Adsorption and Absorption of Collagen Peptides to Polydimethlysiloxane and Its Influence on Platelet Adhesion Flow Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Blood Collection and Preparation
2.3. Glass, PDMS and Silicon Substrate Functionalization
2.4. Microfluidic Patterning Collagen Peptides
2.5. Microspot Patterning Collagen Peptides
2.6. Whole Blood Microfluidic Assays
2.7. Image Analysis
2.8. Atomic Force Microscopy
2.9. Contact Angle Measurement
2.10. Collagen peptide depletion in PDMS
2.11. Visualization of Absorption of Fluorescent Collagen Peptide into PDMS
3. Results
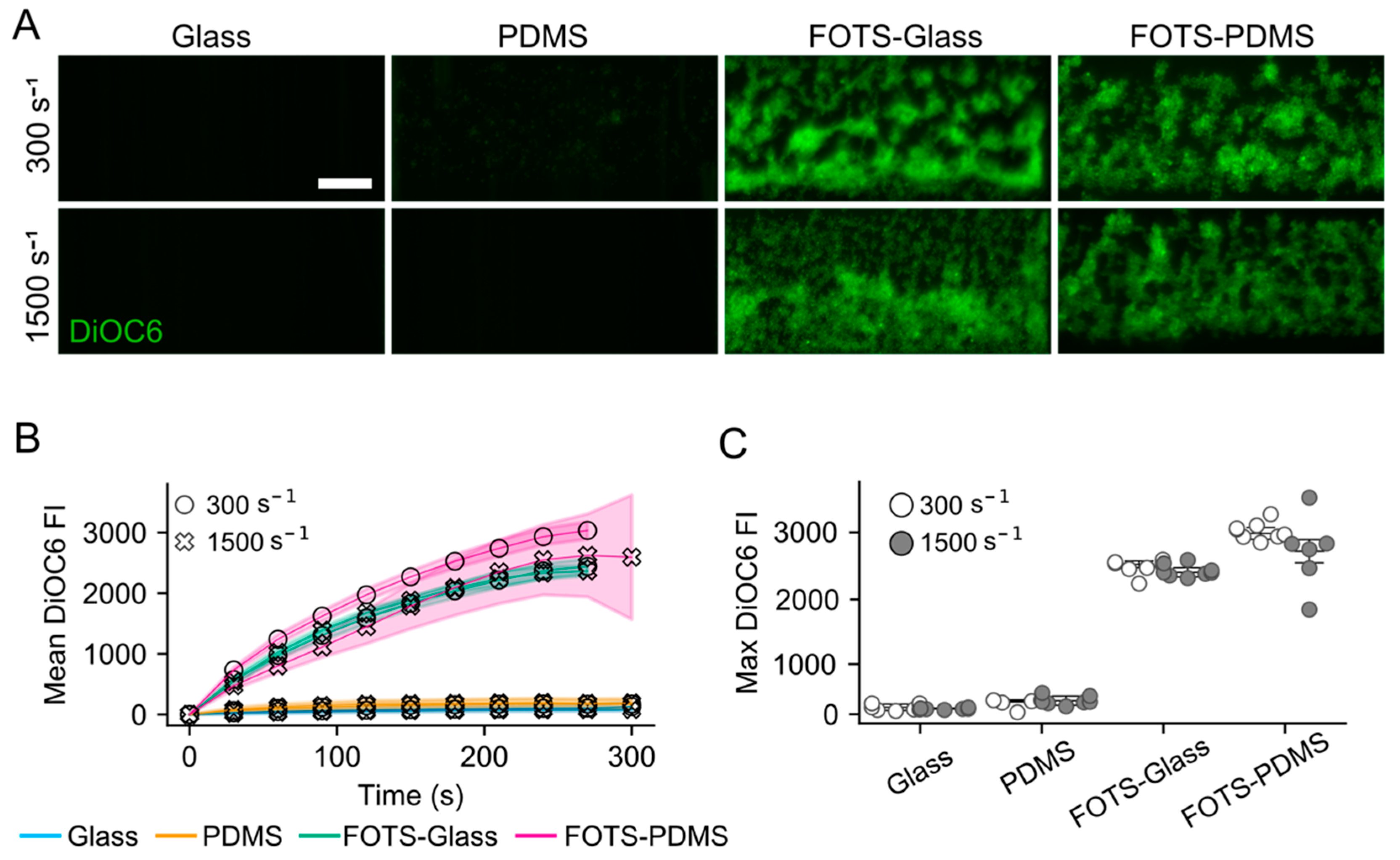
3.1. Flow Assays Reveal Sharply Reduced Platelet Adhesion to Collagen Peptides Patterned on or with PDMS
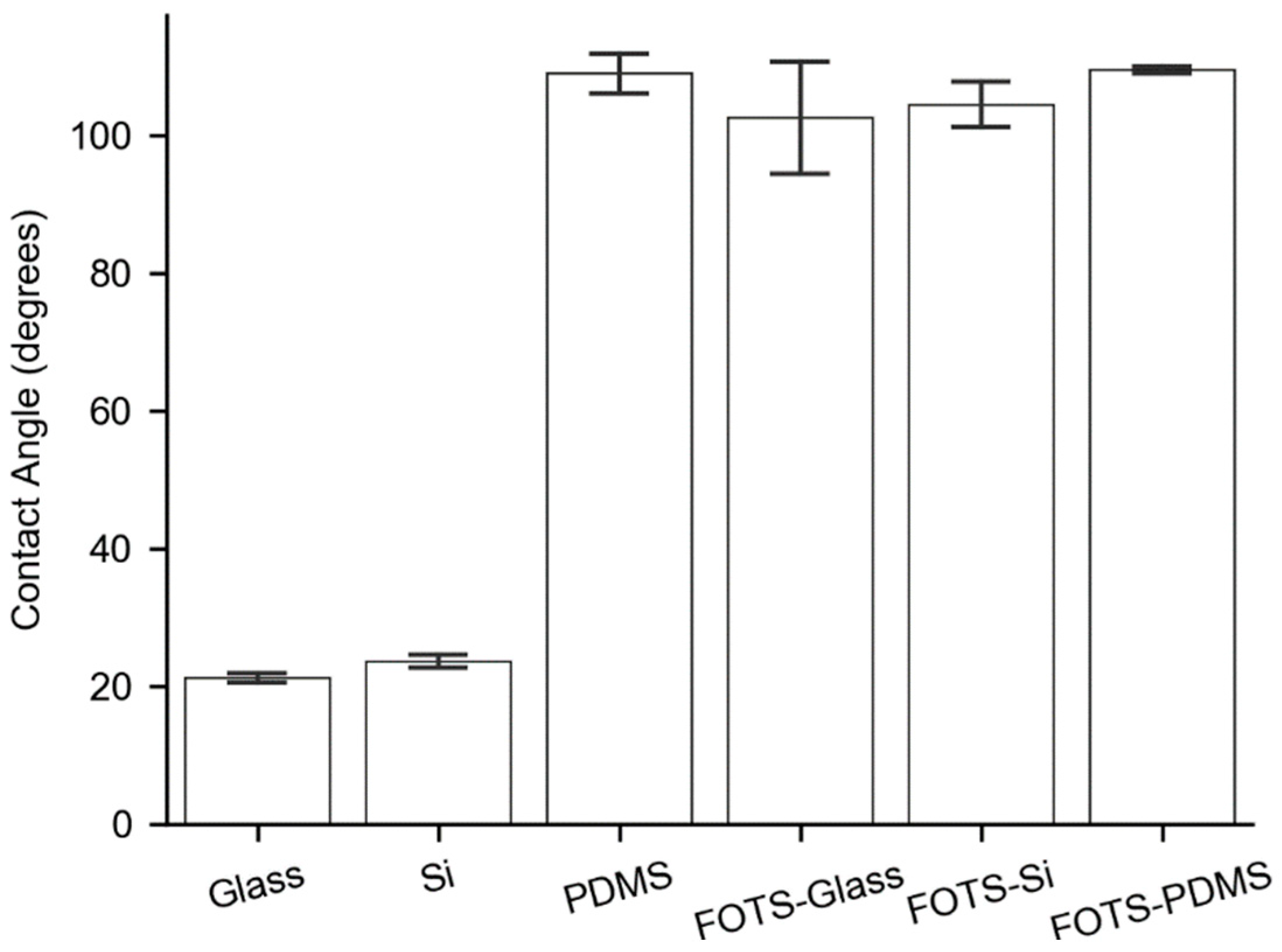
3.2. Substrate Contact Angles
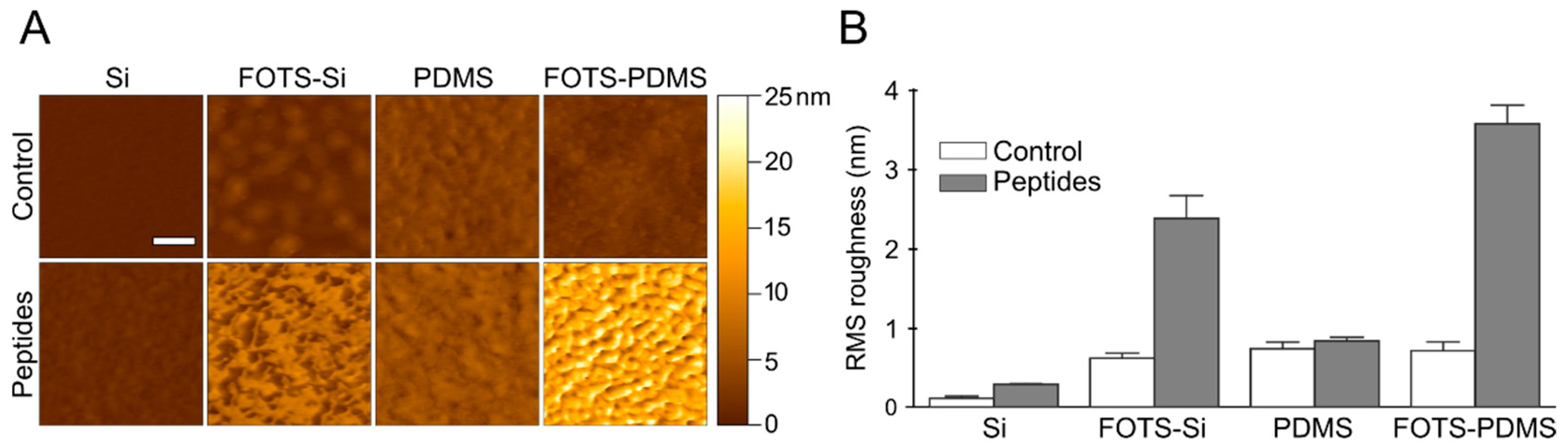
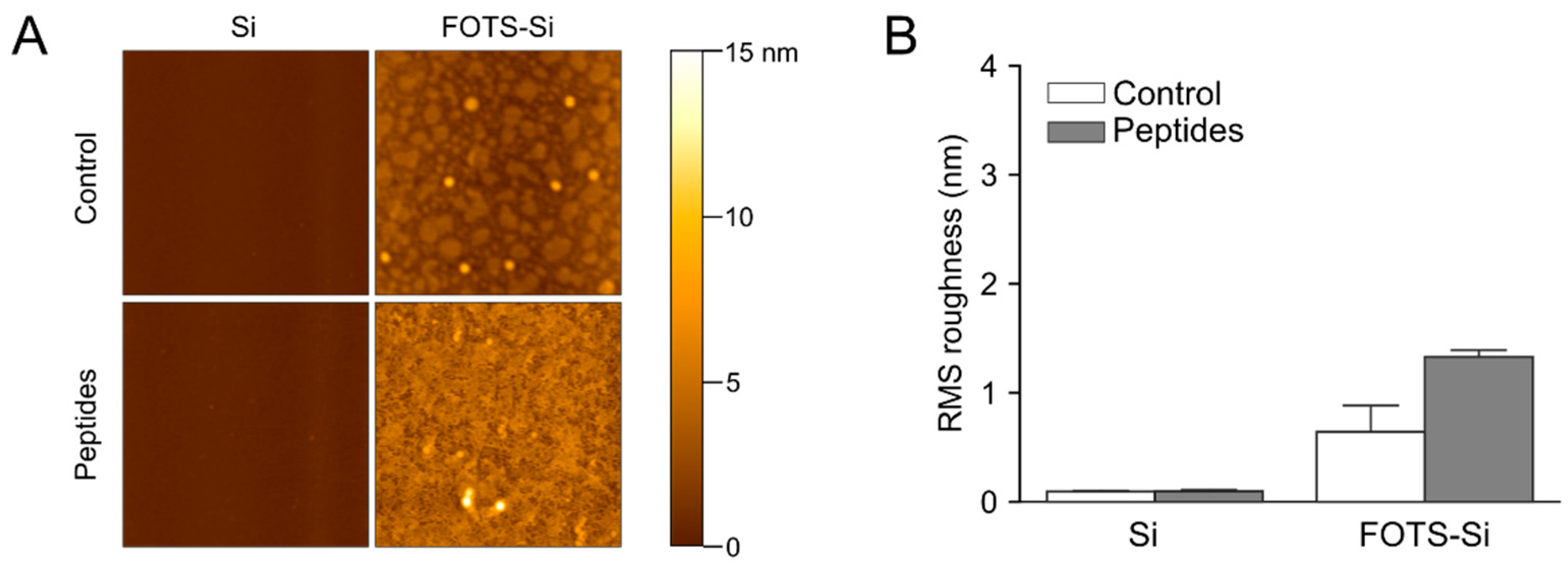
3.3. Atomic Force Microscopy
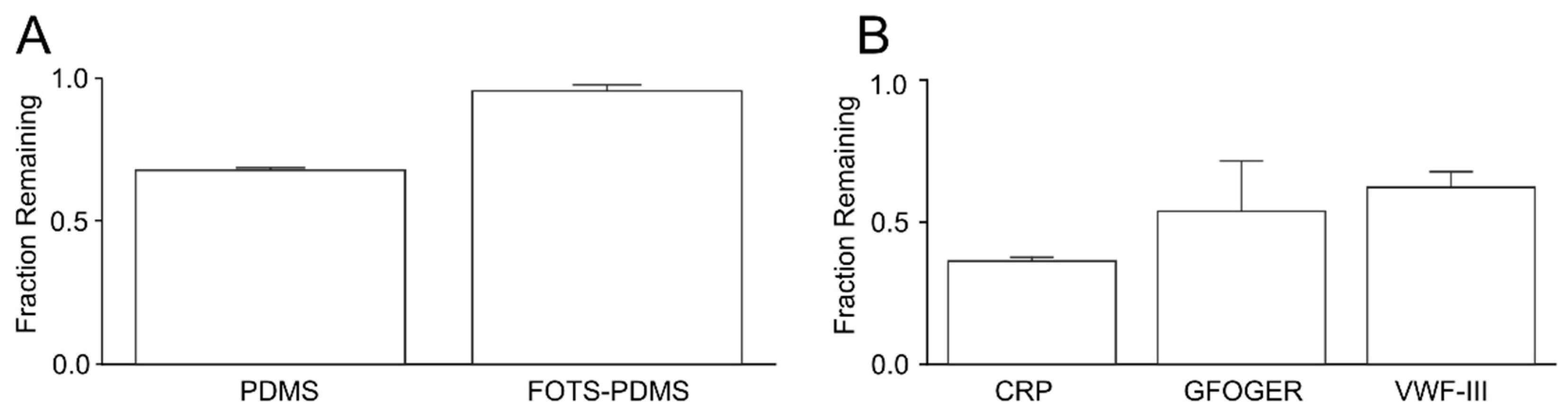
3.4. Depletion of Peptides in Solution
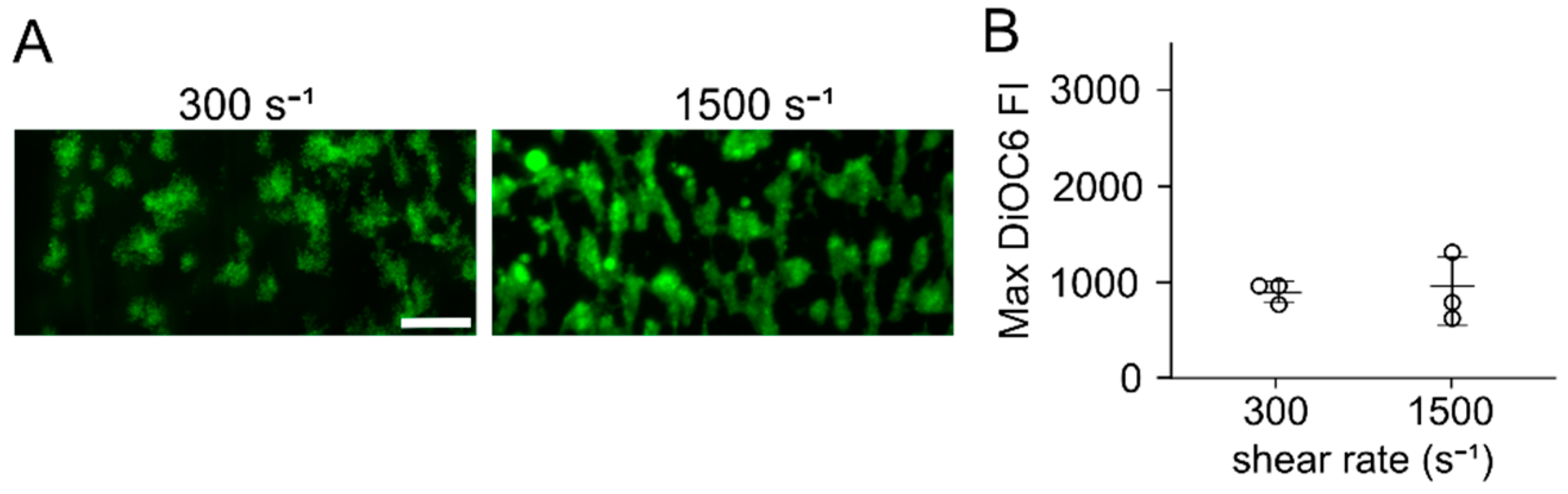
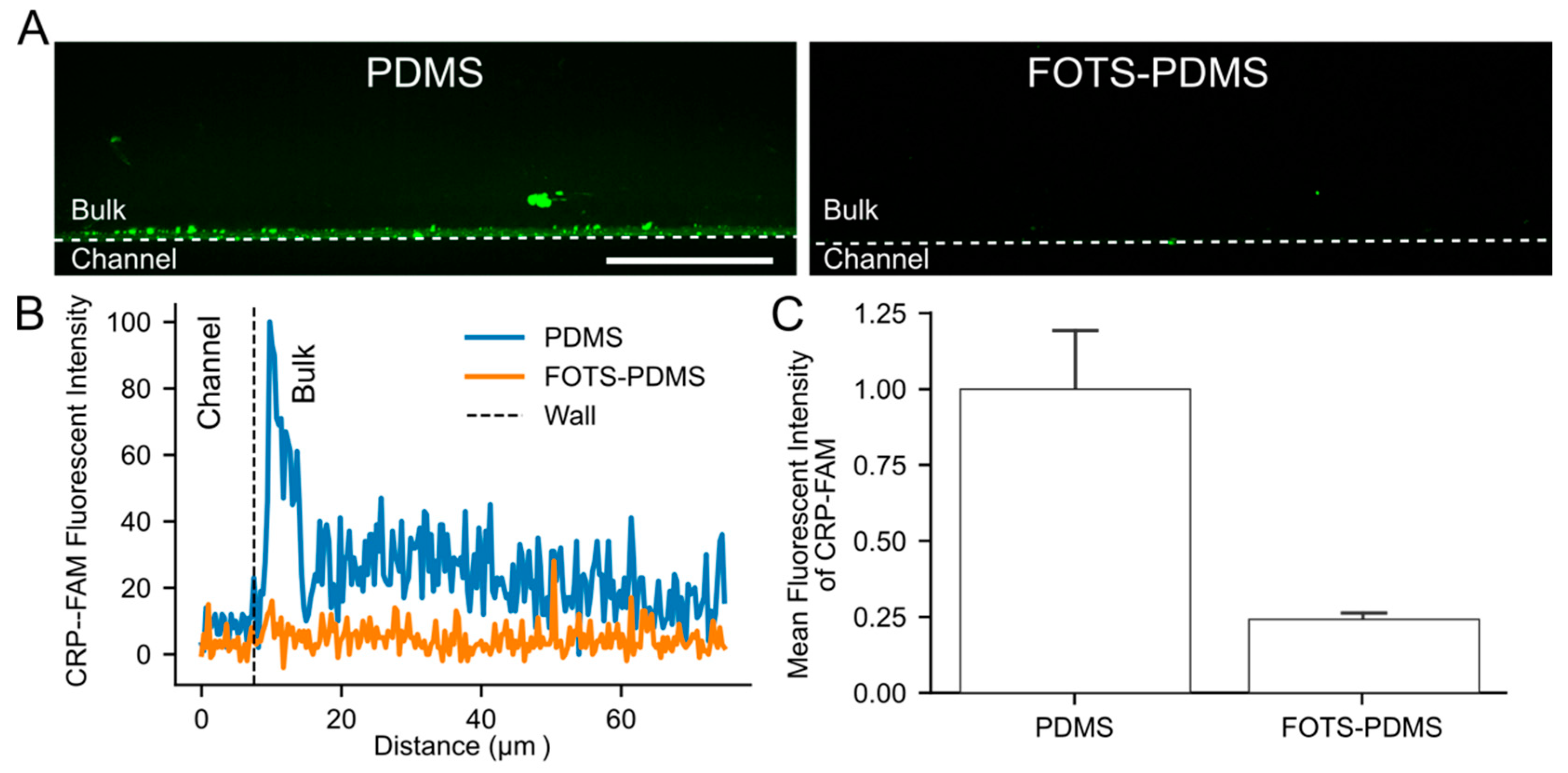
3.5. Visualization of Peptide Absorption into PDMS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fogelson, A.L.; Neeves, K.B. Fluid mechanics of blood clot formation. Annu. Rev. Fluid Mech. 2015, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Neeves, K.B.; Onasogaa, A.A.; Wufsus, A.R. The use of microfluidics in hemostasis: Clinical diagnostics and biomimetic models of vascular injur. Macromol. Mater. Eng. 2011, 296, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Heemskerk, J.W.M.; Sakariassen, K.S.; Zwaginga, J.J.; Brass, L.F.; Jackson, S.P.; Farndale, R.W. Collagen surfaces to measure thrombus formation under flow: Possibilities for standardization. J. Thromb. Haemost. 2011, 9, 856–858. [Google Scholar] [CrossRef] [PubMed]
- Pugh, N.; Simpson, A.M.C.; Smethurst, P.A.; De Groot, P.G.; Raynal, N.; Farndale, R.W. Synergism between platelet collagen receptors defined using receptor-specific collagen-mimetic peptide substrata in flowing blood. Blood 2010, 115, 5069–5079. [Google Scholar] [CrossRef]
- Farndale, R.W.; Sixma, J.J.; Barnes, M.J.; de Groot, P.G. The role of collagen in thrombosis and hemostasis. J. Thromb. Haemost. 2004, 2, 561–573. [Google Scholar] [CrossRef]
- Savage, B.; Saldívar, E.; Ruggeri, Z.M. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell 1996, 84, 289–297. [Google Scholar] [CrossRef]
- Savage, B.; Almus-Jacobs, F.; Ruggeri, Z.M. Specific synergy of multiple substrate-receptor interactions in platelet thrombus formation under flow. Cell 1998, 94, 657–666. [Google Scholar] [CrossRef]
- Goto, S.; Tamura, N.; Handa, S.; Arai, M.; Kodama, K.; Takayama, H. Involvement of glycoprotein VI in platelet thrombus formation on both collagen and von Willebrand factor surfaces under flow conditions. Circulation 2002, 106, 266–272. [Google Scholar] [CrossRef][Green Version]
- Sarratt, K.L.; Chen, H.; Kahn, M.L.; Hammer, D.A. Platelet receptor glycoprotein VI-mediated adhesion to type I collagen under hydrodynamic flow. Ann. Biomed. Eng. 2004, 32, 970–976. [Google Scholar] [CrossRef]
- Sakariassen, K.S.; Turitto, V.T.; Baumgartner, H.R. Recollections of the development of flow devices for studying mechanisms of hemostasis and thrombosis in flowing whole blood. J. Thromb. Haemost. 2004, 2, 1681–1690. [Google Scholar] [CrossRef]
- Hansen, R.R.; Tipnis, A.A.; White-Adams, T.C.; di Paola, J.A.; Neeves, K.B. Characterization of collagen thin films for von Willebrand factor binding and platelet adhesion. Langmuir 2011, 27, 13648–13658. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.S.; Birtles, M.J.; Conway, J.F.; Parry, D.A.D. An estimate of the mean length of collagen fibrils in rat tail-tendon as a function of age. Connect. Tissue. Res. 1989, 19, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Okorie, U.M.; Diamond, S.L. Matrix protein microarrays for spatially and compositionally controlled microspot thrombosis under laminar flow. Biophys. J. 2006, 91, 3474–3481. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lehmann, M.; Ashworth, K.; Manco-Johnson, M.; di Paola, J.; Neeves, K.B.; Ng, C.J. Evaluation of a microfluidic flow assay to screen for von Willebrand disease and low von Willebrand factor levels. J. Thromb. Haemost. 2018, 16, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Neeves, K.B.; Maloney, S.F.; Fong, K.P.; Schmaier, A.A.; Kahn, M.L.; Brass, L.F.; Diamond, S.L. Microfluidic focal thrombosis model for measuring murine platelet deposition and stability: PAR4 signaling enhances shear-resistance of platelet aggregates. J. Thromb. Haemost. 2008, 6, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Sakariassen, K.S.; Joss, R.; Muggli, R.; Kuhn, H.; Tschopp, T.B.; Sage, H.; Baumgartner, H.R. Collagen type III induced ex vivo thrombogenesis in humans. Role of platelets and leukocytes in deposition of fibrin. Arteriosclerosis 1990, 10, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Pugh, N.; Maddox, B.D.; Bihan, D.; Taylor, K.A.; Mahaut-smith, M.P.; Farndale, R.W. Differential integrin activity mediated by platelet collagen receptor engagement under flow conditions. Thromb. Haemost. 2017, 117, 21–23. [Google Scholar] [CrossRef]
- De Witt, S.M.; Swieringa, F.; Cavill, R.; Lamers, M.M.E.; van Kruchten, R.; Mastenbroek, T.; Baaten, C.; Coort, S.; Pugh, N.; Schulz, A.; et al. Identification of platelet function defects by multi-parameter assessment of thrombus formation. Nat. Commun. 2014, 5, 4257. [Google Scholar] [CrossRef]
- Brazilek, R.J.; Tovar-Lopez, F.J.; Wong, A.K.T.; Tran, H.; Davis, A.S.; McFadyen, J.D.; Kaplan, Z.; Chunilal, S.; Jackson, S.P.; Nandurkar, H.; et al. Application of a strain rate gradient microfluidic device to von Willebrand’s disease screening. Lab. Chip. 2017, 17, 2595–2608. [Google Scholar] [CrossRef]
- Zilberman-Rudenko, J.; Sylman, J.L.; Lakshmanan, H.H.S.; McCarty, O.J.T.; Maddala, J. Dynamics of blood flow and thrombus formation in a multi-bypass microfluidic ladder network. Cell. Mol. Bioeng. 2017, 10, 16–29. [Google Scholar] [CrossRef]
- Schoeman, R.M.; Rana, K.; Danes, N.; Lehmann, M.; di Paola, J.A.; Fogelson, A.L.; Leiderman, K.; Neeves, K.B. A microfluidic model of hemostasis sensitive to platelet function and coagulation. Cell. Mol. Bioeng. 2017, 10, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ku, D.N.; Forest, C.R. Microfluidic system for simultaneous optical measurement of platelet aggregation at multiple shear rates in whole blood. Lab. Chip 2012, 12, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Aman, Z.M.; Olcott, K.; Pfeiffer, K.; Sloan, E.D.; Sum, A.K.; Koh, C.A. Surfactant adsorption and interfacial tension investigations on cyclopentane hydrate. Langmuir 2013, 29, 2676–2682. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Chuah, Y.J.; Koh, Y.T.; Lim, K.; Menon, N.V.; Wu, Y.; Kang, Y. Simple surface engineering of polydimethylsiloxane with polydopamine for stabilized mesenchymal stem cell adhesion and multipotency. Sci. Rep. 2015, 5, 18162. [Google Scholar] [CrossRef]
- Bright, L.K.; Baker, C.A.; Agasid, M.T.; Ma, L.; Craig, A. Decreased aperture surface energy enhances electrical, mechanical, and temporal stability of suspended lipid membranes. ACS Appl. Mater. Interfaces 2014, 5, 11918–11926. [Google Scholar] [CrossRef][Green Version]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef]
- Regehr, K.J.; Domenech, M.; Koepsel, J.T.; Carver, K.C.; Ellison-Zelski, S.J.; Murphy, W.L.; Schuler, L.A.; Alarid, E.T.; Beebe, D.J. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009, 9, 2132–2139. [Google Scholar] [CrossRef]
- Wang, J.D.; Douville, N.J.; Takayama, S.; Elsayed, M. Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann. Biomed. Eng. 2012, 40, 1862–1873. [Google Scholar] [CrossRef]
- Zhong, M.; Lee, C.Y.; Croushore, C.A.; Sweedler, J.V. Label-free quantitation of peptide release from neurons in a microfluidic device with mass spectrometry imaging. Lab Chip 2012, 12, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.J.; Payne, M.C.; Ciacchi, L.C. Water structuring and collagen adsorption at hydrophilic and hydrophobic silicon surfaces. Phys. Chem. Chem. Phys. 2009, 11, 11395–11399. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorrells, M.G.; Neeves, K.B. Adsorption and Absorption of Collagen Peptides to Polydimethlysiloxane and Its Influence on Platelet Adhesion Flow Assays. Micromachines 2020, 11, 62. https://doi.org/10.3390/mi11010062
Sorrells MG, Neeves KB. Adsorption and Absorption of Collagen Peptides to Polydimethlysiloxane and Its Influence on Platelet Adhesion Flow Assays. Micromachines. 2020; 11(1):62. https://doi.org/10.3390/mi11010062
Chicago/Turabian StyleSorrells, Matthew G., and Keith B. Neeves. 2020. "Adsorption and Absorption of Collagen Peptides to Polydimethlysiloxane and Its Influence on Platelet Adhesion Flow Assays" Micromachines 11, no. 1: 62. https://doi.org/10.3390/mi11010062
APA StyleSorrells, M. G., & Neeves, K. B. (2020). Adsorption and Absorption of Collagen Peptides to Polydimethlysiloxane and Its Influence on Platelet Adhesion Flow Assays. Micromachines, 11(1), 62. https://doi.org/10.3390/mi11010062




