti-Epithelial Cell Adhesion Molecule Antibodies and the Detection of Circulating Normal-Like Breast Tumor Cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garciablanco, M.A. Circulating Tumor Cells from Patients with Advanced Prostate and Breast Cancer Display Both Epithelial and Mesenchymal Markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef]
- Coumans, F.A.; van Dalum, G.; Beck, M.; Terstappen, L.W. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS ONE 2013, 8, e61770. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Iqbal, S.M.; Wan, Y. Cell detachment: Post-isolation challenges. Biotechnol. Adv. 2013, 31, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xu, X.; Sharma, B.; Wang, W.; Qu, X.; Zhu, L.; Zhang, H.; Song, Y.; Yang, C. Beyond capture: Circulating tumor cell release and single-cell analysis. Small Methods 2019, 3, 1800544. [Google Scholar] [CrossRef]
- Eriksson, O. Method for cytological detection of cancer cells in blood. Cancer 1962, 15, 171–175. [Google Scholar] [CrossRef]
- Shapiro, H.M.; Schildkraut, E.R.; Curbelo, R.; Laird, C.W.; Turner, B.; Hirschfeld, T. Combined blood cell counting and classification with fluorochrome stains and flow instrumentation. J. Histochem. Cytochem. 1976, 24, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Harouaka, R.; Nisic, M.; Zheng, S. Circulating tumor cell enrichment based on physical properties. J. Lab. Autom. 2013, 18, 455–468. [Google Scholar] [CrossRef]
- Gertler, R.; Rosenberg, R.; Fuehrer, K.; Dahm, M.; Nekarda, H.; Siewert, J.R. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003, 162, 149–155. [Google Scholar]
- Shao, C.; Liao, C.P.; Hu, P.; Chu, C.Y.; Zhang, L.; Bui, M.H.T.; Ng, C.S.; Josephson, D.Y.; Knudsen, B.; Tighiouart, M.; et al. Detection of live circulating tumor cells by a class of near-infrared heptamethine carbocyanine dyes in patients with localized and metastatic prostate cancer. PLoS ONE 2014, 9, e88967. [Google Scholar] [CrossRef]
- Ligthart, S.T.; Coumans, F.A.W.; Bidard, F.C.; Simkens, L.H.J.; Punt, C.J.A.; Groot, M.R.D.; Attard, G.; Bono, J.S.D.; Pierga, J.Y.; Terstappen, L. Circulating tumor cells count and morphological features in breast, colorectal and prostate cancer. PLoS ONE 2013, 8, e67148. [Google Scholar] [CrossRef]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.; Schutze, K.; Capron, F.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by Size of Epithelial Tumor Cells: A New Method for the Immunomorphological and Molecular Characterization of Circulating Tumor Cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef]
- Xu, W.W.; Mezencev, R.; Kim, B.; Wang, L.J.; McDonald, J.; Sulchek, T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS ONE 2012, 7, e46609. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Yang, L.; Frazier, A.B. Quantification of the heterogeneity in breast cancer cell lines using whole-cell impedance spectroscopy. Clin. Cancer Res. 2007, 13, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Izabela, D.; Elzbieta, S.; Zbigniew, A.F. Changes in Electric Properties of Human Breast Cancer Cells. J. Membr. Biol. 2013, 246, 161–166. [Google Scholar]
- Zhao, Y.; Zhao, X.T.; Chen, D.Y.; Luo, Y.N.; Jiang, M.; Wei, C.; Long, R.; Yue, W.T.; Wang, J.B.; Chen, J. Tumor cell characterization and classification based on cellular specific membrane capacitance and cytoplasm conductivity. Biosens. Bioelectron. 2014, 57, 245–253. [Google Scholar] [CrossRef]
- Onstenk, W.; Gratama, J.W.; Foekens, J.A.; Sleijfer, S. Towards a personalized breast cancer treatment approach guided by circulating tumor cell (CTC) characteristics. Cancer Treat. Rev. 2013, 39, 691–700. [Google Scholar] [CrossRef]
- Gourdin, T.; Sonpavde, G. Utility of cell-free nucleic acid and circulating tumor cell analyses in prostate cancer. Asian J. Androl. 2018, 20, 230–237. [Google Scholar]
- Cho, H.; Kim, J.; Song, H.; Sohn, K.Y.; Jeon, M.; Han, K.H. Microfluidic technologies for circulating tumor cell isolation. Analyst 2018, 143, 2936–2970. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kozminsky, M.; Nagrath, S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano 2014, 8, 1995–2017. [Google Scholar] [CrossRef]
- Kim, D.J.; Seol, J.K.; Lee, G.; Kim, G.S.; Lee, S.K. Cell adhesion and migration on nanopatterned substrates and their effects on cell-capture yield. Nanotechnology 2012, 23, 395102. [Google Scholar] [CrossRef]
- Wang, L.; Asghar, W.; Demirci, U.; Wan, Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today 2013, 8, 347–387. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Reategui, E.; Li, W.; Tessier, S.N.; Wong, K.H.; Jensen, A.E.; Thapar, V.; Ting, D.; Toner, M.; Stott, S.L.; et al. Enhanced Isolation and Release of Circulating Tumor Cells Using Nanoparticle Binding and Ligand Exchange in a Microfluidic Chip. J. Am. Chem. Soc. 2017, 139, 2741–2749. [Google Scholar] [CrossRef]
- Song, Y.; Shi, Y.; Huang, M.; Wang, W.; Wang, Y.; Cheng, J.; Lei, Z.; Zhu, Z.; Yang, C. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. Engl. 2018, 58, 2236–2240. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Yang, H.; Peng, C.; Zhu, H.; Mei, J.; Huang, S.; Chen, B.; Liu, J.; Wu, W.; Cao, S. Capture and biological release of circulating tumor cells in pancreatic cancer based on peptide-functionalized silicon nanowire substrate. Int. J. Nanomed. 2018, 14, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Tan, Y.; Xu, T.; Yin, D.; Wang, M.; Shen, M.; Chen, X.; Shi, X.; Zhu, X. Hyaluronic acid-functionalized electrospun PLGA nanofibers embedded in a microfluidic chip for cancer cell capture and culture. Biomater. Sci. 2017, 5, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, B.; Zhang, N.; Yin, C.; Chen, H.; Zhang, L.; Cai, B.; He, Z.; Rao, L.; Liu, W.; et al. Capture and Release of Cancer Cells by Combining On-Chip Purification and Off-Chip Enzymatic Treatment. ACS Appl. Mater. Interfaces 2015, 7, 24001–24007. [Google Scholar] [CrossRef]
- Lee, H.J.; Oh, J.H.; Oh, J.M.; Park, J.M.; Lee, J.G.; Kim, M.S.; Kim, Y.J.; Kang, H.J.; Jeong, J.; Kim, S.I.; et al. Efficient Isolation and Accurate In Situ Analysis of Circulating Tumor Cells Using Detachable Beads and a High-Pore-Density Filter. Angew. Chem. Int. Ed. Engl. 2013, 52, 8337–8340. [Google Scholar] [CrossRef]
- Cheng, S.B.; Xie, M.; Chen, Y.; Xiong, J.; Liu, Y.; Chen, Z.; Guo, S.; Shu, Y.; Wang, M.; Yuan, B.; et al. Three-Dimensional Scaffold Chip with Thermosensitive Coating for Capture and Reversible Release of Individual and Cluster of Circulating Tumor Cells. Anal. Chem. 2017, 89, 7924–7932. [Google Scholar] [CrossRef]
- Yan, S.; Chen, P.; Zeng, X.; Zhang, X.; Li, Y.; Xia, Y.; Dai, X.; Wang, J.; Feng, X.; Du, W.; et al. Integrated Multifunctional Electrochemistry Microchip for Highly Efficient Capture, Release, Lysis and Analysis of Circulating Tumor Cells. Anal. Chem. 2017, 89, 12039–12044. [Google Scholar] [CrossRef]
- Naoe, M.; Ogawa, Y.; Morita, J.; Omori, K.; Takeshita, K.; Shichijyo, T.; Okumura, T.; Igarashi, A.; Yanaihara, A.; Iwamoto, S.; et al. Detection of circulating urothelial cancer cells in the blood using the CellSearch System. Cancer 2007, 109, 1439–1445. [Google Scholar] [CrossRef]
- Liu, X.; Wang, S. Three-dimensional nano-biointerface as a new platform for guiding cell fate. Chem. Soc. Rev. 2014, 43, 2385–2401. [Google Scholar] [CrossRef]
- Pankhurst, Q.A.; Connolly, J.S.; Jones, S.K.; Dobson, J.J. Topical review: Applications of magnetic nanoparticles in biomedicine. J. Phys. D Appl. Phys. 2003, 36, R167–R181. [Google Scholar] [CrossRef]
- Hoshino, K.; Huang, Y.Y.; Lane, N.; Huebschman, M.L.; Uhr, J.W.; Frenkel, E.P.; Zhang, X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 2011, 11, 3449–3457. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Huang, Y.Y.; Chen, P.; Hoshino, K.; Liu, H.; Frenkel, E.P.; Zhang, J.X.; Sokolov, K.V. Versatile immunomagnetic nanocarrier platform for capturing cancer cells. ACS Nano 2013, 7, 8816–8823. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Hoshino, K.; Chen, P.; Wu, C.H.; Lane, N.; Huebschman, M.; Liu, H.; Sokolov, K.; Uhr, J.W.; Frenkel, E.P.; et al. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed. Microdevices 2013, 15, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.A.; Lee, T.Y.; Lee, S.H.; Jung, H.I. Two-stage microfluidic chip for selective isolation of circulating tumor cells (CTCs). Biosens. Bioelectron. 2015, 67, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.Y.; Hyun, K.A.; Jung, H.I. An integrated microfluidic chip for one-step isolation of circulating tumor cells. Sens. Actuators B Chem. 2017, 238, 1144–1150. [Google Scholar] [CrossRef]
- Shi, W.; Wang, S.; Maarouf, A.; Uhl, C.G.; He, R.; Yunus, D.E.; Liu, Y. Magnetic particles assisted capture and release of rare circulating tumor cells using wavy-herringbone structured microfluidic devices. Lab Chip 2017, 17, 3291–3299. [Google Scholar] [CrossRef]
- Mohamadi, R.M.; Besant, J.D.; Mepham, A.; Green, B.J.; Mahmoudian, L.; Gibbs, T.; Ivanov, I.; Malvea, A.; Stojcic, J.; Allan, A.L.; et al. Nanoparticle-Mediated Binning and Profiling of Heterogeneous Circulating Tumor Cell Subpopulations. Angew. Chem. Int. Ed. Engl. 2015, 54, 139–143. [Google Scholar] [CrossRef]
- Poudineh, M.; Labib, M.; Ahmed, S.; Nguyen, L.N.; Kermanshah, L.; Mohamadi, R.M.; Sargent, E.H.; Kelley, S.O. Profiling Functional and Biochemical Phenotypes of Circulating Tumor Cells Using a Two-Dimensional Sorting Device. Angew. Chem. Int. Ed. Engl. 2017, 56, 163–168. [Google Scholar] [CrossRef]
- Poudineh, M.; Aldridge, P.M.; Ahmed, S.; Green, B.J.; Kermanshah, L.; Nguyen, V.; Tu, C.; Mohamadi, R.M.; Nam, R.K.; Hansen, A.; et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 2017, 12, 274–281. [Google Scholar] [PubMed]
- Besant, J.D.; Mohamadi, R.M.; Aldridge, P.M.; Li, Y.; Sargent, E.H.; Kelley, S.O. Velocity valleys enable efficient capture and spatial sorting of nanoparticle-bound cancer cells. Nanoscale 2015, 7, 6278–6285. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Dong, Z.; Andarge, H.; Li, W.; Pappas, D. Nanoparticle modification of microfluidic cell separation for cancer cell detection and isolation. Analyst 2019, 145, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Chen, J.F.; Lu, Y.T.; Zhang, Y.; Song, J.; Hou, S.; Ke, Z.; Tseng, H.R. Nanostructure Embedded Microchips for Detection, Isolation, and Characterization of Circulating Tumor Cells. Acc. Chem. Res. 2014, 47, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Lee, G.; Kim, G.S.; Lee, S.K. Statistical analysis of immuno-functionalized tumor-cell behaviors on nanopatterned substrates. Nanoscale Res. Lett. 2012, 7, 1–8. [Google Scholar] [CrossRef]
- Qi, S.; Yi, C.; Ji, S.; Fong, C.C.; Yang, M. Cell Adhesion and Spreading Behavior on Vertically Aligned Silicon Nanowire Arrays. ACS Appl. Mater. Interfaces 2009, 1, 30–34. [Google Scholar] [CrossRef]
- Wang, S.; Liu, K.; Liu, J.; Yu, Z.T.; Xu, X.; Zhao, L.; Lee, T.; Lee, E.K.; Reiss, J.; Lee, Y.K.; et al. Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers. Angew. Chem. Int. Ed. Engl. 2011, 50, 3084–3088. [Google Scholar] [CrossRef]
- Shen, Q.; Xu, L.; Zhao, L.; Wu, D.; Fan, Y.; Zhou, Y.; Ouyang, W.H.; Xu, X.; Zhang, Z.; Song, M.; et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv Mater. 2013, 25, 2368–2373. [Google Scholar] [CrossRef]
- Dong, J.; Jan, Y.J.; Cheng, J.; Zhang, R.Y.; Meng, M.; Smalley, M.; Chen, P.J.; Tang, X.; Tseng, P.; Bao, L.; et al. Covalent chemistry on nanostructured substrates enables noninvasive quantification of gene rearrangements in circulating tumor cells. Sci. Adv. 2019, 5, eaav9186. [Google Scholar] [CrossRef]
- Sun, B.; Long, Y.Z.; Zhang, H.D.; Li, M.M.; Duvail, J.L.; Jiang, X.Y.; Yin, H.L. Advances in three-dimensional nanofibrous macrostructures via electrospinning. Prog. Polym. Sci. 2014, 39, 862–890. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Cai, B.; You, S.; He, Z.; Huang, Q.; Rao, L.; Li, S.; Liu, C.; Sun, W.; et al. Capture and release of cancer cells using electrospun etchable MnO2 nanofibers integrated in microchannels. Appl. Phys. Lett. 2015, 106, 093703. [Google Scholar] [CrossRef]
- Kozminsky, M.; Fouladdel, S.; Chung, J.S.; Wang, Y.; Smith, D.C.; Alva, A.; Azizi, E.; Morgan, T.; Nagrath, S. Detection of CTC Clusters and a Dedifferentiated RNA-Expression Survival Signature in Prostate Cancer. Adv. Sci. 2018, 6, 1801254. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [Google Scholar] [PubMed]
- He, G.; Yang, C.; Feng, J.; Wu, J.; Zhou, L.; Wen, R.; Huang, S.; Wu, Q.; Liu, F.; Chen, H.J.; et al. Hierarchical Spiky Microstraws-Integrated Microfluidic Device for Efficient Capture and In Situ Manipulation of Cancer Cells. Adv. Funct. Mater. 2019, 29, 1806484. [Google Scholar] [CrossRef]
- He, G.; Feng, J.; Zhang, A.; Zhou, L.; Wen, R.; Wu, J.; Yang, C.; Yang, J.; Li, C.; Chen, D.; et al. Multifunctional Branched Nanostraw-Electroporation Platform for Intracellular Regulation and Monitoring of Circulating Tumor Cells. Nano Lett. 2019, 19, 7201–7209. [Google Scholar] [CrossRef]
- Hui, L.; Su, Y.; Ye, T.; Liu, Z.; Tian, Q.; He, C.; Zhao, Y.; Chen, P.; Wang, X.; Han, W.; et al. Self-Sterilizing and Regeneratable Microchip for the Precise Capture and Recovery of Viable Circulating Tumor Cells from Patients with Cancer. ACS Appl. Mater. Interfaces 2018, 10, 207–218. [Google Scholar] [CrossRef]
- Abu-Reesh, I.; Kargi, F. Biological responses of hybridoma cells to defined hydrodynamic shear stress. J. Biotechnol. 1989, 9, 167–178. [Google Scholar] [CrossRef]
- Lu, H.; Koo, L.Y.; Wang, W.M.; Lauffenburger, D.A.; Griffith, L.G.; Jensen, K.F. Microfluidic shear devices for quantitative analysis of cell adhesion. Anal. Chem. 2004, 76, 5257–5264. [Google Scholar] [CrossRef]
- Kwon, K.W.; Choi, S.S.; Lee, S.H.; Kim, B.; Lee, S.N.; Park, M.C.; Kim, P.; Hwang, S.Y.; Suh, K.Y. Label-free, microfluidic separation and enrichment of human breast cancer cells by adhesion difference. Lab Chip 2007, 7, 1461–1468. [Google Scholar] [CrossRef]
- Chowdhury, F.; Na, S.; Li, D.; Poh, Y.C.; Tanaka, T.S.; Wang, F.; Wang, N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010, 9, 82–88. [Google Scholar] [CrossRef]
- Dharmasiri, U.; Njoroge, S.K.; Witek, M.A.; Adebiyi, M.G.; Kamande, J.W.; Hupert, M.L.; Barany, F.; Soper, S.A. High-Throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal. Chem. 2011, 83, 2301–2309. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Gao, F.P.; Li, L.L.; Ma, H.L.; Fan, Y.S.; Liu, W.; Guo, S.S.; Zhao, X.Z.; Wang, H. Gelatin-mesoporous silica nanoparticles as matrix metalloproteinases-degradable drug delivery systems in vivo. Micropor. Mesopor. Mater. 2013, 182, 165–172. [Google Scholar] [CrossRef]
- Shah, A.M.; Yu, M.; Nakamura, Z.; Ciciliano, J.; Ulman, M.; Kotz, K.; Stott, S.L.; Maheswaran, S.; Haber, D.A.; Toner, M. Biopolymer system for cell recovery from microfluidic cell capture devices. Anal. Chem. 2012, 84, 3682–3688. [Google Scholar] [CrossRef] [PubMed]
- Luan, C.; Wang, H.; Han, Q.; Ma, X.; Zhang, D.; Xu, Y.; Chen, B.; Li, M.; Zhao, Y. Folic Acid-Functionalized Hybrid Photonic Barcodes for Capture and Release of Circulating Tumor Cells. ACS Appl. Mater. Interfaces 2018, 10, 21206–21212. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Cai, B.; Chen, B.; Rao, L.; He, Z.; He, R.; Guo, F.; Zhao, L.; Kondamareddy, K.K.; Liu, W.; et al. Efficient purification and release of circulating tumor cells by synergistic effect of biomarker and SiO2 @gel-microbead-based size difference amplification. Adv. Healthc. Mater. 2016, 5, 1554–1559. [Google Scholar] [CrossRef]
- Lv, S.W.; Wang, J.; Xie, M.; Lu, N.N.; Li, Z.; Yan, X.W.; Cai, S.L.; Zhang, P.A.; Dong, W.G.; Huang, W.H. Photoresponsive immunomagnetic nanocarrier for capture and release of rare circulating tumor cells. Chem. Sci. 2015, 6, 6432–6438. [Google Scholar] [CrossRef]
- Bae, Y.H.; Okano, T.; Kim, S.W. Temperature dependence of swelling of crosslinked poly(N,N′-alkyl substituted acrylamides) in water. J. Polym. Sci. Part B Polym. Phys. 1990, 28, 923–936. [Google Scholar] [CrossRef]
- Liu, H.; Liu, X.; Meng, J.; Zhang, P.; Yang, G.; Su, B.; Sun, K.; Chen, L.; Han, D.; Wang, S.; et al. Hydrophobic Interaction-Mediated Capture and Release of Cancer Cells on Thermoresponsive Nanostructured Surfaces. Adv. Mater. 2013, 25, 922–927. [Google Scholar] [CrossRef]
- Lv, S.W.; Liu, Y.; Xie, M.; Wang, J.; Yan, X.W.; Li, Z.; Dong, W.G.; Huang, W.H. Near-Infrared Light-Responsive Hydrogel for Specific Recognition and Photothermal Site-Release of Circulating Tumor Cells. ACS Nano 2016, 10, 6201–6210. [Google Scholar] [CrossRef]
- Zhu, J.; Nguyen, T.; Pei, R.; Stojanovic, M.; Lin, Q. Specific capture and temperature-mediated release of cells in an aptamer-based microfluidic device. Lab Chip 2012, 12, 3504–3513. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, B.; Ji, H.; Han, D.; Chen, S.; Tian, F.; Jiang, X. A Method for Patterning Multiple Types of Cells by Using Electrochemical Desorption of Self-Assembled Monolayers within Microfluidic Channels. Angew. Chem. Int. Ed. 2007, 46, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Moon, J.M.; Lee, E.S.; Kim, Y.H.; Cho, Y. An Electroactive Biotin-Doped Polypyrrole Substrate That Immobilizes and Releases EpCAM-Positive Cancer Cells. Angew. Chem. Int. Ed. Engl. 2014, 53, 4597–4602. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.S.; Ho, B.C.; Yan, H.X.; Kuo, C.W.; Chueh, D.Y.; Yu, H.; Chen, P. Integrated 3D Conducting Polymer-Based Bioelectronics for Capture and Release of Circulating Tumor Cells. J. Mater. Chem. 2015, 3, 5103–5110. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, F.F.; Liu, H.Q.; Wang, Z.X.; Zhang, Z.T.; Wang, Y.; Cui, Y.; Liu, W.; Zhao, X.Z.; Sun, Z.J.; et al. Efficient Capture and High Activity Release of Circulating Tumor Cells by Using TiO2 Nanorod Arrays Coated with Soluble MnO2 Nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 16327–16334. [Google Scholar] [CrossRef]
- Guo, S.; Xu, J.; Xie, M.; Huang, W.; Yuan, E.; Liu, Y.; Fan, L.; Cheng, S.B.; Liu, S.; Wang, F.; et al. Degradable Zinc-Phosphate-Based Hierarchical Nanosubstrates for Capture and Release of Circulating Tumor Cells. ACS Appl. Mater. Interfaces 2016, 8, 15917–15925. [Google Scholar] [CrossRef]
- Xie, M.; Lu, N.N.; Cheng, S.B.; Wang, X.Y.; Wang, M.; Guo, S.; Wen, C.Y.; Hu, J.; Pang, D.W.; Huang, W.H. Engineered Decomposable Multifunctional Nanobioprobes for Capture and Release of Rare Cancer Cells. Anal. Chem. 2014, 86, 4618–4626. [Google Scholar] [CrossRef]
- Shen, M.Y.; Chen, J.F.; Luo, C.H.; Lee, S.; Li, C.H.; Yang, Y.L.; Tsai, Y.H.; Ho, B.C.; Bao, L.R.; Lee, T.J.; et al. Glycan Stimulation Enables Purification of Prostate Cancer Circulating Tumor Cells on PEDOT NanoVelcro Chips for RNA Biomarker Detection. Adv. Healthc. Mater. 2018, 7, 1700701. [Google Scholar] [CrossRef]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Valen, D.V. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- Chen, J.F.; Ho, H.; Lichterman, J.; Lu, Y.T.; Zhang, Y.; Garcia, M.A.; Chen, S.F.; Liang, A.J.; Hodara, E.; Zhau, H.E.; et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer 2015, 121, 3240–3251. [Google Scholar] [CrossRef]
- Christiansen, E.M.; Yang, S.J.; Ando, D.M.; Javaherian, A.; Skibinski, G.; Lipnick, S.; Mount, E.; O’Neil, A.; Shah, K.; Lee, A.K.; et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell 2018, 173, 792–803. [Google Scholar] [CrossRef]
- Hou, S.; Zhao, L.; Shen, Q.; Yu, J.; Ng, C.; Kong, X.; Wu, D.; Song, M.; Shi, X.; Xu, X.; et al. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew. Chem. Int. Ed. Engl. 2013, 52, 3379–3383. [Google Scholar] [CrossRef] [PubMed]
- Court, C.M.; Ankeny, J.S.; Sho, S.; Hou, S.; Li, Q.; Hsieh, C.; Song, M.; Liao, X.; Rochefort, M.M.; Wainberg, Z.A.; et al. Reality of single circulating tumor cell sequencing for molecular diagnostics in pancreatic cancer. J. Mol. Diagn. 2016, 18, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, H.; Cui, Y.; Lei, Y.; Wang, Y.; Xu, D.; Jiang, N.; Chen, Y.; Sun, Y.; Zhang, Y.; et al. Polymer nanofiber-based microchips for EGFR mutation analysis of circulating tumor cells in lung adenocarcinoma. Int. J. Nanomed. 2018, 13, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Lin, M.; Chen, J.F.; Choi, J.S.; Zhang, Y.; Fong, A.; Liang, A.J.; Chen, S.F.; Li, Q.; Fang, W.; et al. Programming Thermoresponsiveness of NanoVelcro Substrates Enables Effective Purification of Circulating Tumor Cells in Lung Cancer Patients. ACS Nano 2015, 9, 62–71. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.C.; Behr, B.; Quake, S.R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 2012, 150, 402–412. [Google Scholar] [CrossRef]
- Wang, Y.; Waters, J.; Leung, M.L.; Unruh, A.; Roh, W.; Shi, X.; Chen, K.; Scheet, P.; Vattathil, S.; Liang, H.; et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 2014, 512, 155–160. [Google Scholar] [CrossRef]
- Jiang, R.; Lu, Y.T.; Ho, H.; Li, B.; Chen, J.F.; Lin, M.; Li, F.; Wu, K.; Wu, H.; Lichterman, J.; et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget 2015, 6, 44781–44793. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Barrette, T.R.; Ghosh, D.; Chinnaiyan, A.M. Mining for regulatory programs in the cancer transcriptome. Nat. Genet. 2005, 37, 579–583. [Google Scholar] [CrossRef]
- Park, S.M.; Wong, D.J.; Ooi, C.C.; Kurtz, D.M.; Vermesh, O.; Aalipour, A.; Suh, S.; Pian, K.L.; Chabon, J.J.; Lee, S.H.; et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc. Natl. Acad. Sci. USA 2016, 113, E8379–E8386. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Chen, Y.C.; Lin, E.; Brien, R.; Jung, S.; Chen, Y.T.; Lee, W.; Hao, Z.; Sahoo, S.; Kang, H.M.; et al. Hydro-Seq enables contamination-free highthroughput single-cell RNA-sequencing for circulating tumor cells. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Baek, S.; Green, R.; Granville, A.; Martens, P.; Poole-Warren, L. Thin film hydrophilic electroactive polymer coatings for bioelectrodes. J. Mater. Chem. B 2013, 1, 3803–3810. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.L.; Tang, M.; Zhang, Z.L.; Qi, C.B.; Hu, J.; Ma, X.Y.; Pang, D.W. Chip-assisted single-cell biomarker profiling of heterogeneous circulating tumor cells using multifunctional nanospheres. Anal. Chem. 2018, 90, 10518–10526. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Wu, L.; Zong, S.; Yun, B.; Cui, Y. Combining Multiplex SERS Nanovectors and Multivariate Analysis for In Situ Profiling of Circulating Tumor Cell Phenotype Using a Microfluidic Chip. Small 2018, 14, e1704433. [Google Scholar] [CrossRef] [PubMed]
- Law, W.C.; Yong, K.T.; Baev, A.; Prasad, P.N. Sensitivity improved surface plasmon resonance biosensor for cancer biomarker detection based on plasmonic enhancement. ACS Nano 2011, 5, 4858–4864. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Steeg, P.S. Metastasis suppressors alter the signal transduction of cancer cells. Nat. Rev. Cancer 2003, 3, 55–63. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Millon, S.R.; Ostrander, J.H.; Brown, J.Q.; Raheja, A.; Seewaldt, V.L.; Ramanujam, N. Uptake of 2-NBDG as a method to monitor therapy response in breast cancer cell lines. Breast Cancer Res. Treat. 2011, 126, 55–62. [Google Scholar] [CrossRef]
- Biersack, H.-J.; Bender, H.; Palmedo, H. FDG-PET in monitoring therapy of breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2004, 31, S112–S117. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, Y.; Sun, S.; Wang, Z.; Wu, W.; Zhao, X.; Czajkowsky, D.M.; Li, Y.; Tian, J.; Xu, L.; et al. Single-cell codetection of metabolic activity, intracellular functional proteins, and genetic mutations from rare circulating tumor cells. Anal. Chem. 2015, 87, 9761–9768. [Google Scholar] [CrossRef]
- Green, B.J.; Kermanshah, L.; Labib, M.; Ahmed, S.; Silva, P.N.; Mahmoudian, L.; Chang, I.; Mohamadi, R.M.; Rocheleau, J.V.; Kelley, S.O. Isolation of phenotypically distinct cancer cells using nanoparticle-mediated sorting. ACS Appl. Mater. Interfaces 2017, 9, 20435–20443. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Choudhury, A.D.; Yamanaka, Y.J.; Adalsteinsson, V.A.; Gierahn, T.M.; Williamson, C.A.; Lamb, C.R.; Taplin, M.E.; Nakabayashi, M.; Chabot, M.S.; et al. Functional analysis of single cells identifies a rare subset of circulating tumor cells with malignant traits. Integr. Biol. 2014, 6, 388–398. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Liu, Y.; Zhao, Y.; Zhang, L.; Zhang, L.; Mao, H.; Huang, C. Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices. Micromachines 2020, 11, 774. https://doi.org/10.3390/mi11080774
Cheng J, Liu Y, Zhao Y, Zhang L, Zhang L, Mao H, Huang C. Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices. Micromachines. 2020; 11(8):774. https://doi.org/10.3390/mi11080774
Chicago/Turabian StyleCheng, Jie, Yang Liu, Yang Zhao, Lina Zhang, Lingqian Zhang, Haiyang Mao, and Chengjun Huang. 2020. "Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices" Micromachines 11, no. 8: 774. https://doi.org/10.3390/mi11080774
APA StyleCheng, J., Liu, Y., Zhao, Y., Zhang, L., Zhang, L., Mao, H., & Huang, C. (2020). Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices. Micromachines, 11(8), 774. https://doi.org/10.3390/mi11080774







