Fabrication of Blood Capillary Models for Live Imaging Microarray Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Characterization of the Collagen Microfibers (CMFs)
2.2. Fabrication of the 3D-Blood Capillary Models in 48-Well Plate
2.3. Immunofluorescence Staining and Histological Analysis
2.4. Analysis of Blood Capillaries Formation
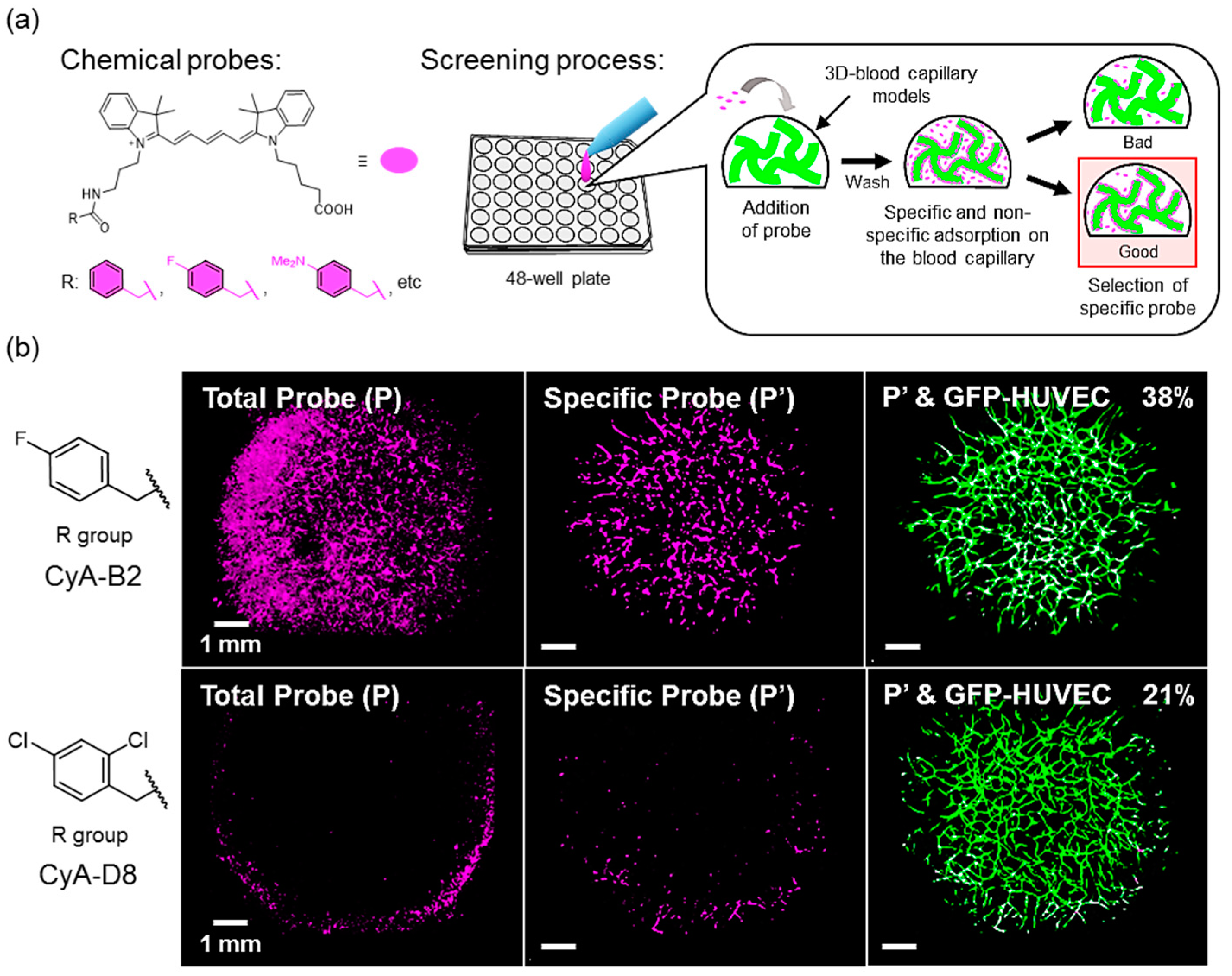
2.5. Application of Microarray Analysis for Screening of Chemical Probes
2.6. Statistical Analysis
3. Results and Discussion
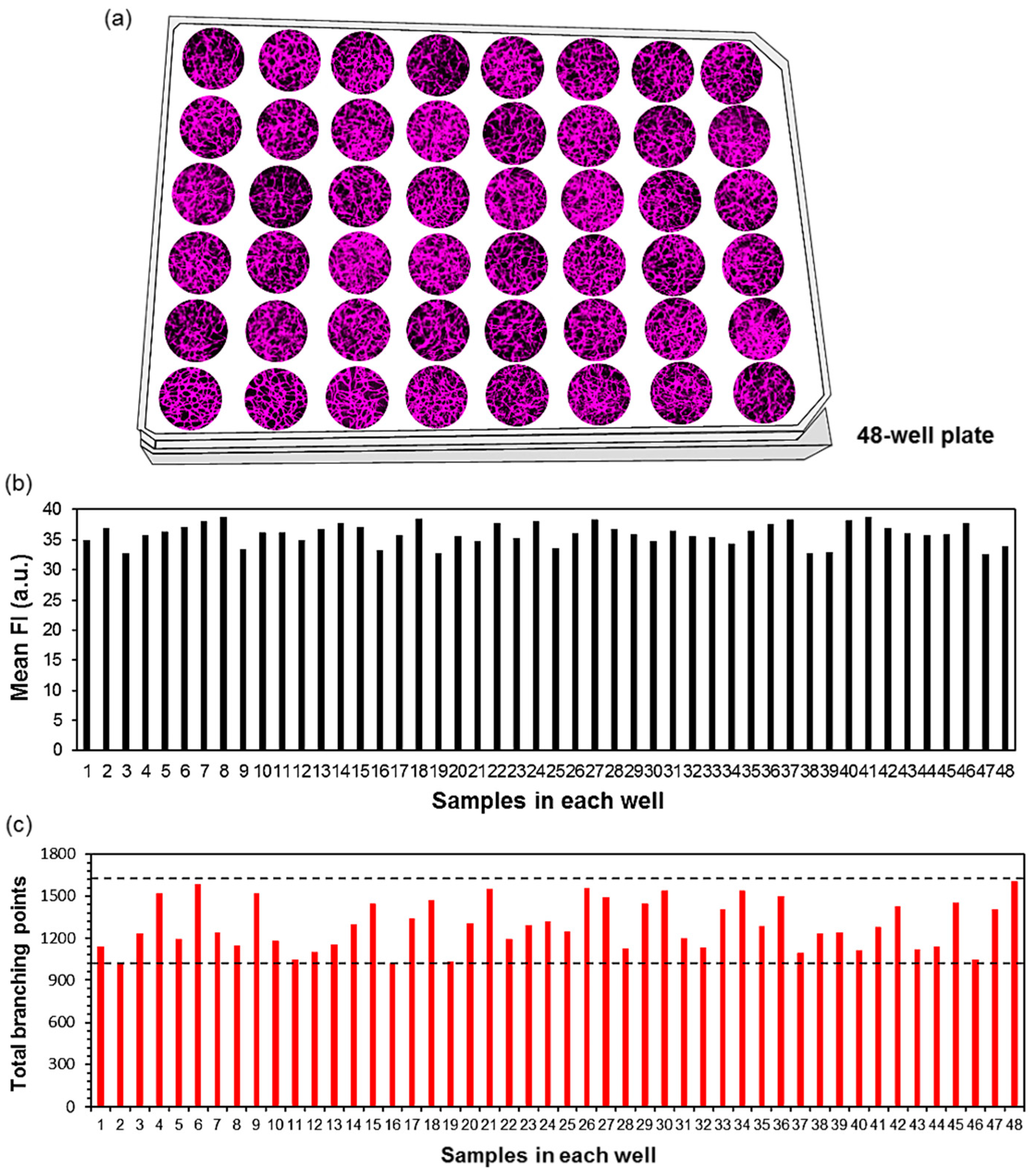
3.1. D-Blood Capillary Models in 48-Well Plate
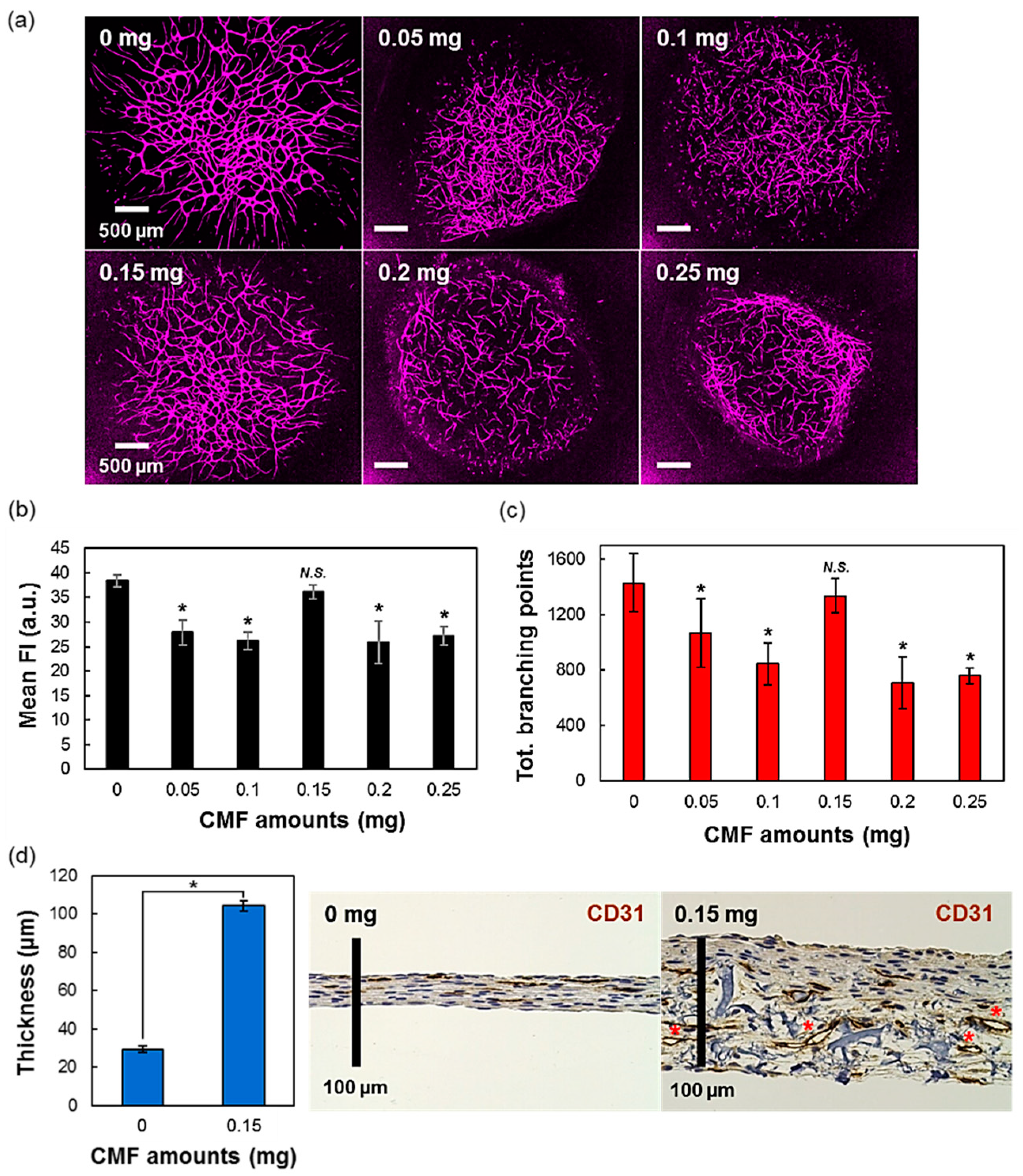
3.2. The Effect of Different Amounts of CMF in the 3D-Tissues
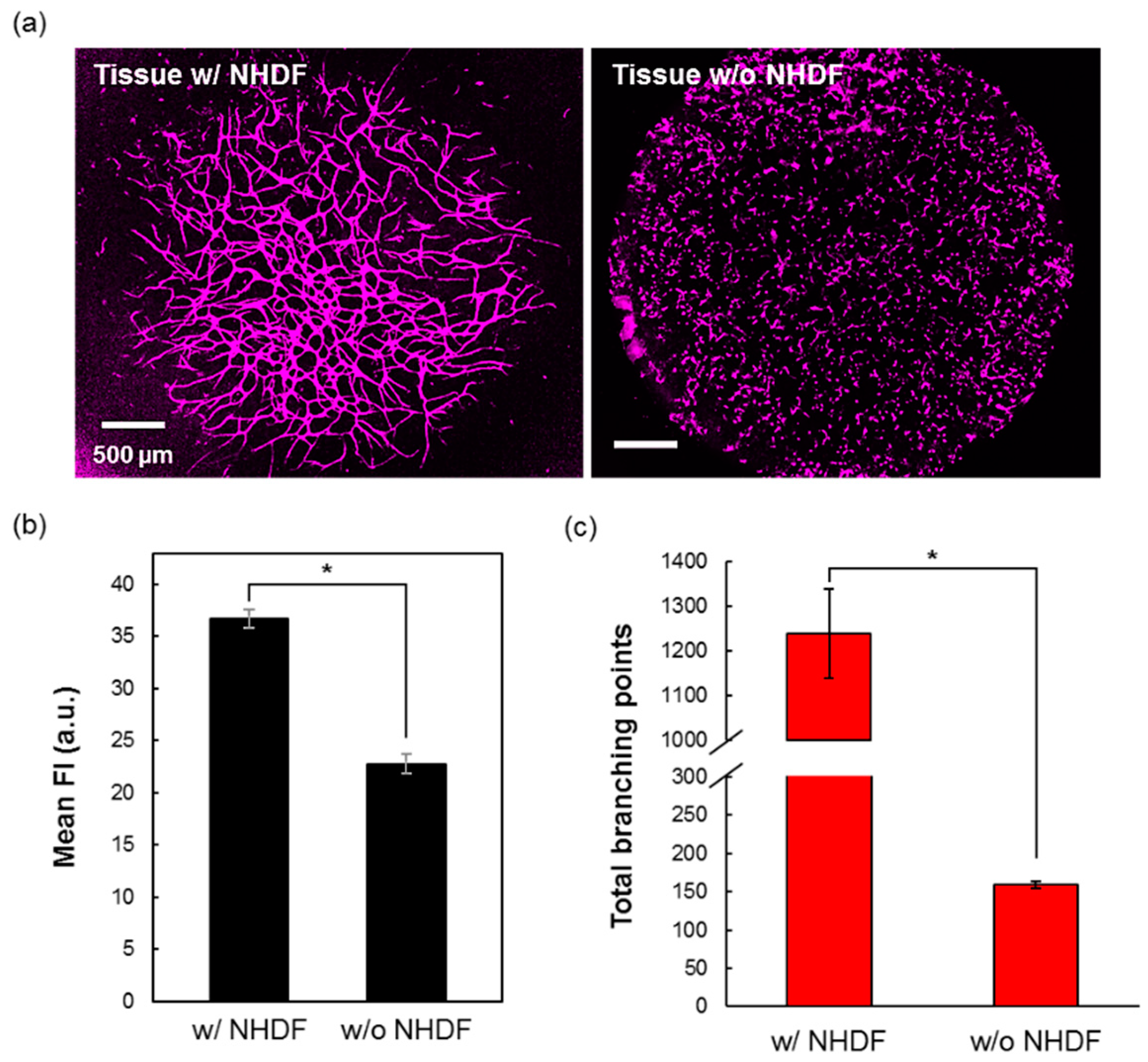
3.3. The Role of NHDF for the Vasculature Formation
3.4. Importance of the Cell Culture Medium Volume during the Culture
3.5. Comparison of Different Volumes of Seeded Drops of 3D-Blood Capillary Model
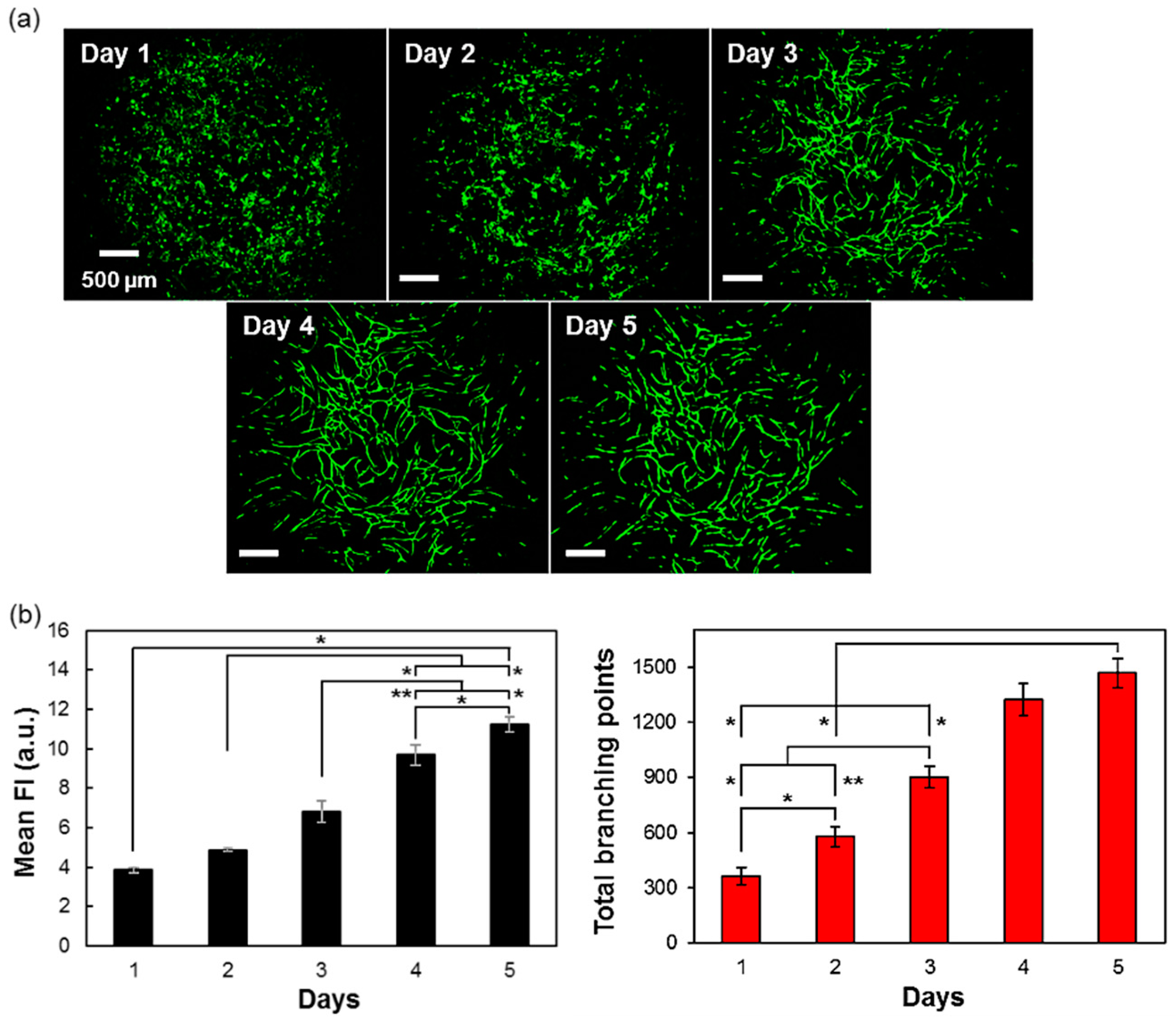
3.6. Time-Related Formation of Vascular Networks
3.7. Application of the Model in a Microarray Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Kola, I. The State of Innovation in Drug Development. Clin. Pharmacol. Ther. 2008, 83, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Bhadriraju, K.; Chen, C.S. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov. Today 2002, 7, 612–620. [Google Scholar] [CrossRef]
- Niklason, L.E.; Langer, R.S. Advances in tissue engineering of blood vessels and other tissues. Transpl. Immunol. 1997, 5, 303–306. [Google Scholar] [CrossRef]
- Sarker, M.D.; Naghieh, S.; Sharma, N.K.; Chen, X. 3D biofabrication of vascular networks for tissue regeneration: A report on recent advances. J. Pharm. Anal. 2018, 8, 277–296. [Google Scholar] [CrossRef]
- Abdul Sisak, M.A.; Louis, F.; Matsusaki, M. In vitro fabrication and application of engineered vascular hydrogels. Polym. J. 2020, 52, 871–881. [Google Scholar] [CrossRef]
- Tomasina, C.; Bodet, T.; Mota, C.; Moroni, L.; Camarero-Espinosa, S. Bioprinting vasculature: Materials, cells and emergent techniques. Materials (Basel). 2019, 12. [Google Scholar] [CrossRef]
- Ouyang, L.; Armstrong, J.P.K.; Chen, Q.; Lin, Y.; Stevens, M.M. Void-Free 3D Bioprinting for In Situ Endothelialization and Microfluidic Perfusion. Adv. Funct. Mater. 2020, 30. [Google Scholar] [CrossRef]
- Brown, A.; He, H.; Trumper, E.; Valdez, J.; Hammond, P.; Griffith, L.G. Engineering PEG-based hydrogels to foster efficient endothelial network formation in free-swelling and confined microenvironments. Biomaterials 2020, 243, 119921. [Google Scholar] [CrossRef]
- Tanaka, R.I.; Sakaguchi, K.; Umezu, S. Printing support hydrogels for creating vascular-like structures in stacked cell sheets. Artif. Life Robot. 2020, 25, 199–203. [Google Scholar] [CrossRef]
- Juliar, B.A.; Beamish, J.A.; Busch, M.E.; Cleveland, D.S.; Nimmagadda, L.; Putnam, A.J. Cell-mediated matrix stiffening accompanies capillary morphogenesis in ultra-soft amorphous hydrogels. Biomaterials 2020, 230. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.; Memic, A.; Annabi, N.; Hossain, M.; Paul, A.; Dokmeci, M.R.; Dehghani, F.; Khademhosseini, A. Electrospun scaffolds for tissue engineering of vascular grafts. Acta Biomater. 2014, 10, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Sakai, Y.; Fujii, T. Organ/body-on-a-chip based on micro fluidic technology for drug discovery. Drug Metab. Pharmacokinet. 2018, 33, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Yong, K.W.; Choi, J.Y.; Cowie, A.C. Recent advances in photo-crosslinkable hydrogels for biomedical applications. BioTechnique 2019, 66, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, Q.; Shi, X.; Han, D. Hierarchical Hydrogel Composite Interfaces with Robust Mechanical Properties for Biomedical Applications. Adv. Mater. 2019, 31, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef]
- Drury, J.L.; Mooney, D.J. Hydrogels for tissue engineering: Scaffold design variables and applications. Biomaterials 2003, 24, 4337–4351. [Google Scholar] [CrossRef]
- Litvinov, R.I.; Weisel, J.W. Fibrin mechanical properties and their structural origins. Matrix Biol. 2017, 60–61, 110–123. [Google Scholar] [CrossRef]
- Swartz, D.D.; Russell, J.A.; Andreadis, S.T. Engineering of fibrin-based functional and implantable small-diameter blood vessels. Am. J. Physiol. -Hear. Circ. Physiol. 2005, 288, 1451–1460. [Google Scholar] [CrossRef]
- Yao, L.; Liu, J.; Andreadis, S.T. Composite fibrin scaffolds increase mechanical strength and preserve contractility of tissue engineered blood vessels. Pharm. Res. 2008, 25, 1212–1221. [Google Scholar] [CrossRef]
- Eyrich, D.; Brandl, F.; Appel, B.; Wiese, H.; Maier, G.; Wenzel, M.; Staudenmaier, R.; Goepferich, A.; Blunk, T. Long-term stable fibrin gels for cartilage engineering. Biomaterials 2007, 28, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Swartz, D.D.; Gugino, S.F.; Russell, J.A.; Andreadis, S.T. Fibrin-based tissue-engineered blood vessels: Differential effects of biomaterial and culture parameters on mechanical strength and vascular reactivity. Tissue Eng. 2005, 11, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kitano, S.; Irie, S.; Levato, R.; Matsusaki, M. Collagen Microfibers Induce Blood Capillary Orientation and Open Vascular Lumen. Adv. Biosyst. 2020, 4, 2000038. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Teoh, C.L.; Park, S.-J.; Kim, J.-J.; Samanta, A.; Bi, R.; Dinish, U.S.; Olivo, M.; Piantino, M.; Louis, F.; et al. Seeing Elastin: A Near-Infrared Zwitterionic Fluorescent Probe for In Vivo Elastin Imaging. Chem 2018, 4, 1128–1138. [Google Scholar] [CrossRef]
- Louis, F.; Kitano, S.; Mano, J.F.; Matsusaki, M. 3D collagen microfibers stimulate the functionality of preadipocytes and maintain the phenotype of mature adipocytes for long term cultures. Acta Biomater. 2019, 84, 194–207. [Google Scholar] [CrossRef]
- Naka, Y.; Kitano, S.; Irie, S.; Matsusaki, M. Wholly vascularized millimeter-sized engineered tissues by cell-sized microscaffolds. Mater. Today Bio 2020, 6, 100054. [Google Scholar] [CrossRef]
- Abdul Sisak, M.A.; Louis, F.; Hyeok Lee, S.; Chang, Y.T.; Matsusaki, M. Unbiased Throughput Screening of A Near-Infrared Organic Fluorophore for In Vivo Endothelium Imaging via In Vitro Engineered 3D-Blood Capillaries. To be submitted.
- Richardson, D.S.; Lichtman, J.W. Clarifying Tissue Clearing. Cell 2015, 162, 246–257. [Google Scholar] [CrossRef]
- Kniebs, C.; Kreimendahl, F.; Köpf, M.; Fischer, H.; Jockenhoevel, S.; Thiebes, A.L. Influence of Different Cell Types and Sources on Pre-Vascularisation in Fibrin and Agarose-Collagen Gels. Organogenesis 2019, 16, 1–13. [Google Scholar] [CrossRef]
- Chwalek, K.; Tsurkan, M.V.; Freudenberg, U.; Werner, C. Glycosaminoglycan-based hydrogels to modulate heterocellular communication in in vitro angiogenesis models. Sci. Rep. 2014, 4, 4–11. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Sisak, M.A.; Louis, F.; Hyeok Lee, S.; Chang, Y.-T.; Matsusaki, M. Fabrication of Blood Capillary Models for Live Imaging Microarray Analysis. Micromachines 2020, 11, 727. https://doi.org/10.3390/mi11080727
Abdul Sisak MA, Louis F, Hyeok Lee S, Chang Y-T, Matsusaki M. Fabrication of Blood Capillary Models for Live Imaging Microarray Analysis. Micromachines. 2020; 11(8):727. https://doi.org/10.3390/mi11080727
Chicago/Turabian StyleAbdul Sisak, Muhammad Asri, Fiona Louis, Sun Hyeok Lee, Young-Tae Chang, and Michiya Matsusaki. 2020. "Fabrication of Blood Capillary Models for Live Imaging Microarray Analysis" Micromachines 11, no. 8: 727. https://doi.org/10.3390/mi11080727
APA StyleAbdul Sisak, M. A., Louis, F., Hyeok Lee, S., Chang, Y.-T., & Matsusaki, M. (2020). Fabrication of Blood Capillary Models for Live Imaging Microarray Analysis. Micromachines, 11(8), 727. https://doi.org/10.3390/mi11080727





