An Interphase Microfluidic Culture System for the Study of Ex Vivo Intestinal Tissue
Abstract
1. Introduction
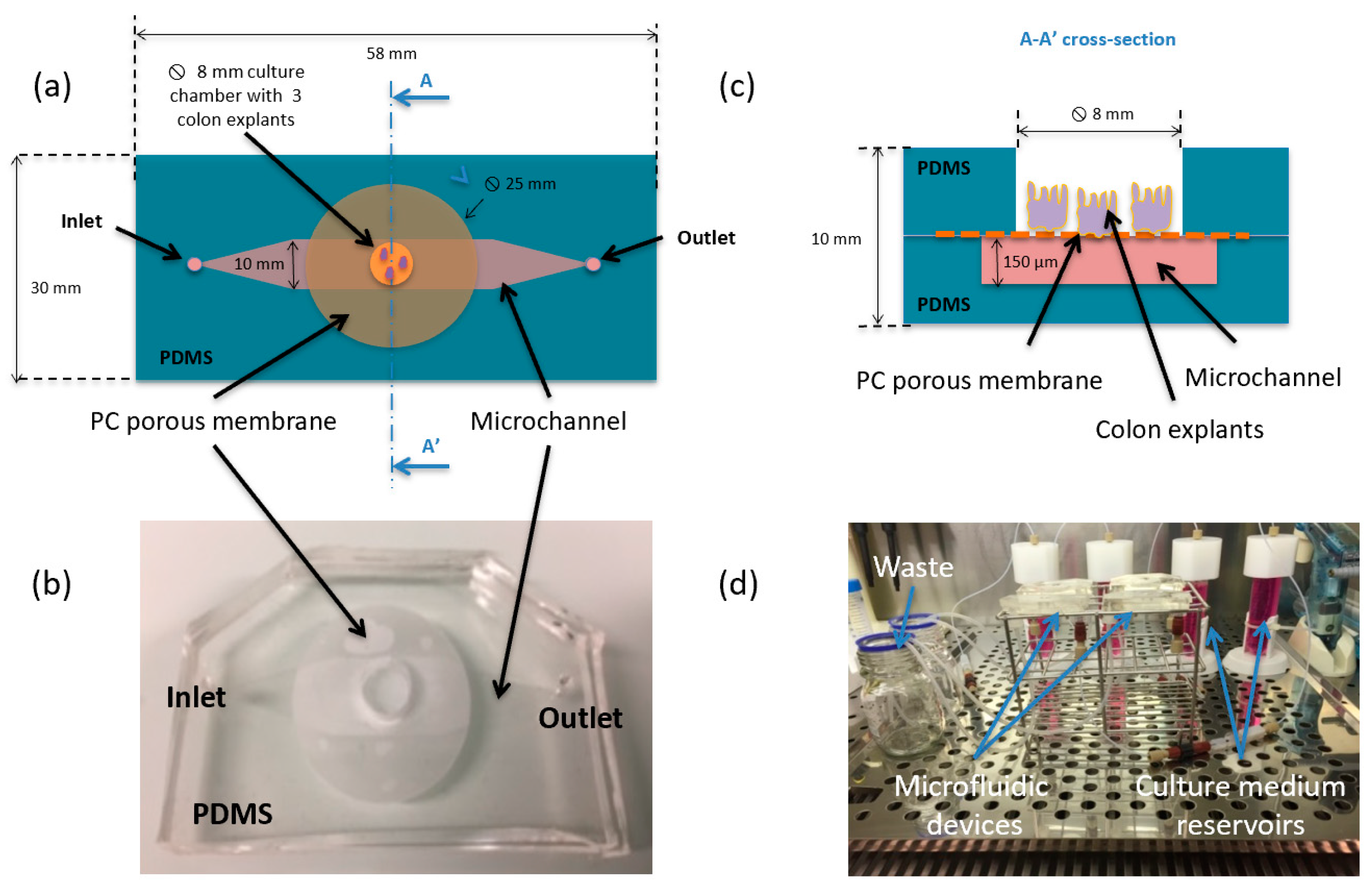
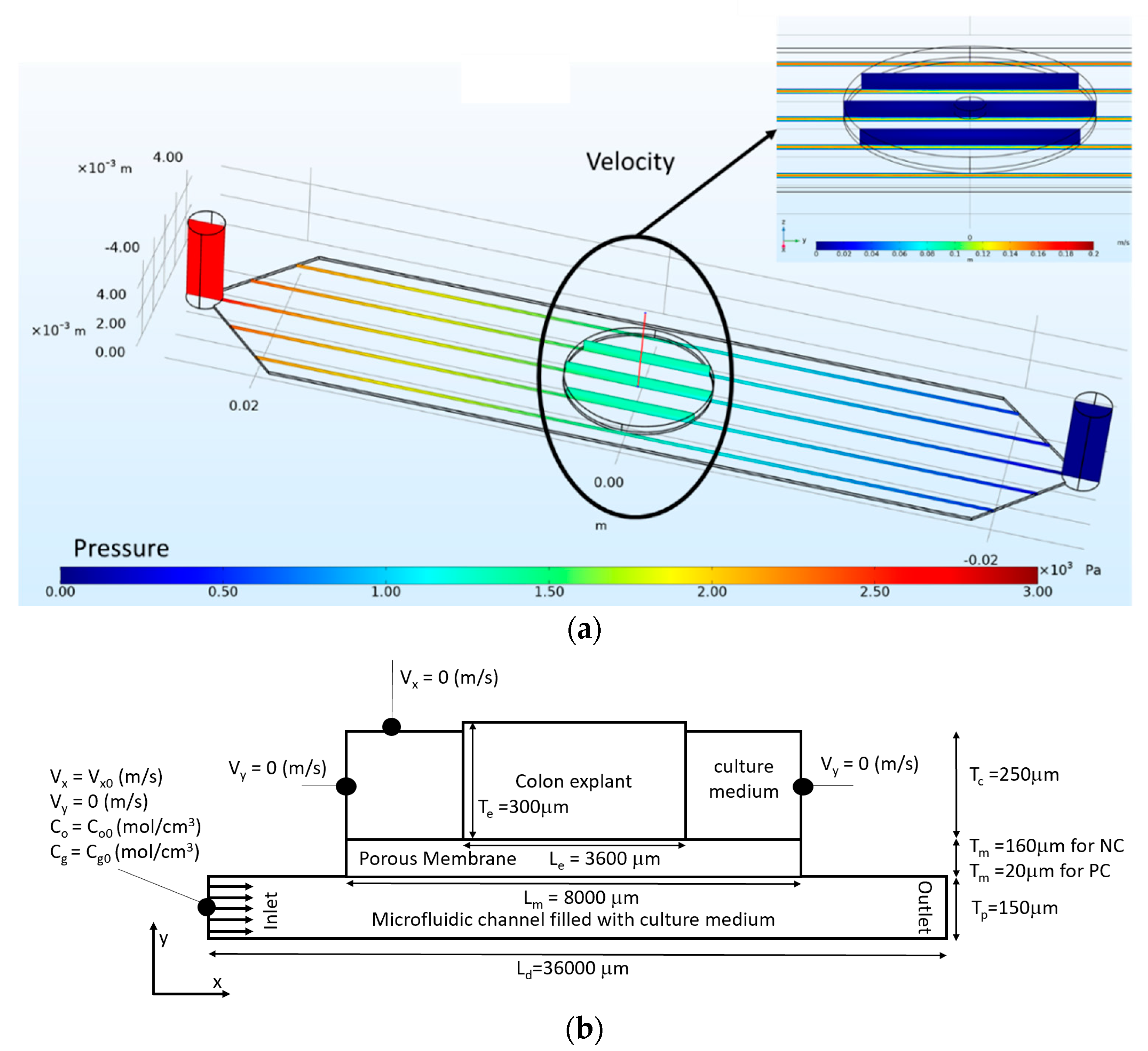
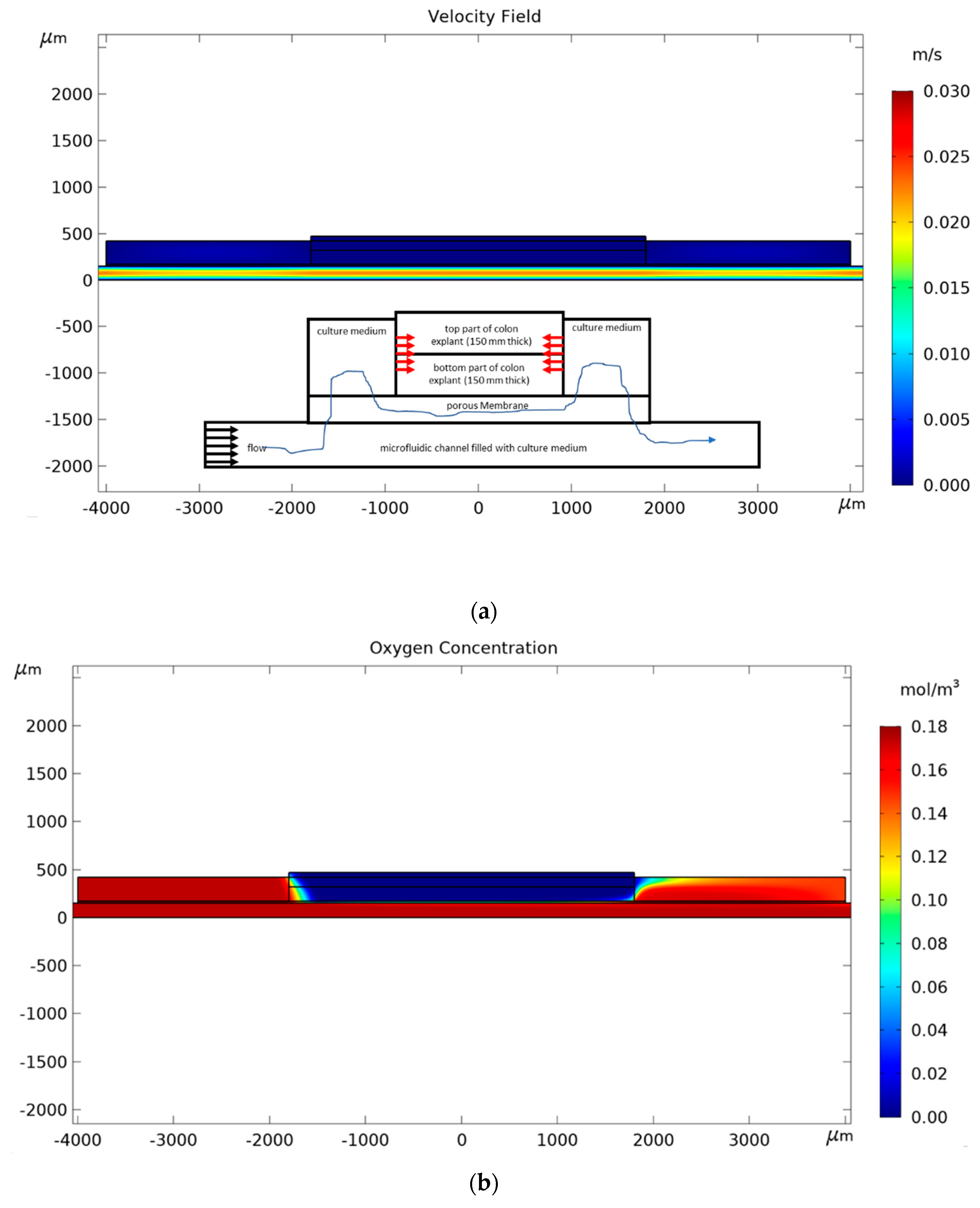
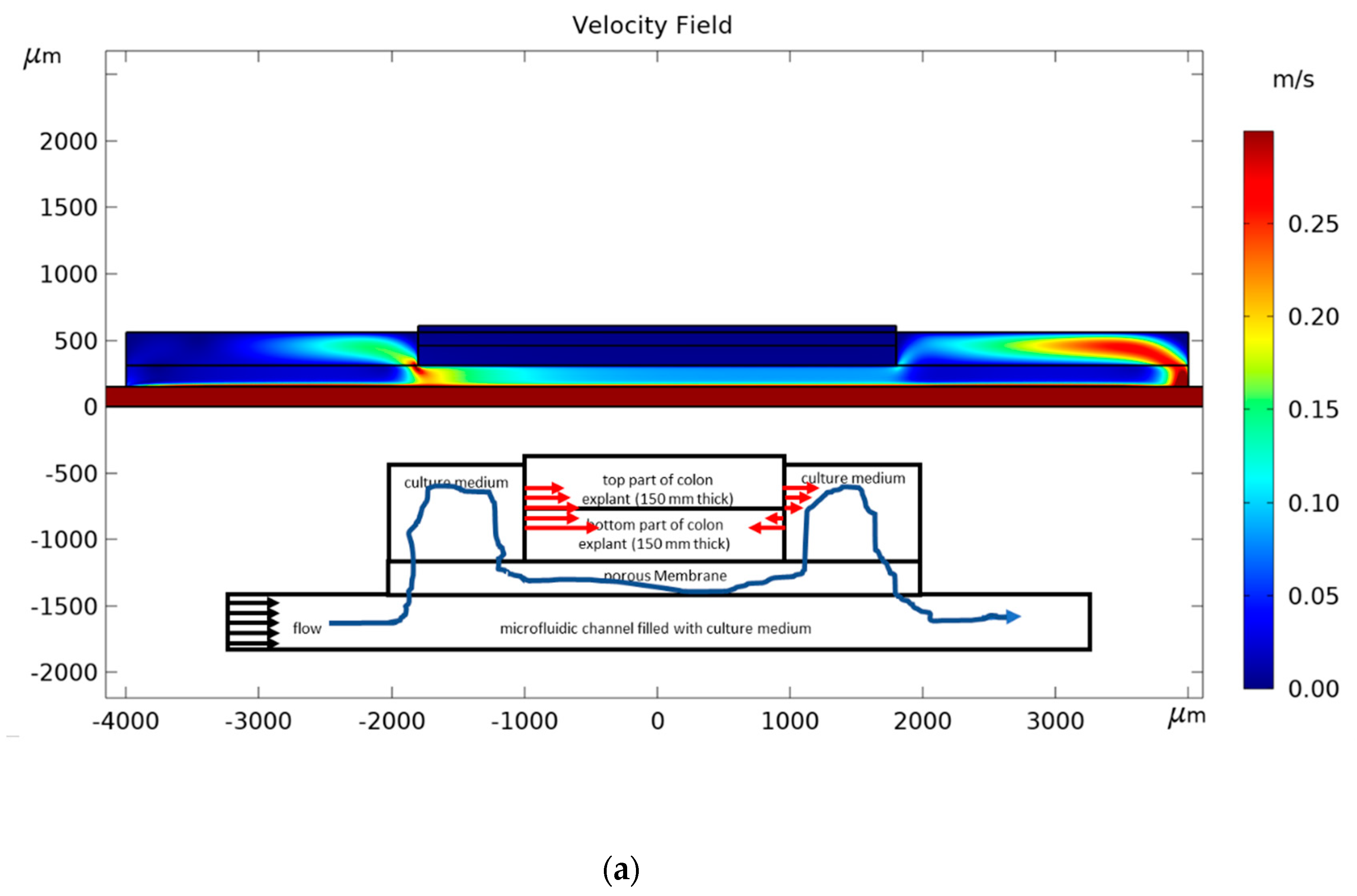
2. Materials and Methods
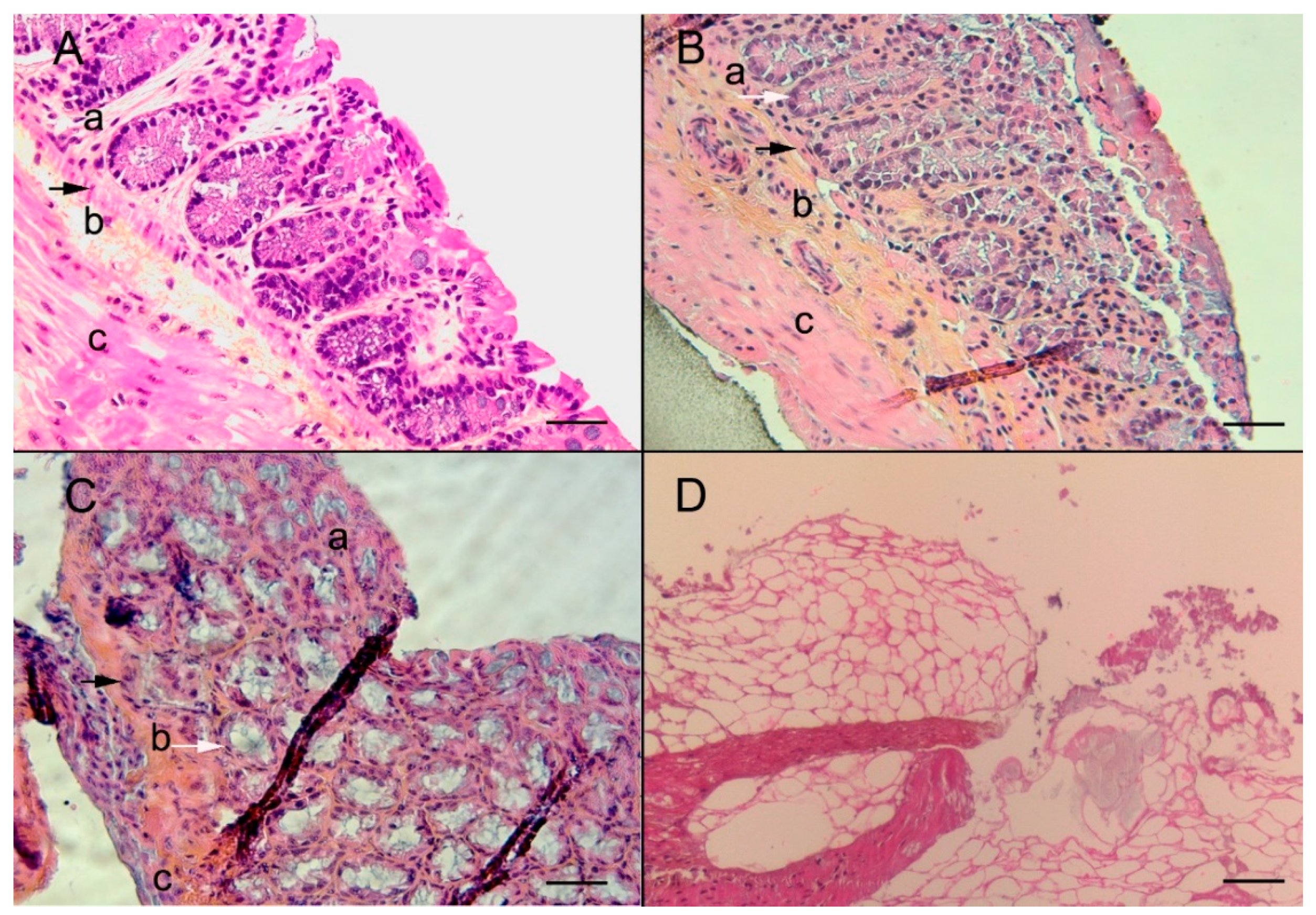
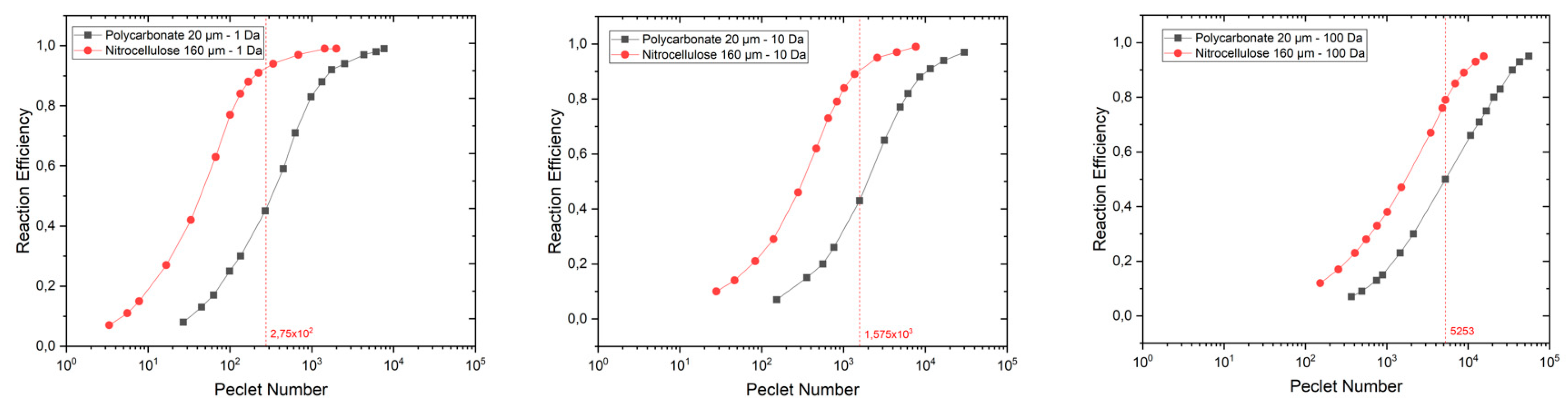
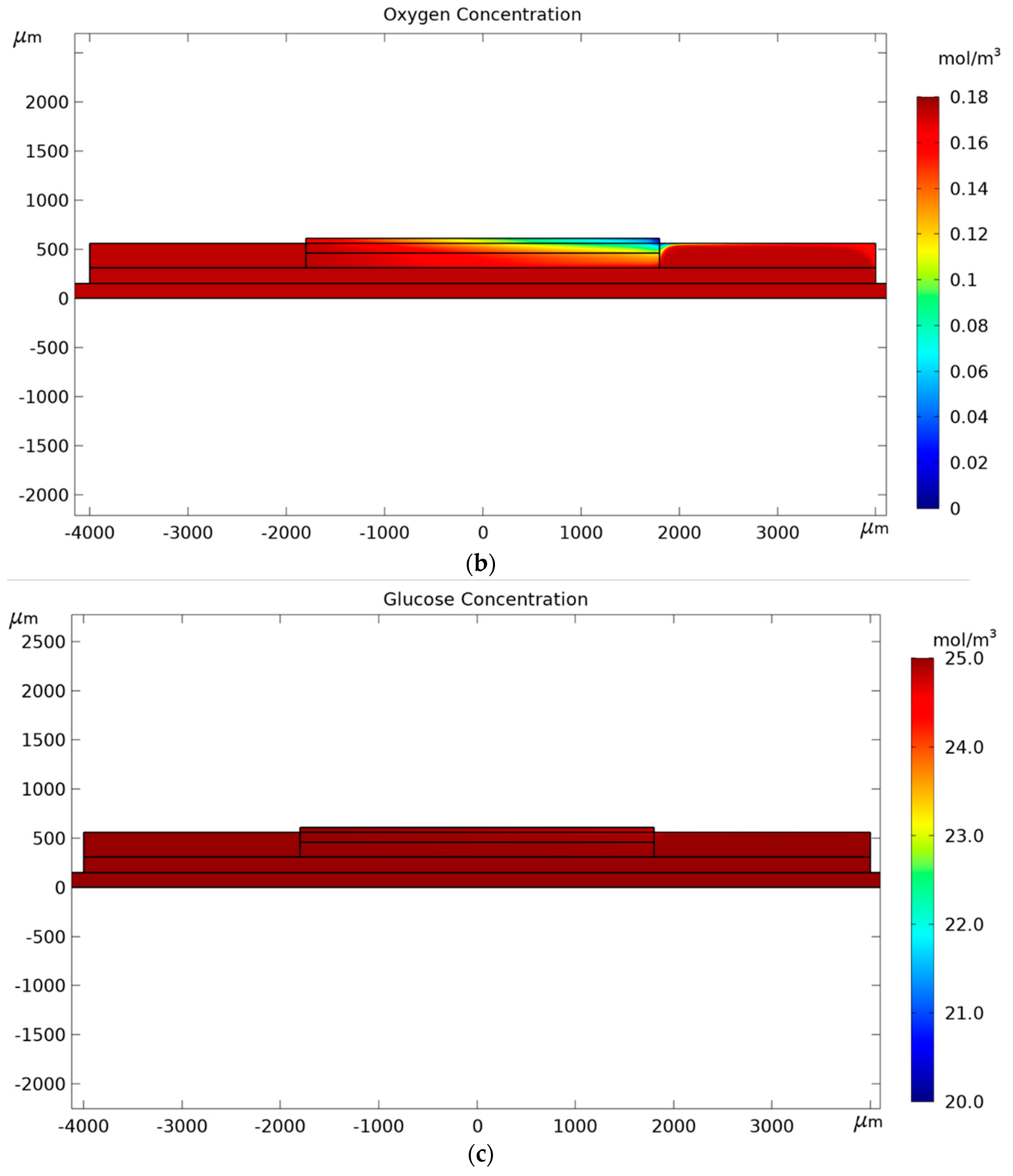
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
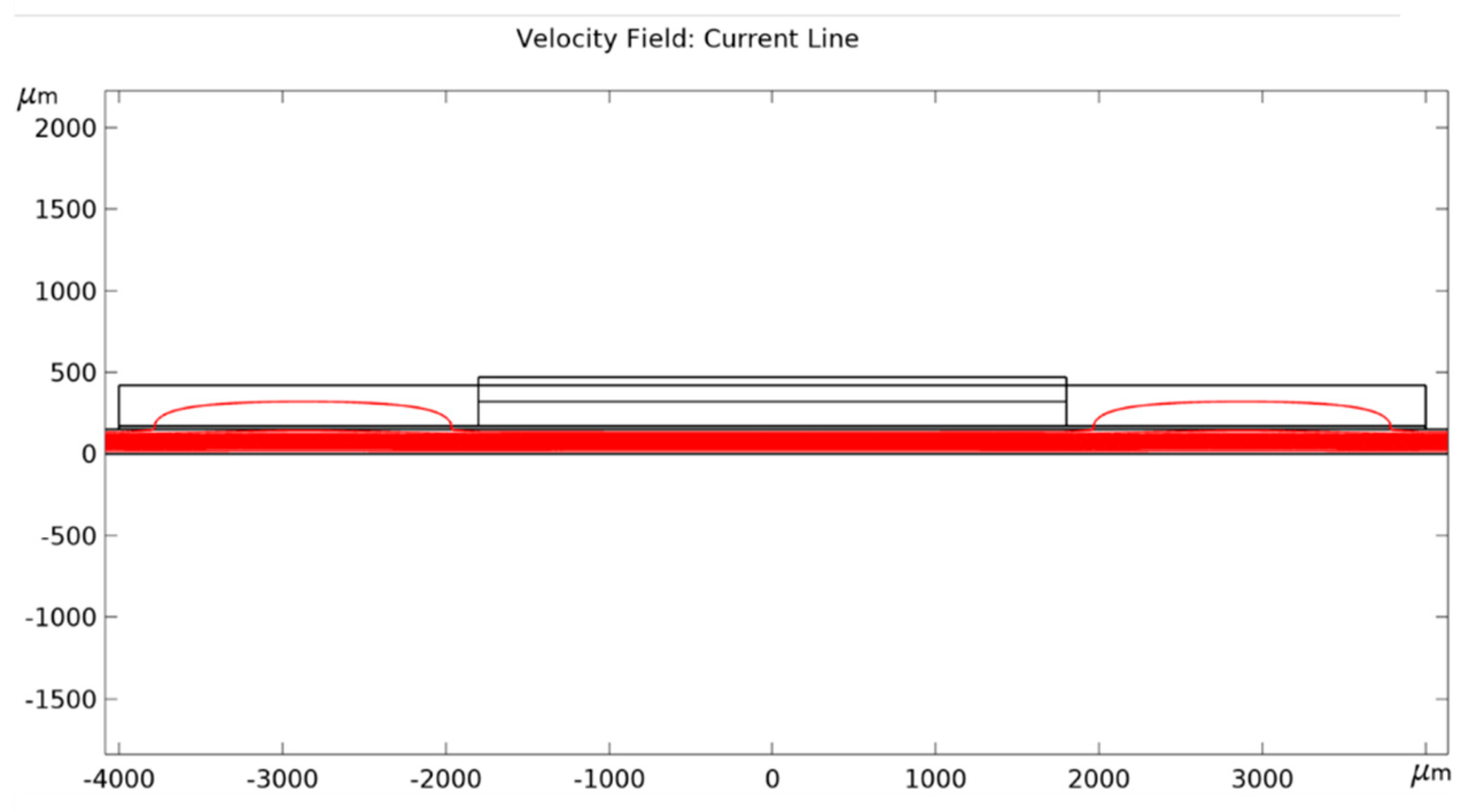
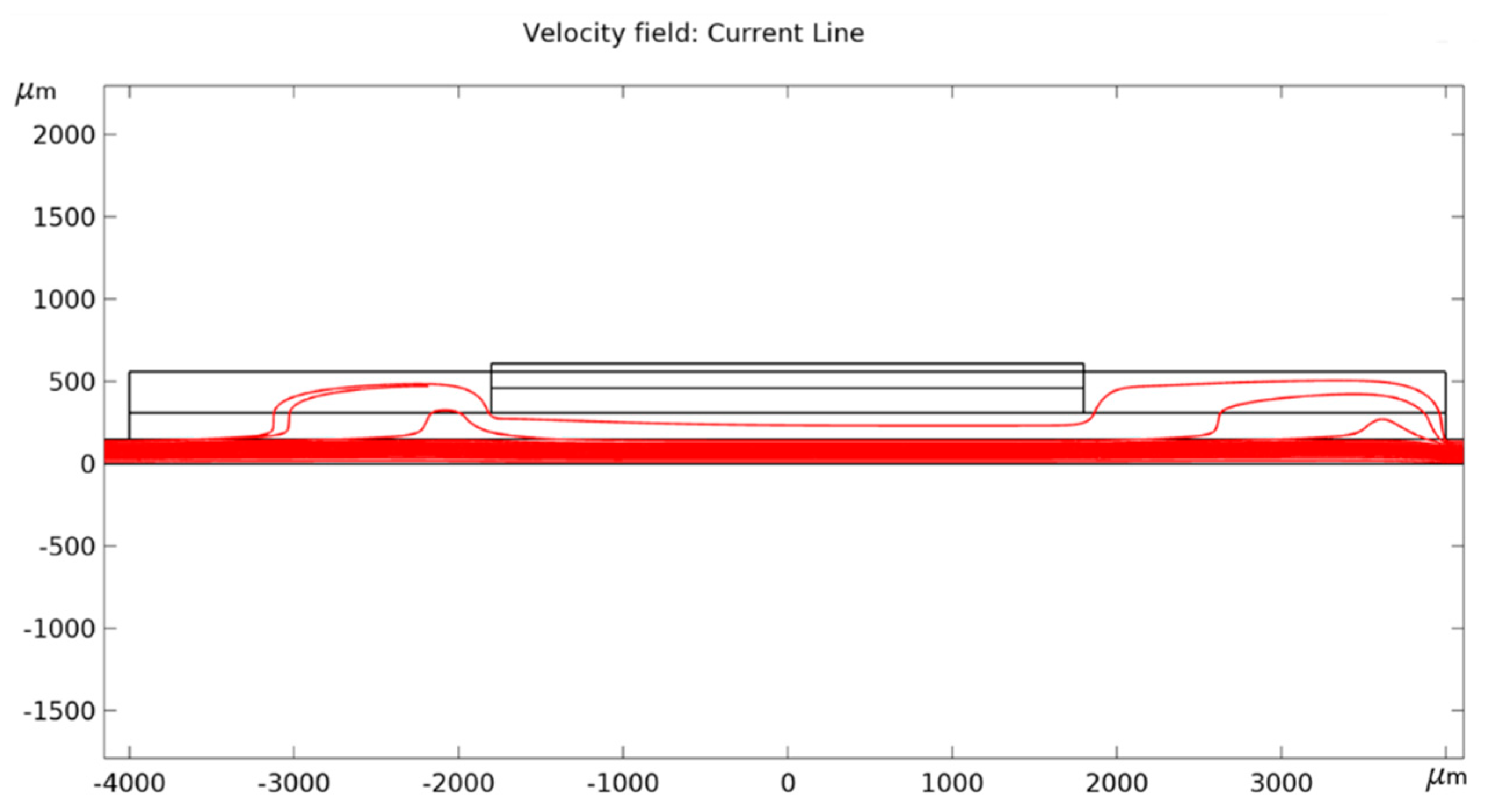
References
- Thomson, D.; Elin, C.E.; Cantab, D.P. Some further researches on the cultivation of tissues in vitro. Proc. R Soc. Med. 1914, 7, 21–46. [Google Scholar] [CrossRef][Green Version]
- Trowell, O.A. The culture of mature organs in a synthetic medium. Exp. Cell Res. 1959, 16, 118–147. [Google Scholar] [CrossRef]
- Shamir, E.R.; Ewald, A.J. Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Astolfi, M.B.; Péant, B.; Lateef, M.A.; Rousset, N.; Kendall-Dupont, J.; Carmona, E.; Monet, F.; Saad, F.; Provencher, D.; Mes-Masson, A.-M.; et al. Micro-dissected tumor tissues on chip: An ex vivo method for drug testing and personalized therapy. Lab Chip. 2015, 16, 312–325. [Google Scholar] [CrossRef]
- Von Martels, J.Z.H.; Sadaghian Sadabad, M.; Bourgonje, A.R.; Blokzijl, T.; Dijkstra, G.; Nico Faber, K.; Harmsen, H.J.M. The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe. 2017, 44, 3–12. [Google Scholar] [CrossRef]
- May, S.; Evans, S.; Parry, L. Organoids, organs-on-chips and other systems, and microbiota. Emerg. Top. Life Sci. 2017, 1, 385–400. [Google Scholar]
- Dedhia, P.H.; Bertaux-Skeirik, N.; Zavros, Y.; Spence, J.R. Organoid models of human gastrointestinal development and disease. Gastroenterology 2016, 150, 1098–1112. [Google Scholar] [CrossRef]
- Li, M.; de Graaf, I.A.M.; Groothuis, G.M. Precision-cut intestinal slices: Alternative model for drug transport, metabolism, and toxicology research. Expert opin. drug metab. toxicol. 2016, 12, 175–190. [Google Scholar] [CrossRef]
- Williams, C.F.; Walton, G.E.; Jiang, L.; Plummer, S.; Garaiova, I.; Gibson, G.R. Comparative analysis of intestinal tract models. Annu. Rev. Food Sci. Technol. 2015, 6, 329–350. [Google Scholar] [CrossRef]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Hee Park, M.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Jung Kim, H.; Ingber, D.E. Microfluidic organ-on-a-chip models of human intestine. Cell. Molecule. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef]
- Groothuis, G.M.M.; Casini, A.; Meurs, H.; Olinga, P. Translational research in pharmacology and toxicology using precision-cut tissue slices. In Human-Based Systems for Translational Research, Chapter, Chapter 3; Coleman, R., Ed.; The Royal Society of Chemistry: London, UK, 2015; pp. 38–65. [Google Scholar]
- Baydoun, M.; Vanneste, S.B.; Creusy, C.; Guyot, K.; Gantois, N.; Chabe, M.; Delaire, B.; Mouray, A.; Baydoun, A.; Forzy, G.; et al. Three-dimensional (3D) culture of adult murine colon as an in vitro model of cryptosporidiosis: Proof of concept. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- McLean, I.C.; Schwerdtfeger, L.A.; Tobet, S.A.; Henry, C.S. Powering ex vivo tissue models in microfluidic systems. Lab Chip. 2018, 18, 1399–1410. [Google Scholar] [CrossRef]
- Bettinger, C.J.; Langer, R.; Borenstein, J.T. Engineering substrate topography at the micro-and nanoscale to control cell function. Angew. Chem. Int. Ed. 2009, 48, 5406–5415. [Google Scholar] [CrossRef]
- Chen, M.B.; Whisler, J.A.; Fröse, J.; Yu, C.; Shin, Y.; Kamm, R.D. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat. Protoc. 2017, 12, 865–880. [Google Scholar] [CrossRef]
- Dvir, T.; Timko, B.P.; Kohane, D.; Langer, R. Nanotechnological strategies for engineering complex tissues. Nat. Nanotechnol. 2010, 6, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Young, E.W.; Beebe, D.J. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010, 39, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Van Midwoud, P.M.; Groothuis, G.M.; Merema, M.T.; Verpoorte, E. Microfluidic biochip for the perifusion of precision-cut rat liver slices for metabolism and toxicology studies. Biotechnol. Bioeng. 2010, 105, 184–194. [Google Scholar] [CrossRef]
- Komeya, M.; Kimura, H.; Nakamura, H.; Yokonishi, T.; Sato, T.; Kojima, K.; Hayashi, K.; Katagiri, K.; Yamanaka, H.; Sanjo, H.; et al. Long-term ex vivo maintenance of testis tissues producing fertile sperm in a microfluidic device. Sci. Rep. 2016, 6, 21472. [Google Scholar] [CrossRef]
- Nery, F.C.; da Hora, C.C.; Yaqub, U.; Zhang, X.; McCarthy, D.M.; Bhide, P.G.; Irimia, D.; Breakefield, X.O. New methods for investigation of neuronal migration in embryonic brain explants. J. Neurosci. Method. 2015, 239, 80–84. [Google Scholar] [CrossRef]
- Ionescu, A.; Zahavi, E.E.; Gradus, T.; Ben-Yaakov, K.; Perlson, E. Compartmental microfluidic system for studying muscle–neuron communication and neuromuscular junction maintenance. Eur. J. Cell Biol. 2016, 95, 69–88. [Google Scholar] [CrossRef]
- Kim, Y.T.; Joshi, S.D.; Messner, W.C.; LeDuc, P.R.; Davidson, L.A. Detection of dynamic spatiotemporal response to periodic chemical stimulation in a Xenopus embryonic tissue. PLoS ONE 2011, 6, e14624. [Google Scholar] [CrossRef]
- Ross, A.E.; Belanger, M.C.; Woodroof, J.F.; Pompano, R.R. Spatially resolved microfluidic stimulation of lymphoid tissue ex vivo. Analyst 2017, 142, 649–659. [Google Scholar] [CrossRef]
- Yasotharan, S.; Pinto, S.; Sled, J.G.; Bolz, S.-S.; Günther, A. Artery-on-a-chip platform for automated, multimodal assessment of cerebral blood vessel structure and function. Lab Chip 2015, 15, 2660–2669. [Google Scholar] [CrossRef]
- Bower, R.; Green, V.L.; Kuvshinova, E.; Kuvshinov, D.; Karsai, L.; Crank, S.T.; Stafford, N.D.; Greenman, J. Maintenance of head and neck tumor on-chip: Gateway to personalized treatment? Future Sci. OA 2017, 3, FSO174. [Google Scholar] [CrossRef]
- Van Midwoud, P.M.; Merema, M.T.; Verpoort, E.; Groothuis, G.M.M. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip 2010, 10, 2778–2786. [Google Scholar] [CrossRef]
- Costa, M.O.; Nosach, R.; Harding, J.C.S. Development of a 3D printed device to support long term intestinal culture as an alternative to hyperoxic chamber methods. 3D Print. Med. 2017, 3, 9. [Google Scholar] [CrossRef]
- Yissachar, N.; Zhou, Y.; Ung, L.; Lai, N.Y.; Mohan, J.F.; Ehrlicher, A.; Weitz, D.A.; Kasper, D.L.; Chiu, I.M.; Mathis, D.; et al. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell 2017, 168, 1135–1148. [Google Scholar] [CrossRef]
- Dawson, A.; Dyer, C.; Macfie, J.; Davies, J.; Karsai, L.; Greenman, J.; Jacobsen, M. A microfluidic chip based model for the study of full thickness human intestinal tissue using dual flow. Biomicrofluidics 2016, 10, 064101. [Google Scholar] [CrossRef]
- Browning, T.H.; Trier, J.S. Organ culture of mucosal biopsies of human small intestine. J. Clin. Invest. 1969, 48, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Zahorodny-Burke, M.; Nearingburg, B.; Elias, A.L. Finite element analysis of oxygen transport in microfluidic cell culture devices with varying channel architectures, perfusion rates, and materials. Chem. Eng. Sci. 2011, 66, 6244–6253. [Google Scholar] [CrossRef]
- Casciari, J.J.; Sotirchos, S.V.; Sutherland, R.M. Glucose diffusivity in multicellular tumor spheroids. Cancer Res. 1988, 48, 3905–3909. [Google Scholar]
- Jain, R.K. Transport of molecules in the tumor interstitium: A review. Cancer Res. 1987, 47, 3039–3051. [Google Scholar]
- Avgoustiniatos, E.S.; Colton, C.K. Effect of external oxygen mass transfer resistances on viability of immunoisolated tissue. Ann. N.Y. Acad. Sci. 1997, 831, 145–167. [Google Scholar] [CrossRef]
- Buladi, B.M.; Chang, C.C.; Belovich, J.M.; Gatica, J.E. Transport phenomena and kinetics in an extravascular bioartificial pancreas. AIChE J. 1996, 42, 2668–2682. [Google Scholar] [CrossRef]
- Place, T.L.; Domann, F.E.; Case, A.J. Limitations of oxygen delivery to cells in culture: An underappreciated problem in basic and translational research. Free Radic. Biol. Med. 2017, 113, 311–322. [Google Scholar] [CrossRef]
- Tomadakis, M.M.; Robertson, J.T. Viscous permeability of random fiber structures: Comparison of electrical and diffusional estimates with experimental and analytical results. J. Compos. Mater. 2005, 39, 163–188. [Google Scholar] [CrossRef]
- Fell, H.B. Studies of development in organ culture: Summary, correlation, and speculation. J. Natl. Cancer Inst. 1963, 11, 73–80. [Google Scholar]
- Autrup, H.; Barrett, L.A.; Jackson, F.E.; Jesudason, M.L.; Stoner, G.; Phelps, P.; Trump, B.F.; Harris, C.C. Explant culture of human colon. Gastroenterology 1978, 74, 1248–1257. [Google Scholar] [CrossRef]
- Abud, H.E.; Watson, N.; Heath, J.K. Growth of intestinal epithelium in organ culture is dependent on EGF signaling. Expe. Cell Res. 2005, 303, 252–262. [Google Scholar] [CrossRef]
- Mitchell, J.D.; Mitchell, J.; Peters, T.J. Enzyme changes in human small bowel mucosa during culture in vitro. Gut 1974, 15, 805–811. [Google Scholar] [CrossRef]
- Quinlan, J.M.; Yu, W.Y.; Hornsey, M.A.; Tosh, D.; Slack, J.M.W. In vitro culture of embryonic mouse intestinal epithelium: Cell differentiation and introduction of reporter genes. BMC Dev. Biol. 2006, 6, 24–33. [Google Scholar] [CrossRef]
- Randall, K.J.; Turton, J.; Foster, J.R. Explant culture of gastrointestinal tissue: A review of methods and applications. Cell Biol. Toxicol. 2011, 27, 267–284. [Google Scholar] [CrossRef]
- Resau, J.H.; Sakamoto, K.; Cottrell, J.R.; Hudson, E.A.; Meltzer, S.J. Explant organ culture: A review. Cytotechnology 1991, 7, 137–149. [Google Scholar] [CrossRef]
- Clarke, L.L. A guide to Using chamber studies of mouse intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G1151–G1166. [Google Scholar] [CrossRef]
- Shamsuddin, A.K.M.; Barrett, L.A.; Autrup, H.; Harris, C.C.; Trump, B.F. Long-term organ culture of adult rat colon. Pathol. Res. Pract. 1978, 163, 362–372. [Google Scholar] [CrossRef]
- Byers, S.W.; Hadley, M.A.; Djakiew, D.; Dym, M. Growth and characterization of polarized monolayers of epididymal epithelial cells and Sertoli cells in dual environment culture chambers. J. Androl. 1986, 7, 59–68. [Google Scholar] [CrossRef]
- Chen, X.; Shen, J. Review of membranes in microfluidics. J. Chem. Technol. Biotechnol. 2017, 92, 271–282. [Google Scholar] [CrossRef]
- Karatay, E.; Lammertink, R.G.H. Oxygenation by a superhydrophobic slip G/L contactor. Lab Chip 2012, 12, 2922–2929. [Google Scholar] [CrossRef]
- Epshteyn, A.; Maher, S.; Taylor, A.J.; Holton, A.B.; Borenstein, J.T.; Cuiffi, J.D. Membrane-integrated microfluidic device for high-resolution live cell imaging. Biomicrofluidics 2011, 5, 46501–465016. [Google Scholar] [CrossRef]
- Snakenborg, D.; Klank, H.; Kutter, J.P. Polymer microvalve with pre-stressed membranes for tunable flow–pressure characteristics. Microfluid. Nanofluid. 2010, 10, 381–388. [Google Scholar] [CrossRef]
- Qiu, J.; Gao, Q.; Zhao, H.; Fu, J.; He, Y. Rapid Customization of 3D Integrated Microfluidic Chips via Modular Structure-Based Design. ACS Biomater. Sci. Eng. 2017, 3, 2606–2616. [Google Scholar] [CrossRef]
- Russo, P.S.; Retterer, T.; Spence, A.J.; Isaacson, M.S.; Lepak, L.A.; Spencer, M.G.; Martin, D.L.; MacColl, R.; Turner, J.N. Direct Casting of Polymer Membranes into Microfluidic Devices. Sep. Sci. Technol. 2010, 39, 2515–2530. [Google Scholar] [CrossRef]
- Tokuyama, T.; Fujii, S.; Sato, K.; Abo, M.; Okubo, A. Microbioassay system for antiallergic drug screening using suspension cells retaining in a poly (dimethylsiloxane) microfluidic device. Anal. Chem. 2005, 77, 3309–3314. [Google Scholar] [CrossRef]
- Sakuta, Y.; Tsunoda, K.; Sato, K. Development of a Multichannel Dialysis Microchip for Bioassay of Drug Efficacy and Retention. Anal. Sci. 2017, 33, 391–394. [Google Scholar] [CrossRef]
- Chueh, B.; Huh, D.; Kyrtsos, C.R.; Houssin, T.; Futai, N.; Takayama, S. Leakage-free bonding of porous membranes into layered microfluidic array systems. Anal. Chem. 2007, 79, 3504–3508. [Google Scholar] [CrossRef]

| Parameter | Unit | Aqueous Media | Porous Membrane | Tissue Explant | Reference |
|---|---|---|---|---|---|
| Temperature | °K | 310 | 310 | 310 | – |
| Density | kg/m3 | 993 | N/Ac | N/Ac | – |
| Viscosity | Pa·s | 0.7 × 10-3 | N/Ac | N/Ac | – |
| Porosity | % | N/Ac | 79 a or 15 b | N/Ac | – |
| Permeability | Darcy | N/Ac | 1 to 100 | N/Ac | – |
| Oxygen diffusivity | m2/s | 2.6 × 10-9 | 1.1× 10-9 a or 9.4 × 10-11 b | 2.0 × 10-9 | [33] |
| Glucose diffusivity | m2/s | 0.7 × 10-9 | 0,3 × 10-9 a or 2.5 × 10-11 b | 0.3 × 10-9 | [34] |
| Retardation effect (β) | – | 1 | 0,407 a or 0.036 b | 1 | [35] |
| Max oxygen reaction rate | mol/m3/s | N/Ac | N/Ac | −0.034 | [36] |
| Max glucose reaction rate | mol/m3/s | N/Ac | N/Ac | −0.028 | [37] |
| Critical oxygen conc. | mol/m3 | N/Ac | N/Ac | 1 × 10-4 | [36] |
| Critical glucose conc. | mol/m3 | N/Ac | N/Ac | 0.1 | [37] |
| Initial oxygen conc. | mol/m3 | 0.174 | 0.174 | 0.174 | [38] |
| Initial glucose conc. | mol/m3 | 25 | 25 | 25 | – |
| M.M.d constant (oxygen) | mol/m3 | N/Ac | N/Ac | 1 × 10-3 | [36] |
| M.M.d constant (glucose) | mol/m3 | N/Ac | N/Ac | 1 × 10-2 | [37] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baydoun, M.; Treizeibré, A.; Follet, J.; Vanneste, S.B.; Creusy, C.; Dercourt, L.; Delaire, B.; Mouray, A.; Viscogliosi, E.; Certad, G.; et al. An Interphase Microfluidic Culture System for the Study of Ex Vivo Intestinal Tissue. Micromachines 2020, 11, 150. https://doi.org/10.3390/mi11020150
Baydoun M, Treizeibré A, Follet J, Vanneste SB, Creusy C, Dercourt L, Delaire B, Mouray A, Viscogliosi E, Certad G, et al. An Interphase Microfluidic Culture System for the Study of Ex Vivo Intestinal Tissue. Micromachines. 2020; 11(2):150. https://doi.org/10.3390/mi11020150
Chicago/Turabian StyleBaydoun, Martha, Anthony Treizeibré, Jérôme Follet, Sadia Benamrouz Vanneste, Colette Creusy, Lucie Dercourt, Baptiste Delaire, Anthony Mouray, Eric Viscogliosi, Gabriela Certad, and et al. 2020. "An Interphase Microfluidic Culture System for the Study of Ex Vivo Intestinal Tissue" Micromachines 11, no. 2: 150. https://doi.org/10.3390/mi11020150
APA StyleBaydoun, M., Treizeibré, A., Follet, J., Vanneste, S. B., Creusy, C., Dercourt, L., Delaire, B., Mouray, A., Viscogliosi, E., Certad, G., & Senez, V. (2020). An Interphase Microfluidic Culture System for the Study of Ex Vivo Intestinal Tissue. Micromachines, 11(2), 150. https://doi.org/10.3390/mi11020150





