Graphene-Based Biosensors for Detection of Biomarkers
Abstract
1. Introduction
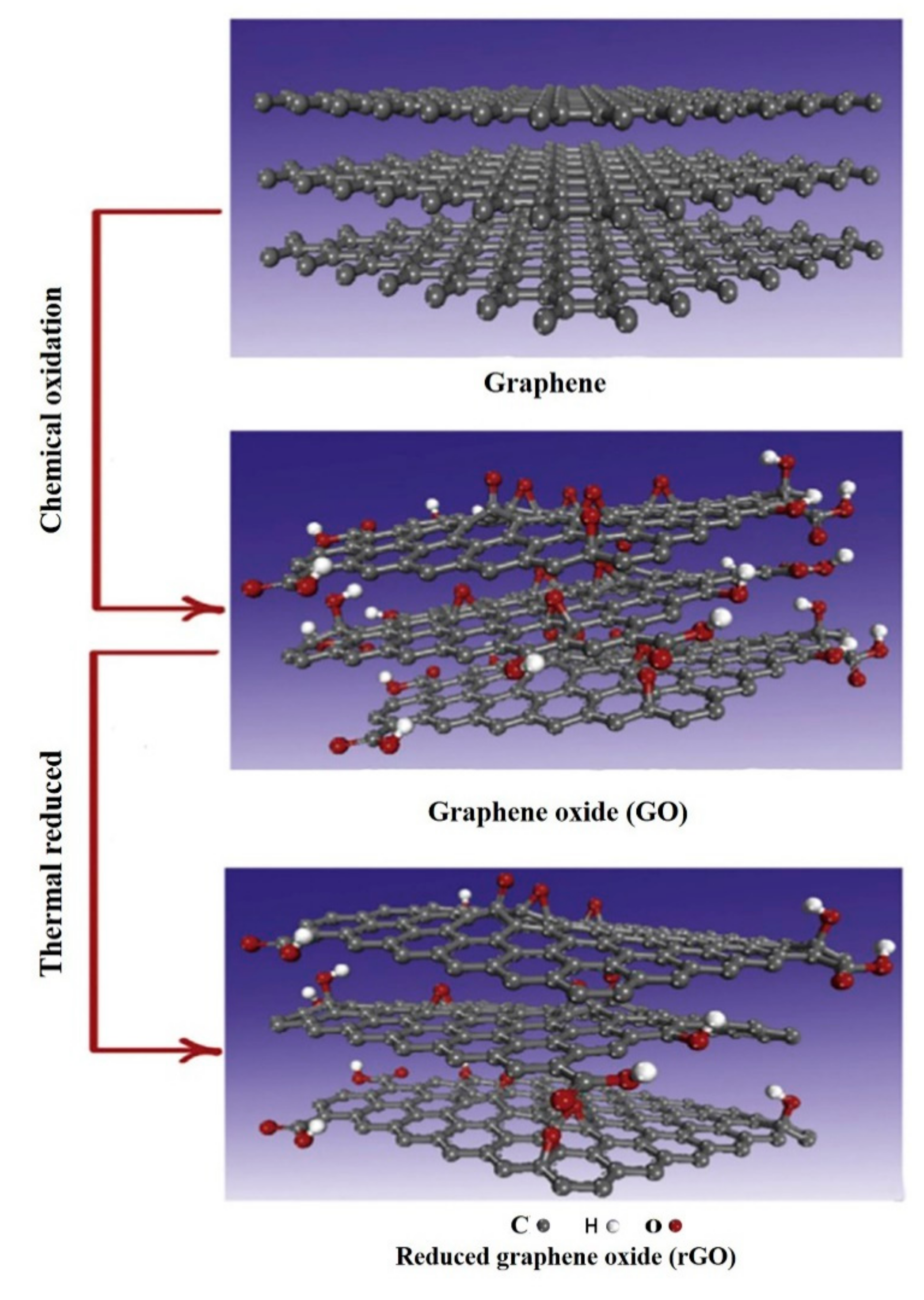
2. Characteristics and Classification of Graphene
- (1)
- High specific surface area. 2630 m2/g for single-layer graphene theoretically, which gives rise to high densities of attached recognition component or analyte molecules. It contributes to high detection sensitivity and the miniaturization of the device.
- (2)
- Excellent electronic properties and electron transport capabilities. The carbon atoms of graphene hybridized in the form of sp2 constitute a huge π–π conjugate system in which the electrons are freely moving. These properties make graphene a candidate in the field of electrochemical sensing.
- (3)
- Strong mechanical strength and pliability. Single-layer graphene possesses a thickness of ~0.335 nm, the hardness of which is higher than diamond due to strong C=C bonding in the atom plane; while opposite to diamond, the interlayer bonding via Van der Waals forces makes it a soft material. This will greatly benefit the development of wearable sensor devices.
3. Applications of Different Types of Graphene-Based Biosensors
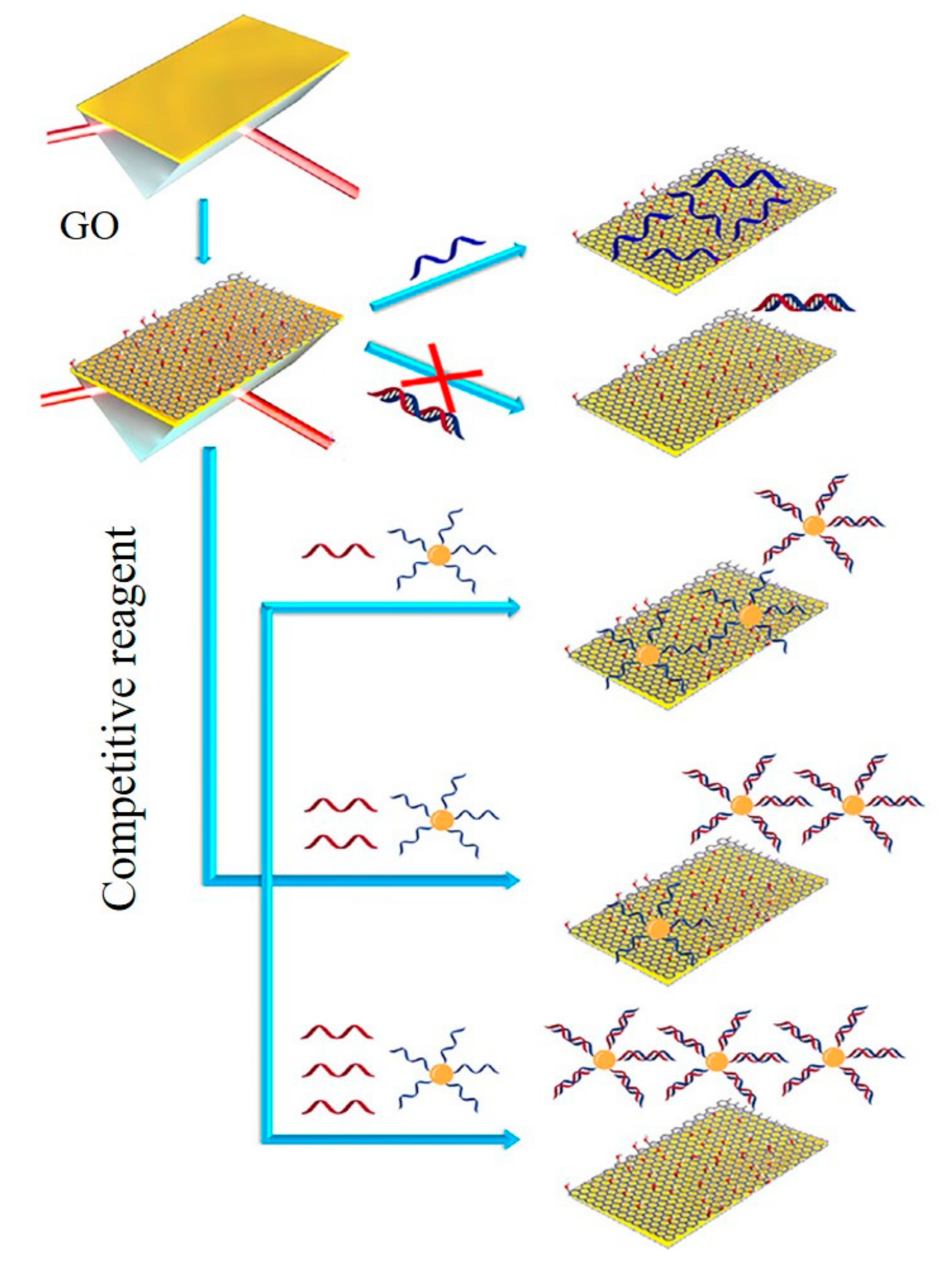
3.1. Graphene-Based Fluorescent Biosensors
3.2. Graphene-Based Electrochemical Biosensors
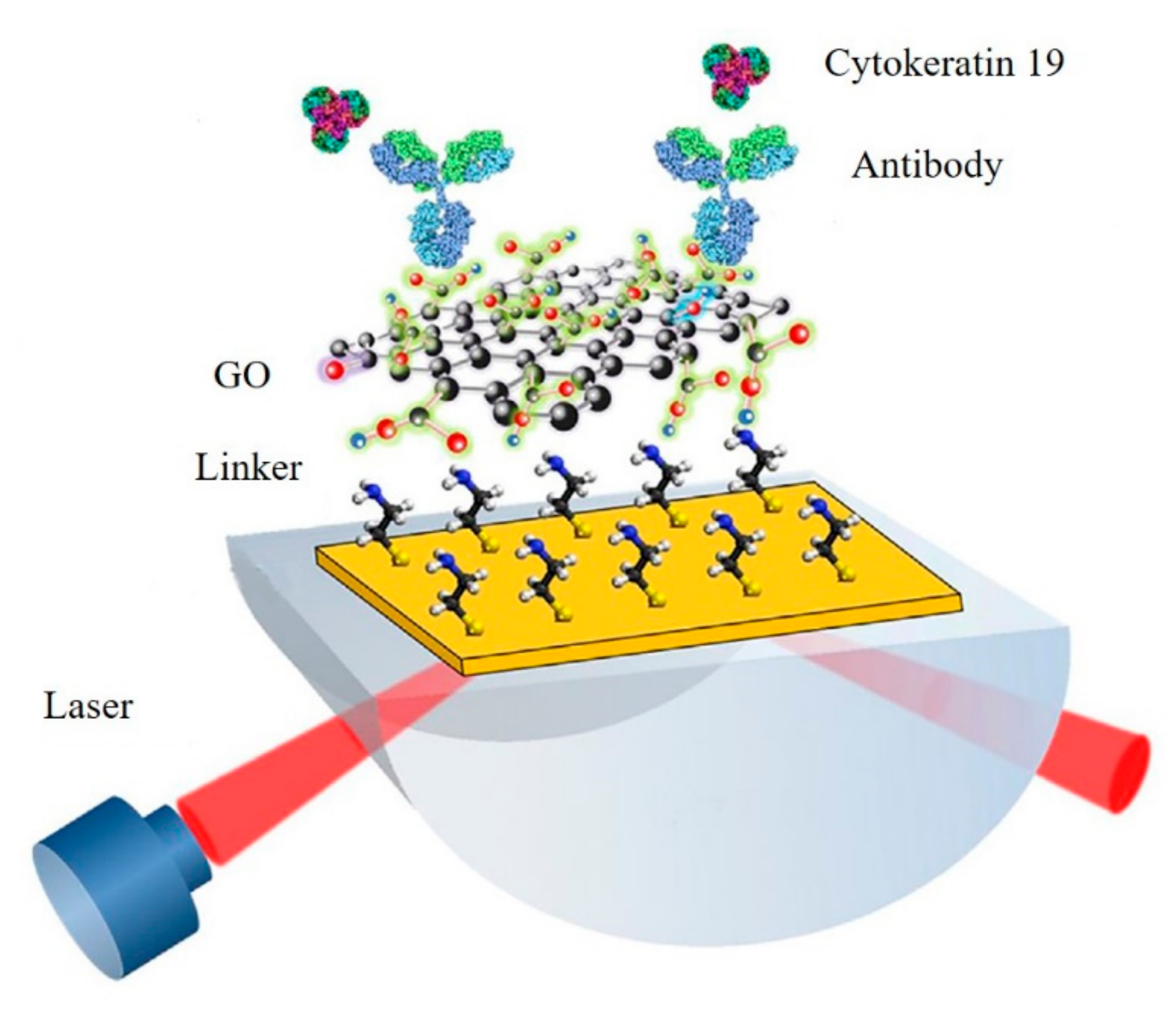
3.3. Graphene-Based SPR Biosensors
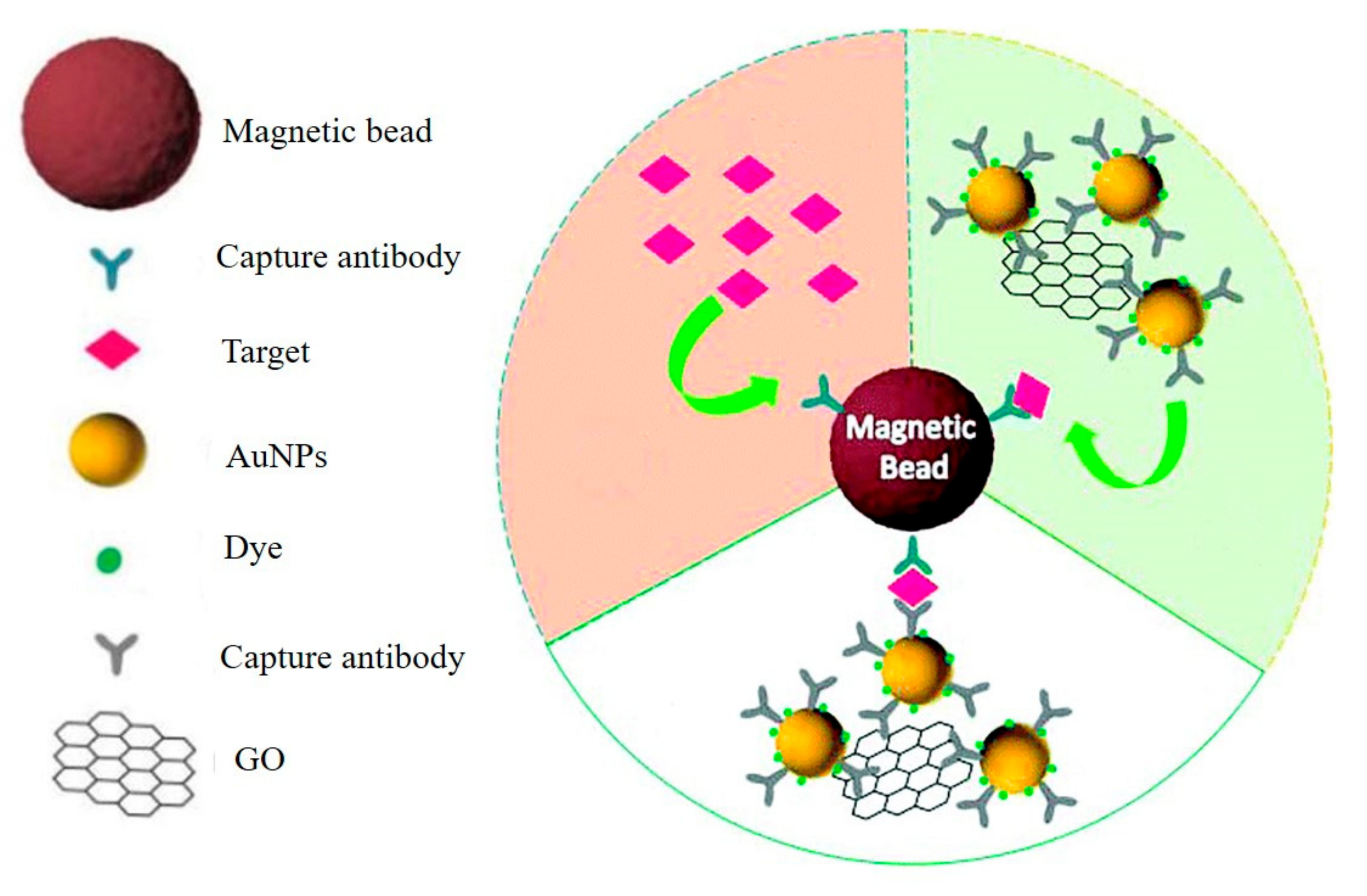
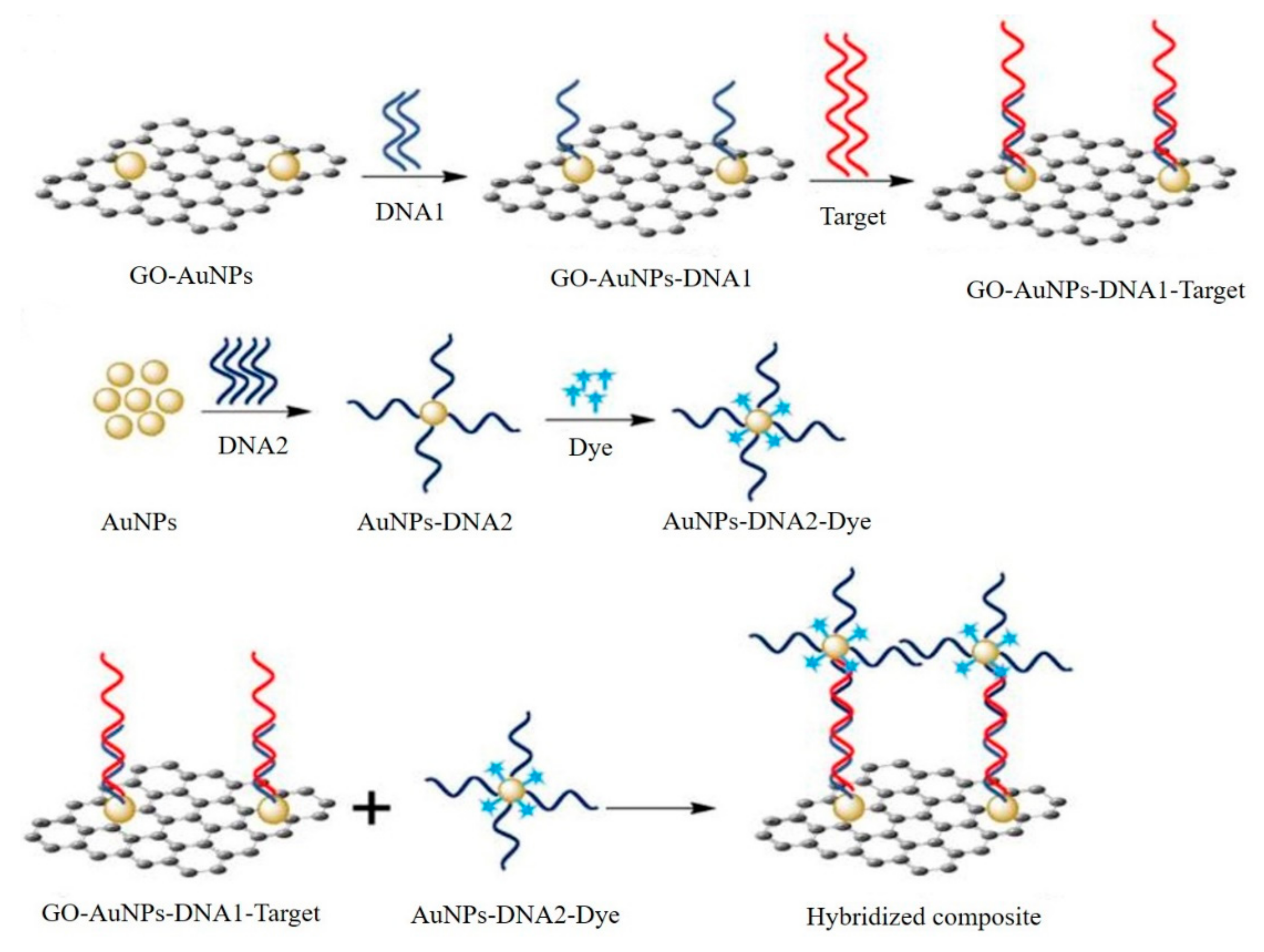
3.4. Graphene-Based SERS Biosensors
3.5. Graphene-Based Multifunction Detection Sensor
4. Conclusions and Outlooks
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Liu, H.-W.; Chen, L.; Xu, C.; Li, Z.; Zhang, H.; Zhang, X.-B.; Tan, W. Recent progresses in small-molecule enzymatic fluorescent probes for cancer imaging. Chem. Soc. Rev. 2018, 47, 7140–7180. [Google Scholar] [CrossRef]
- Ye, F.; Zhao, Y.; Ei-Sayed, R.; Muhammed, M.; Hassan, M. Advances in nanotechnology for cancer biomarkers. Nano Today 2018, 18, 103–123. [Google Scholar] [CrossRef]
- Perfezou, M.; Turner, A.; Merkoci, A. Cancer detection using nanoparticle-based sensors. Chem. Soc. Rev. 2012, 41, 2606–2622. [Google Scholar] [CrossRef]
- Miquel-Cases, A.; Schouten, P.C.; Steuten, L.M.; Retel, V.P.; Linn, S.C.; van Harten, W.H. (Very) Early technology assessment and translation of predictive biomarkers in breast cancer. Cancer Treat. Rev. 2017, 52, 117–127. [Google Scholar] [CrossRef]
- Jayanthi, V.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef]
- Xu, T.; Scafa, N.; Xu, L.-P.; Su, L.; Li, C.; Zhou, S.; Liu, Y.; Zhang, X. Electrochemical Sensors for Nitric Oxide Detection in Biological Applications. Electroanalysis 2014, 26, 449–468. [Google Scholar] [CrossRef]
- Huang, R.; He, N.; Li, Z. Recent progresses in DNA nanostructure-based biosensors for detection of tumor markers. Biosens. Bioelectron. 2018, 109, 27–34. [Google Scholar] [CrossRef]
- Xu, T.; Scafa, N.; Xu, L.P.; Zhou, S.; Abdullah Al-Ghanem, K.; Mahboob, S.; Fugetsu, B.; Zhang, X. Electrochemical hydrogen sulfide biosensors. Analyst 2016, 141, 1185–1195. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Costa-Rama, E.; Fernandez-Abedul, M.T.; Merkoci, A.; Manz, A.; Urban, G.A.; Guder, F. Disposable Sensors in Diagnostics, Food, and Environmental Monitoring. Adv. Mater. 2019, 31, e1806739. [Google Scholar] [CrossRef]
- De La Franier, B.; Thompson, M. Early stage detection and screening of ovarian cancer: A research opportunity and significant challenge for biosensor technology. Biosens. Bioelectron. 2019, 135, 71–81. [Google Scholar] [CrossRef]
- Pirsaheb, M.; Mohammadi, S.; Salimi, A. Current advances of carbon dots based biosensors for tumor marker detection, cancer cells analysis and bioimaging. Trac Trends Anal. Chem. 2019, 115, 83–99. [Google Scholar] [CrossRef]
- Xu, T.; Xu, L.-P.; Zhang, X.; Wang, S. Bioinspired superwettable micropatterns for biosensing. Chem. Soc. Rev. 2019, 48, 3153–3165. [Google Scholar] [CrossRef]
- Georgakilas, V.; Otyepka, M.; Bourlinos, A.B.; Chandra, V.; Kim, N.; Kemp, K.C.; Hobza, P.; Zboril, R.; Kim, K.S. Functionalization of Graphene: Covalent and Non-Covalent Approaches, Derivatives and Applications. Chem. Rev. 2012, 112, 6156–6214. [Google Scholar] [CrossRef]
- Kuila, T.; Bose, S.; Khanra, P.; Mishra, A.K.; Kim, N.H.; Lee, J.H. Recent advances in graphene-based biosensors. Biosens. Bioelectron. 2011, 26, 4637–4648. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.A.; Lin, Y. Graphene Based Electrochemical Sensors and Biosensors: A Review. Electroanalysis 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Justino, C.I.L.; Gomes, A.R.; Freitas, A.C.; Duarte, A.C.; Rocha-Santos, T.A.P. Graphene based sensors and biosensors. Trac Trends Anal. Chem. 2017, 91, 53–66. [Google Scholar] [CrossRef]
- Lawal, A.T. Graphene-based nano composites and their applications. A review. Biosens. Bioelectron. 2019, 141, 111384. [Google Scholar] [CrossRef]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The chemistry of graphene oxide. Chem. Soc. Rev. 2010, 39, 228–240. [Google Scholar] [CrossRef]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.C.; Perman, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent Functionalization of Graphene and Graphene Oxide for Energy Materials, Biosensing, Catalytic, and Biomedical Applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar] [CrossRef]
- Chen, D.; Feng, H.; Li, J. Graphene oxide: Preparation, functionalization, and electrochemical applications. Chem. Rev. 2012, 112, 6027–6053. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ruoff, R.S. Chemical methods for the production of graphenes. Nat. Nanotechnol. 2009, 4, 217–224. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Qu, L.; Tan, Y.; Liu, F.; Wang, Y.; Zhang, W.; Cai, Z.; Mou, L.; Jiang, Y. A fluorescent aptasensor with product-triggered amplification by exonuclease III digestion for highly sensitive ATP detection. Anal. Methods 2017, 9, 4837–4842. [Google Scholar] [CrossRef]
- Pothipor, C.; Wiriyakun, N.; Putnin, T.; Ngamaroonchote, A.; Jakmunee, J.; Ounnunkad, K.; Laocharoensuk, R.; Aroonyadet, N. Highly sensitive biosensor based on graphene–poly (3-aminobenzoic acid) modified electrodes and porous-hollowed-silver-gold nanoparticle labelling for prostate cancer detection. Sens. Actuator B Chem. 2019, 296, 126657. [Google Scholar] [CrossRef]
- He, L.; Wang, Q.; Mandler, D.; Li, M.; Boukherroub, R.; Szunerits, S. Detection of folic acid protein in human serum using reduced graphene oxide electrodes modified by folic-acid. Biosens. Bioelectron. 2016, 75, 389–395. [Google Scholar] [CrossRef]
- Senthilkumar, E.; Shanmugharaj, A.M.; Suresh Babu, R.; Sivagaami Sundari, G.; Thileep Kumar, K.; Raghu, S.; Kalaivani, R. Development of constructed nanoporous graphene-modified electrode for electrical detection of folic acid. J. Mater. Sci. Mater. Electron. 2019, 30, 13488–13496. [Google Scholar] [CrossRef]
- Salahandish, R.; Ghaffarinejad, A.; Omidinia, E.; Zargartalebi, H.; Majidzadeh-A, K.; Naghib, S.M.; Sanati-Nezhad, A. Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens. Bioelectron. 2018, 120, 129–136. [Google Scholar] [CrossRef]
- Trindade, E.K.G.; Silva, B.V.M.; Dutra, R.F. A probeless and label-free electrochemical immunosensor for cystatin C detection based on ferrocene functionalized-graphene platform. Biosens. Bioelectron. 2019, 138, 111311. [Google Scholar] [CrossRef]
- Batool, R.; Akhtar, M.A.; Hayat, A.; Han, D.; Niu, L.; Ahmad, M.A.; Nawaz, M.H. A nanocomposite prepared from magnetite nanoparticles, polyaniline and carboxy-modified graphene oxide for non-enzymatic sensing of glucose. Microchim. Acta 2019, 186, 267. [Google Scholar] [CrossRef]
- Assari, P.; Rafati, A.A.; Feizollahi, A.; Joghani, R.A. An electrochemical immunosensor for the prostate specific antigen based on the use of reduced graphene oxide decorated with gold nanoparticles. Microchim. Acta 2019, 186, 484. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, X.; Li, Y.; Ren, Y.; Zhang, C.; Wang, P.; Xu, Z.; Li, X.; Chen, Z.; Dong, Y. An electrochemical sandwich immunosensor for cardiac troponin I by using nitrogen/sulfur co-doped graphene oxide modified with Au@Ag nanocubes as amplifiers. Microchim. Acta 2019, 186, 416. [Google Scholar] [CrossRef] [PubMed]
- Mazloum-Ardakani, M.; Tavakolian-Ardakani, Z.; Sahraei, N.; Moshtaghioun, S.M. Fabrication of an ultrasensitive and selective electrochemical aptasensor to detect carcinoembryonic antigen by using a new nanocomposite. Biosens. Bioelectron. 2019, 129, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.H.; Kuo, S.H.; Lin, T.Y.; Lin, C.W.; Fang, P.Y.; Yang, H.W. An electrochemical biosensor to simultaneously detect VEGF and PSA for early prostate cancer diagnosis based on graphene oxide/ssDNA/PLLA nanoparticles. Biosens. Bioelectron. 2017, 89, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, S.; Velusamy, V.; Ramaraj, S.; Chen, S.W.; Yang, T.C.K.; Balu, S.; Banks, C.E. Facile synthesis of cellulose microfibers supported palladium nanospindles on graphene oxide for selective detection of dopamine in pharmaceutical and biological samples. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 98, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, A.; Gaspar, S.; Gheorghiu, M.; David, S.; Dinca, V.; Peteu, S.; Wang, Q.; Li, M.; Boukherroub, R.; Szunerits, S. Surface Plasmon Resonance based sensing of lysozyme in serum on Micrococcus lysodeikticus-modified graphene oxide surfaces. Biosens. Bioelectron. 2017, 89, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Q.; Yang, X.; Wang, K.; Zhang, H.; Nie, W. High sensitivity surface plasmon resonance biosensor for detection of microRNA and small molecule based on graphene oxide-gold nanoparticles composites. Talanta 2017, 174, 521–526. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, B.-T. Surface plasmon resonance biosensor based on graphene oxide/silver coated polymer cladding silica fiber. Sens. Actuator B Chem. 2018, 275, 332–338. [Google Scholar] [CrossRef]
- He, L.; Pagneux, Q.; Larroulet, I.; Serrano, A.Y.; Pesquera, A.; Zurutuza, A.; Mandler, D.; Boukherroub, R.; Szunerits, S. Label-free femtomolar cancer biomarker detection in human serum using graphene-coated surface plasmon resonance chips. Biosens. Bioelectron. 2017, 89, 606–611. [Google Scholar] [CrossRef]
- Ng, S.P.; Qiu, G.; Ding, N.; Lu, X.; Wu, C.L. Label-free detection of 3-nitro-l-tyrosine with nickel-doped graphene localized surface plasmon resonance biosensor. Biosens. Bioelectron. 2017, 89, 468–476. [Google Scholar] [CrossRef]
- Chiu, N.-F.; Lin, T.-L.; Kuo, C.-T. Highly sensitive carboxyl-graphene oxide-based surface plasmon resonance immunosensor for the detection of lung cancer for cytokeratin 19 biomarker in human plasma. Sens. Actuator B Chem. 2018, 265, 264–272. [Google Scholar] [CrossRef]
- Ali, A.; Hwang, E.Y.; Choo, J.; Lim, D.W. PEGylated nanographene-mediated metallic nanoparticle clusters for surface enhanced Raman scattering-based biosensing. Analyst 2018, 143, 2604–2615. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhen, S.J.; Li, Y.F.; Huang, C.Z. Silver nanoparticles deposited on graphene oxide for ultrasensitive surface-enhanced Raman scattering immunoassay of cancer biomarker. Nanoscale 2018, 10, 11942–11947. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Yu, Z.; Yu, W.; Liu, H.; Zhang, H.; Guo, C. Split aptamer-based detection of adenosine triphosphate using surface enhanced Raman spectroscopy and two kinds of gold nanoparticles. Microchim. Acta 2019, 186, 251. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wang, Y.; Liu, Y.; Liu, H.; Fu, L.; Wen, J.; Li, J.; Wei, P.; Chen, L. A graphene oxide/gold nanoparticle-based amplification method for SERS immunoassay of cardiac troponin I. Analyst 2019, 144, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Khalil, I.; Yehye, W.A.; Julkapli, N.M.; Rahmati, S.; Ibn Sina, A.A.; Basirun, W.J.; Johan, M.R. Graphene oxide and gold nanoparticle based dual platform with short DNA probe for the PCR free DNA biosensing using surface-enhanced Raman scattering. Biosens. Bioelectron. 2019, 131, 214–223. [Google Scholar] [CrossRef]
- Ouyang, L.; Zhang, Q.; Ma, G.; Zhu, L.; Wang, Y.; Chen, Z.; Wang, Y.; Zhao, L. New Dual-Spectroscopic Strategy for the Direct Detection of Aristolochic Acids in Blood and Tissue. Anal. Chem. 2019, 91, 8154–8161. [Google Scholar] [CrossRef]
- Chinen, A.B.; Guan, C.M.; Ferrer, J.R.; Barnaby, S.N.; Merkel, T.J.; Mirkin, C.A. Nanoparticle Probes for the Detection of Cancer Biomarkers, Cells, and Tissues by Fluorescence. Chem. Rev. 2015, 115, 10530–10574. [Google Scholar] [CrossRef]
- Lu, C.-H.; Yang, H.-H.; Zhu, C.-L.; Chen, X.; Chen, G.-N. A Graphene Platform for Sensing Biomolecules. Angew. Chem. Int. Ed. 2009, 48, 4785–4787. [Google Scholar] [CrossRef]
- Wang, S.E.; Si, S. A fluorescent nanoprobe based on graphene oxide fluorescence resonance energy transfer for the rapid determination of oncoprotein vascular endothelial growth factor (VEGF). Appl. Spectrosc. 2013, 67, 1270–1274. [Google Scholar] [CrossRef]
- Chen, J.L.; Yan, X.P.; Meng, K.; Wang, S.F. Graphene oxide based photoinduced charge transfer label-free near-infrared fluorescent biosensor for dopamine. Anal. Chem. 2011, 83, 8787–8793. [Google Scholar] [CrossRef]
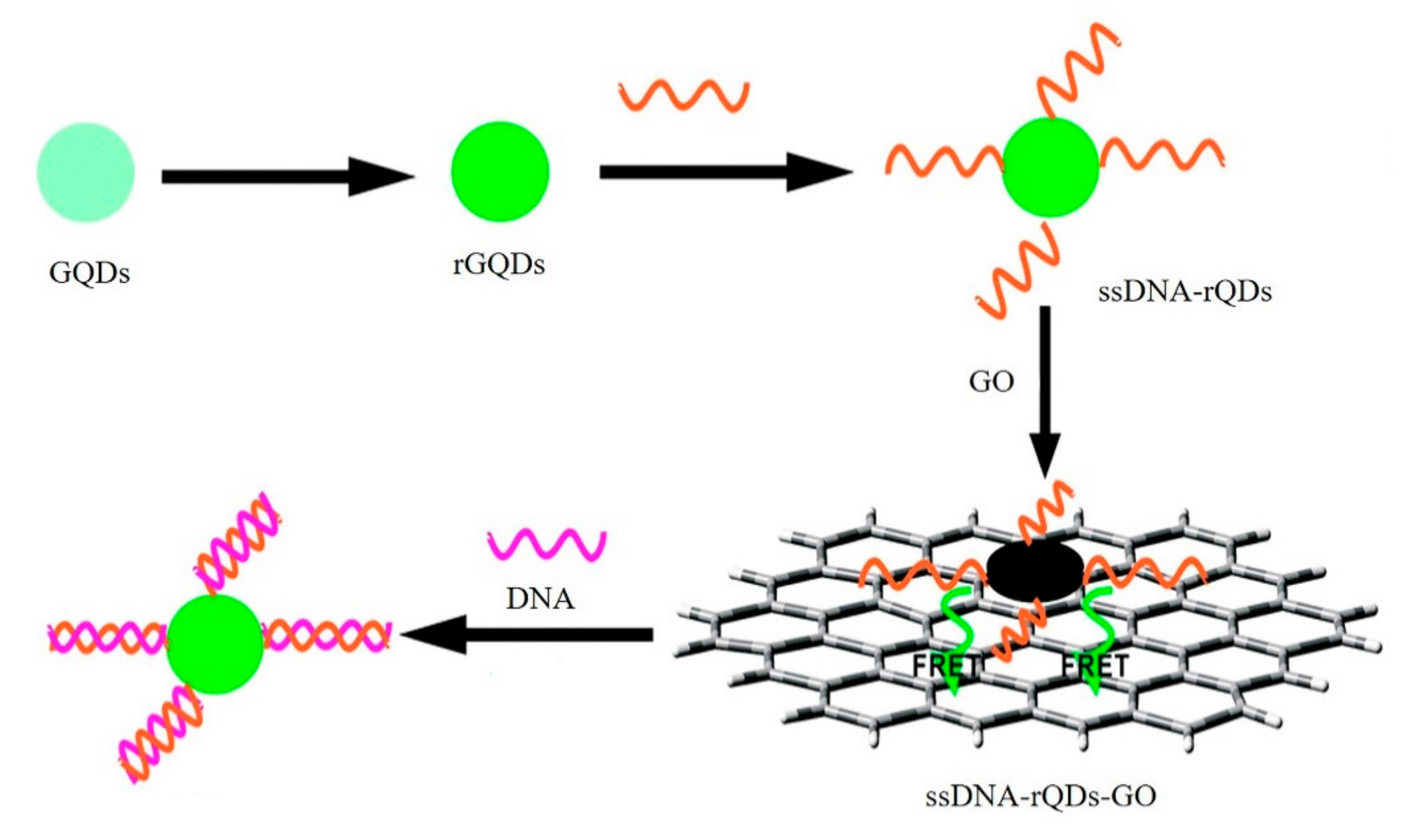
- Qian, Z.S.; Shan, X.Y.; Chai, L.J.; Ma, J.J.; Chen, J.R.; Feng, H. A universal fluorescence sensing strategy based on biocompatible graphene quantum dots and graphene oxide for the detection of DNA. Nanoscale 2014, 6, 5671–5674. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Zhan, S.; Sun, C.; Cheng, Y.; Wang, X.; Liu, B.; Zhai, T.; Lou, X.; Xia, F. Simultaneous detection of telomerase and miRNA with graphene oxide-based fluorescent aptasensor in living cells and tissue samples. Biosens. Bioelectron. 2019, 124-125, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Jameson, D.M.; Ross, J.A. Fluorescence Polarization/Anisotropy in Diagnostics and Imaging. Chem. Rev. 2010, 110, 2685–2708. [Google Scholar] [CrossRef] [PubMed]
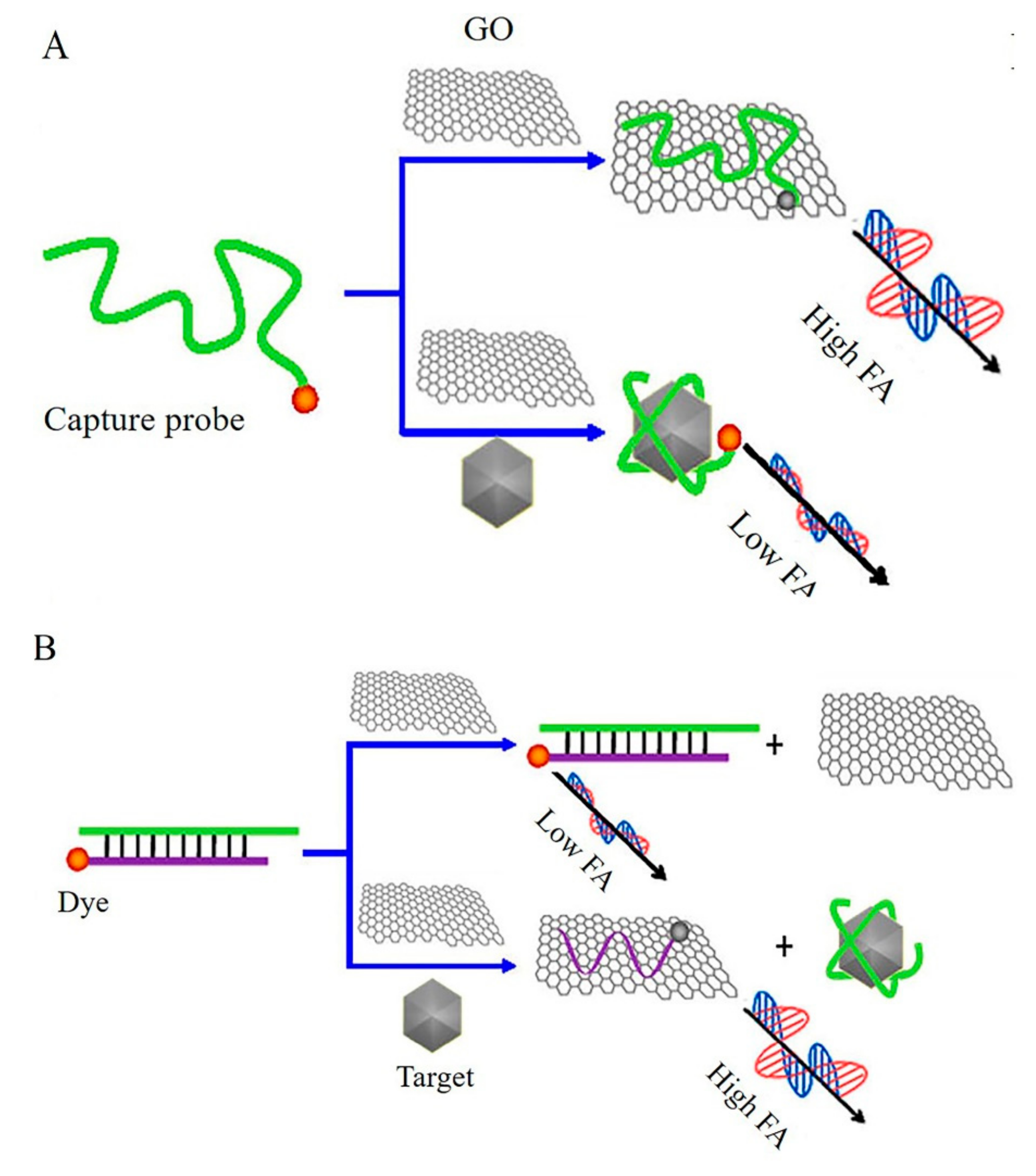
- Liu, J.; Wang, C.; Jiang, Y.; Hu, Y.; Li, J.; Yang, S.; Li, Y.; Yang, R.; Tan, W.; Huang, C.Z. Graphene Signal Amplification for Sensitive and Real-Time Fluorescence Anisotropy Detection of Small Molecules. Anal. Chem. 2013, 85, 1424–1430. [Google Scholar] [CrossRef]
- Xiao, X.; Li, Y.F.; Huang, C.Z.; Zhen, S.J. A novel graphene oxide amplified fluorescence anisotropy assay with improved accuracy and sensitivity. Chem. Commun. 2015, 51, 16080–16083. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, X.; Zhang, L.; Hu, K.; Zhao, S.; Fang, B.; Chen, Z.-F.; Liang, H. Nicking enzyme and graphene oxide-based dual signal amplification for ultrasensitive aptamer-based fluorescence polarization assays. Biosens. Bioelectron. 2015, 63, 178–184. [Google Scholar] [CrossRef]
- Wongkaew, N.; Simsek, M.; Griesche, C.; Baeumner, A.J. Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-on-a-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem. Rev. 2019, 119, 120–194. [Google Scholar] [CrossRef]
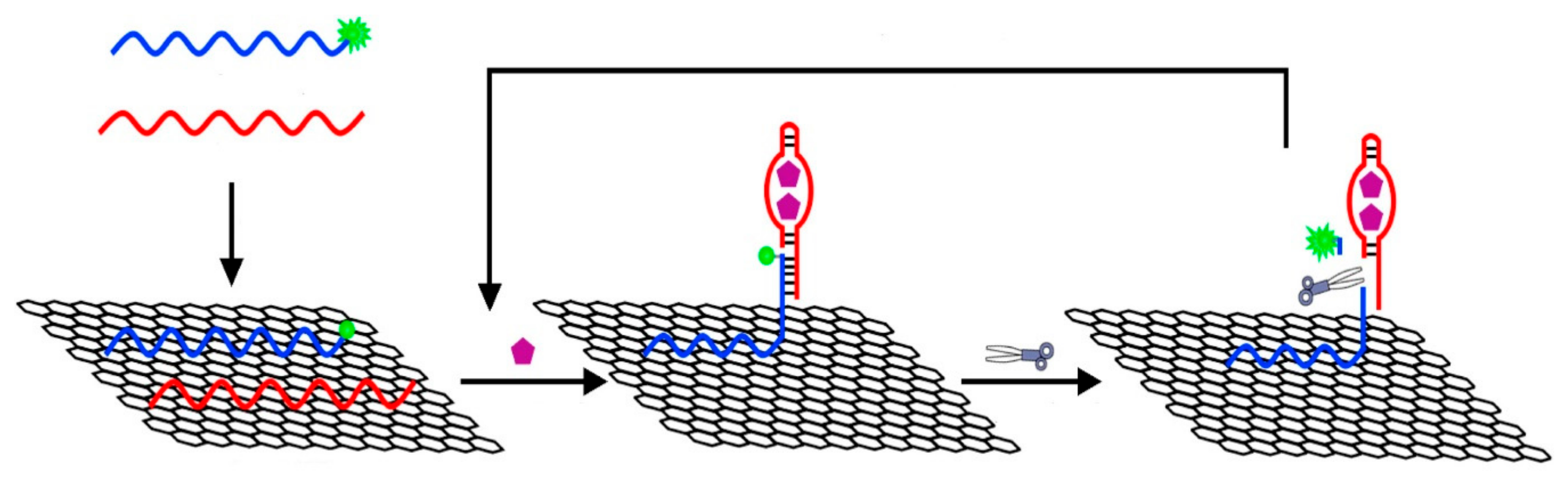
- Akhavan, O.; Ghaderi, E.; Rahighi, R. Toward single-DNA electrochemical biosensing by graphene nanowalls. ACS Nano 2012, 6, 2904–2916. [Google Scholar] [CrossRef]
- Peng, H.-P.; Hu, Y.; Liu, P.; Deng, Y.-N.; Wang, P.; Chen, W.; Liu, A.-L.; Chen, Y.-Z.; Lin, X.-H. Label-free electrochemical DNA biosensor for rapid detection of mutidrug resistance gene based on Au nanoparticles/toluidine blue–graphene oxide nanocomposites. Sens. Actuator B Chem. 2015, 207, 269–276. [Google Scholar] [CrossRef]
- Lin, C.W.; Wei, K.C.; Liao, S.S.; Huang, C.Y.; Sun, C.L.; Wu, P.J.; Lu, Y.J.; Yang, H.W.; Ma, C.C. A reusable magnetic graphene oxide-modified biosensor for vascular endothelial growth factor detection in cancer diagnosis. Biosens. Bioelectron. 2015, 67, 431–437. [Google Scholar] [CrossRef]
- Castillo, J.J.; Svendsen, W.E.; Rozlosnik, N.; Escobar, P.; Martinez, F.; Castillo-Leon, J. Detection of cancer cells using a peptide nanotube-folic acid modified graphene electrode. Analyst 2013, 138, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jia, X.; Han, J.; Ma, J.; Ma, Z. Electrochemical immunosensor for simultaneous detection of multiplex cancer biomarkers based on graphene nanocomposites. Biosens. Bioelectron. 2013, 50, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yue, R.; Ren, F.; Yao, Z.; Jiang, F.; Yang, P.; Du, Y. Novel graphene flowers modified carbon fibers for simultaneous determination of ascorbic acid, dopamine and uric acid. Biosens. Bioelectron. 2014, 53, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.O.; Pandey, G.R.; Patil, A.G.; Borse, V.B.; Deshmukh, P.K.; Patil, D.R.; Tade, R.S.; Nangare, S.N.; Khan, Z.G.; Patil, A.M.; et al. Graphene-based nanocomposites for sensitivity enhancement of surface plasmon resonance sensor for biological and chemical sensing: A review. Biosens. Bioelectron. 2019, 139, 111324. [Google Scholar] [CrossRef] [PubMed]
- Szunerits, S.; Maalouli, N.; Wijaya, E.; Vilcot, J.P.; Boukherroub, R. Recent advances in the development of graphene-based surface plasmon resonance (SPR) interfaces. Anal. Bioanal. Chem. 2013, 405, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Baillargeat, D.; Ho, H.P.; Yong, K.T. Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications. Chem. Soc. Rev. 2014, 43, 3426–3452. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, C.; Han, L.; Jin, L.; Zhou, M.; Dong, S. Label-free, regenerative and sensitive surface plasmon resonance and electrochemical aptasensors based on graphene. Chem. Commun. 2011, 47, 7794–7796. [Google Scholar] [CrossRef] [PubMed]
- Zagorodko, O.; Spadavecchia, J.; Serrano, A.Y.; Larroulet, I.; Pesquera, A.; Zurutuza, A.; Boukherroub, R.; Szunerits, S. Highly sensitive detection of DNA hybridization on commercialized graphene-coated surface plasmon resonance interfaces. Anal. Chem. 2014, 86, 11211–11216. [Google Scholar] [CrossRef]
- Xue, T.; Cui, X.; Guan, W.; Wang, Q.; Liu, C.; Wang, H.; Qi, K.; Singh, D.J.; Zheng, W. Surface plasmon resonance technique for directly probing the interaction of DNA and graphene oxide and ultra-sensitive biosensing. Biosens. Bioelectron. 2014, 58, 374–379. [Google Scholar] [CrossRef]
- Li, J.F.; Zhang, Y.J.; Ding, S.Y.; Panneerselvam, R.; Tian, Z.Q. Core-Shell Nanoparticle-Enhanced Raman Spectroscopy. Chem. Rev. 2017, 117, 5002–5069. [Google Scholar] [CrossRef]
- Wang, L.; Wu, A.; Wei, G. Graphene-based aptasensors: From molecule-interface interactions to sensor design and biomedical diagnostics. Analyst 2018, 143, 1526–1543. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Huang, S.; Deng, S.; Mao, N.; Kong, J.; Dresselhaus, M.S.; Zhang, J. Lighting up the Raman signal of molecules in the vicinity of graphene related materials. Acc. Chem. Res. 2015, 48, 1862–1870. [Google Scholar] [CrossRef] [PubMed]
- Demeritte, T.; Nellore, B.P.; Kanchanapally, R.; Sinha, S.S.; Pramanik, A.; Chavva, S.R.; Ray, P.C. Hybrid graphene oxide based plasmonic-magnetic multifunctional nanoplatform for selective separation and label-Free identification of Alzheimer’s disease biomarkers. ACS Appl. Mater. Interfaces 2015, 7, 13693–13700. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xue, T.; Li, F.; Liu, W.; Song, Y. Graphene: Diversified flexible 2D material for wearable vital signs monitoring. Adv. Mater. Technol. 2019, 4, 1800574. [Google Scholar] [CrossRef]
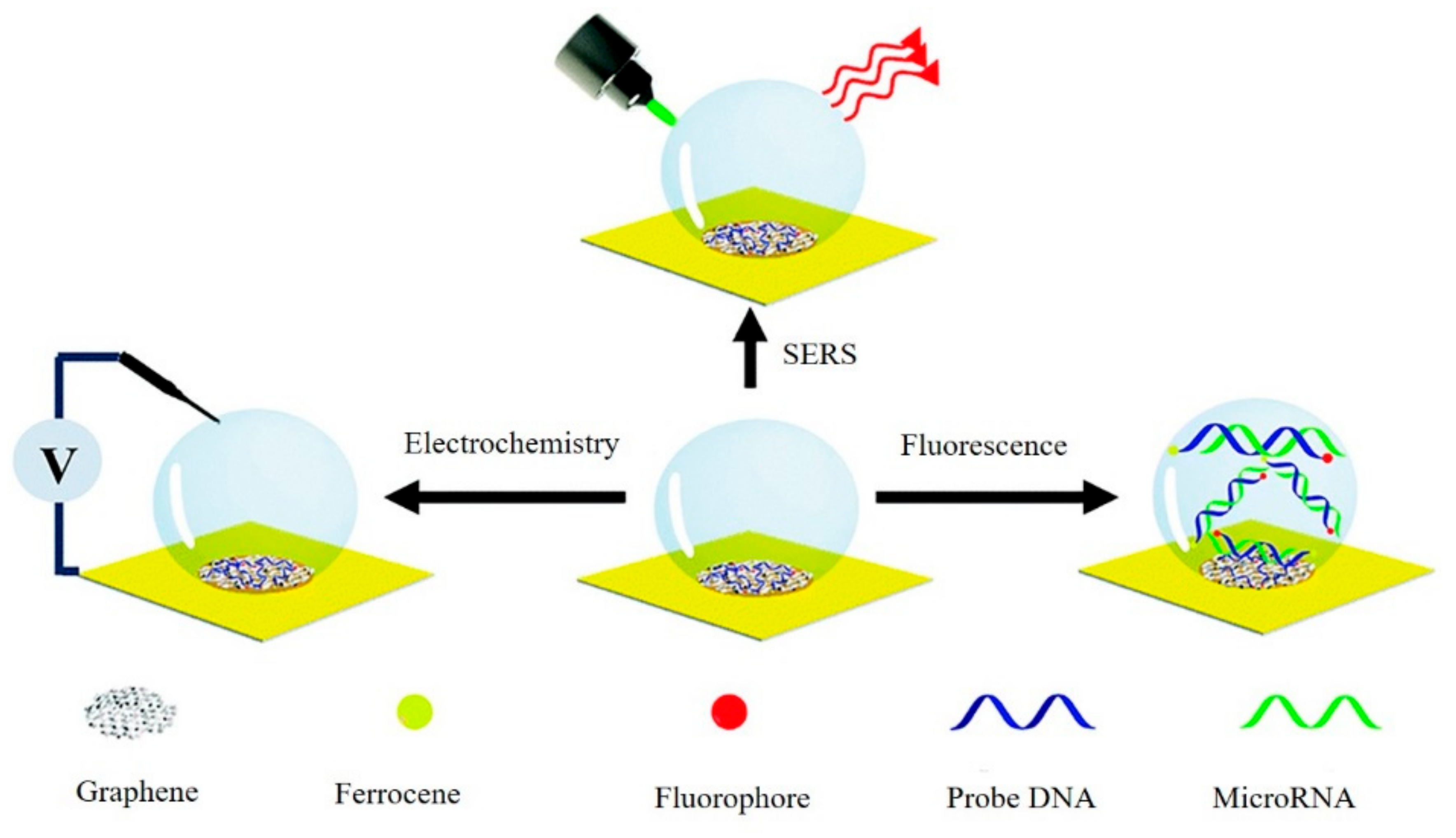
- Song, Y.; Xu, T.; Xu, L.-P.; Zhang, X. Nanodendritic gold/graphene-based biosensor for tri-mode miRNA sensing. Chem. Commun. 2019, 55, 1742–1745. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xu, T.; Wang, W.; Wen, Y.; Li, K.; Qian, L.; Zhang, X.; Liu, G. Lateral flow biosensors based on the use of micro- and nanomaterials: A review on recent developments. Microchim. Acta 2019, 187, 70. [Google Scholar] [CrossRef]


| Sensor Type | Design Method | Target | Detection Limit | Dynamic Range | Ref |
|---|---|---|---|---|---|
| Fluorescence | GO-aptamer, enzyme | ATP | 31 nM | 50–500 nM | [23] |
| Electrochemistry | Graphene-poly (3-aminobenzoic acid) | PSA | 0.13 pg | 0.01–80 ng/mL | [24] |
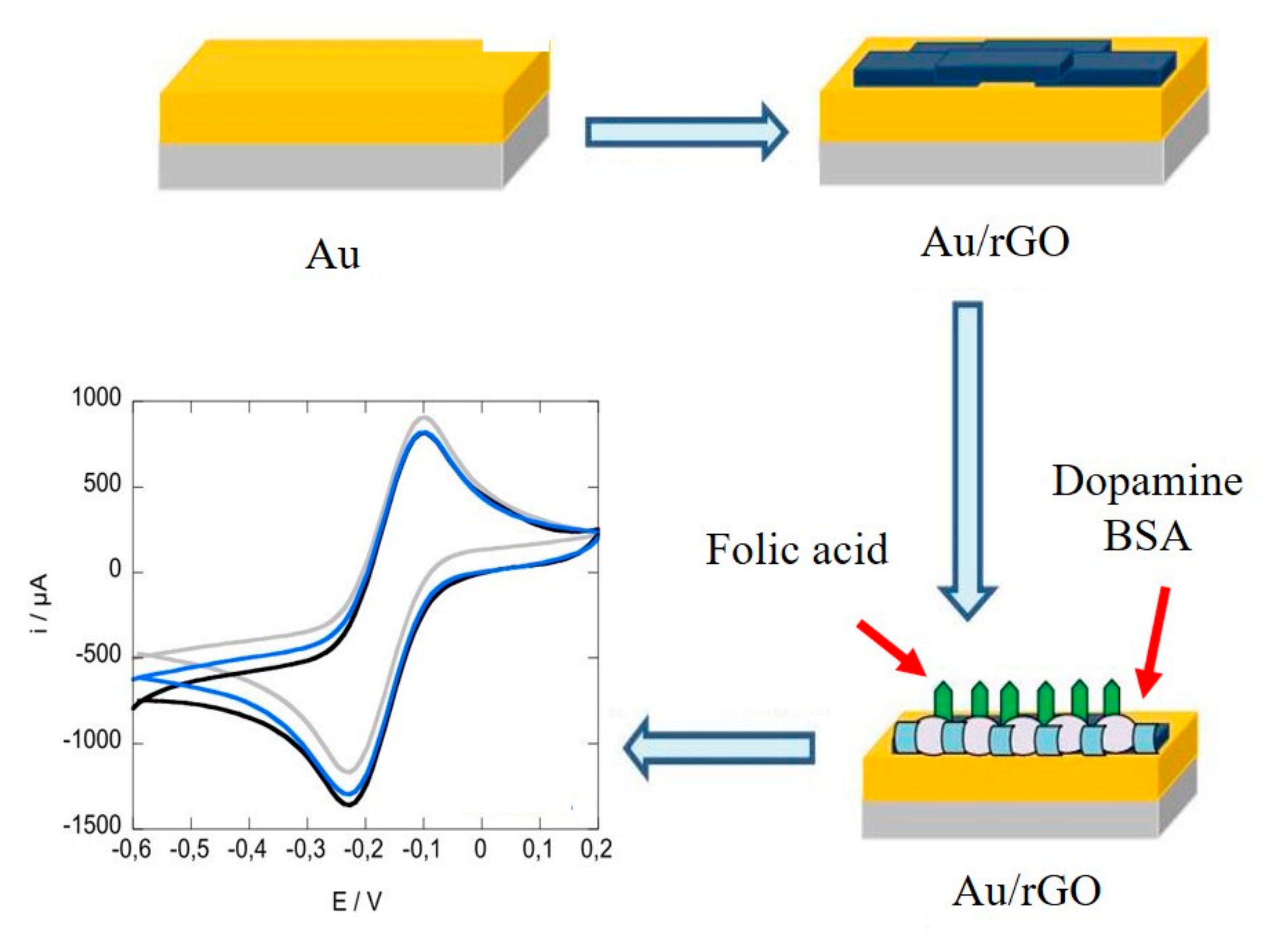
| Electrochemistry | rGO-Au | Folic acid | 1 pM | 1–200 pM | [25] |
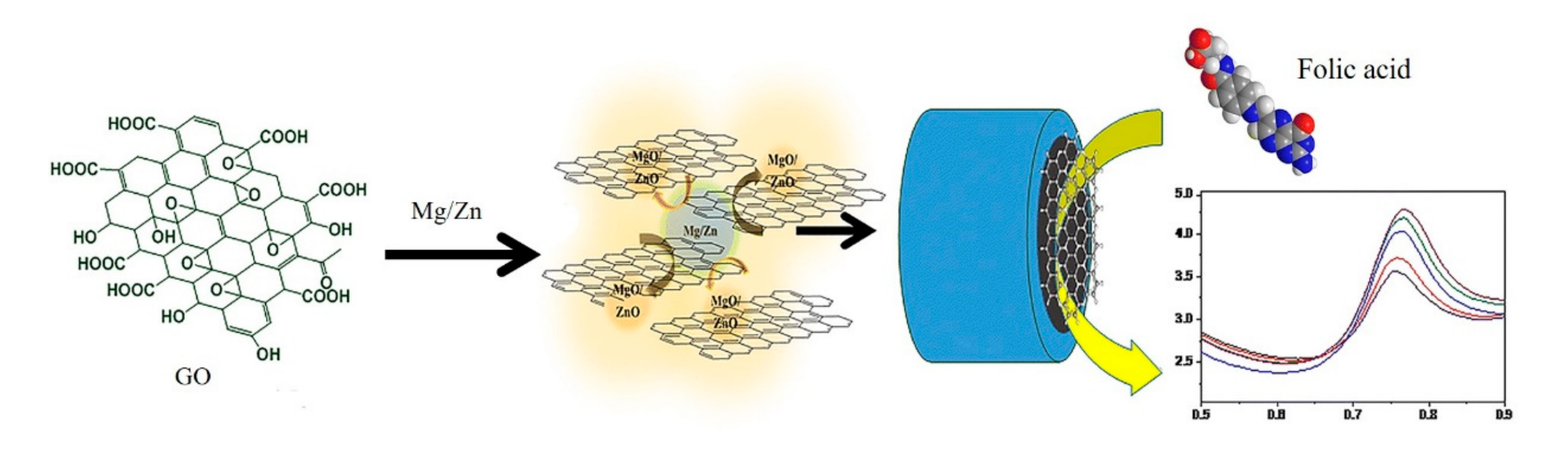
| Electrochemistry | rGO (Mg/Zn) | Folic acid | 25 nM | 0.902–8.52 μM | [26] |
| Electrochemistry | N-graphene-polyaniline AgNPs-ssDNA | MicroRNA | 0.2 fM | 10 fM–10 µM | [27] |
| Electrochemistry | Graphene-antibody | Cystatin C | 0.03 ng/mL | 0.1–1000 ng/mL | [28] |
| Electrochemistry | GO-Fe3O4 | Glucose | 0.01 μM | 0.05–50 μM | [29] |
| Electrochemistry | rGO | PSA | 2 pg/mL 0.06 ng/mL | 1–36 ng/mL 0.0018–41.15 ng/mL | [30] |
| Electrochemistry | N, S-rGO-antibody-AuNPs- AgNPs | cTnI | 33 fg/mL | 100 fg/mL–250 ng/mL | [31] |
| Electrochemistry | GO-carbon nanotubes-aptamer-hemin- | CEA | 0.82 fg/mL | 1 fg/mL–10 μg/mL | [32] |
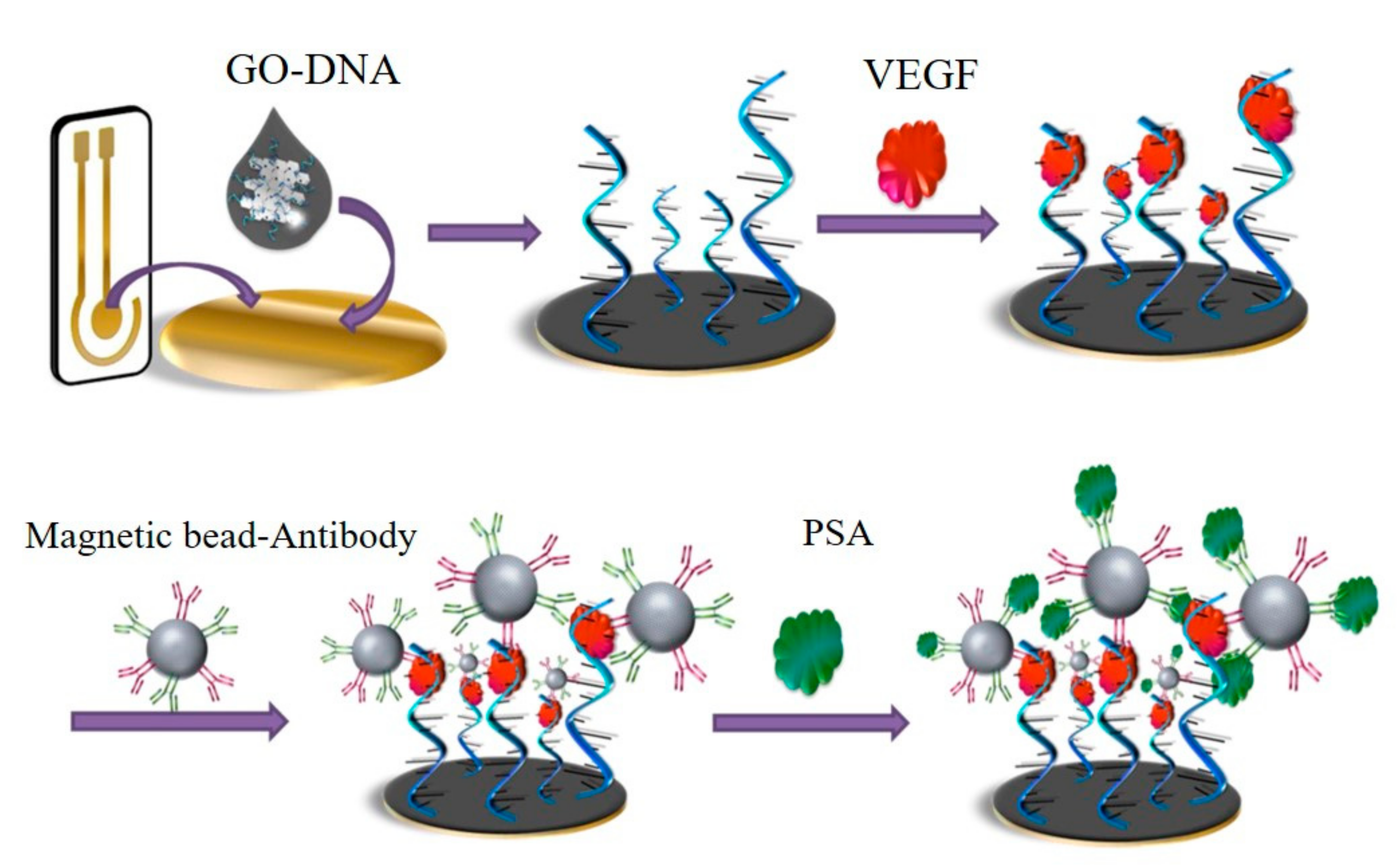
| Electrochemistry | GO-ssDNA-poly-l-lactide | VEGF PSA | 50 ng/mL 1 ng/mL | 0.05–100 ng/mL 1–100 ng/mL | [33] |
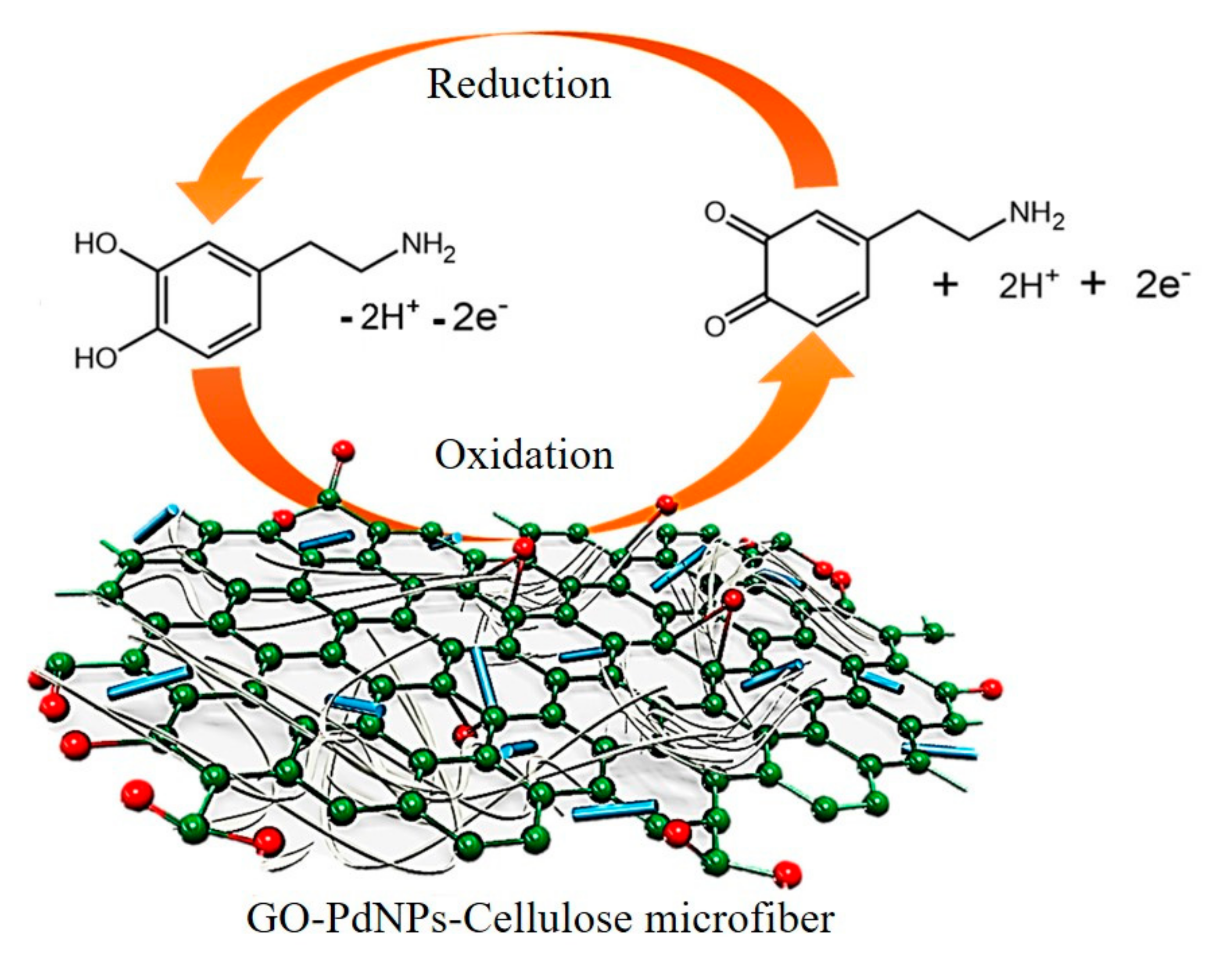
| Electrochemistry | GO-PdNPs | Dopamine | 23 nM | 0.3–196.3 μM | [34] |
| SPR | GO-bacterial | Lysozyme | 0.05 mg/mL | 0.2–40 μg/mL | [35] |
| SPR | GO-AuNPs-ss DNAGO-AuNPs-aptamer | MicroRNA Adenosine | 0.1 fM 0.1 pM | - 0.1 pM–2 nM | [36] |
| SPR | silver-GO silica fiber | IgG | 0.04 μg/mL | - | [37] |
| SPR | Graphene | Folic acid | 5 fM | 5–500 fM | [38] |
| SPR | Nickel-graphene | 3-nitro-l-tyrosine | 0.13 pg/mL | 0.5 pg/mL–1 ng/mL | [39] |
| SPR | GO-antibody | Cytokeratin 19 | 1 fg/mL | 1 fg/mL–1 ng/mL | [40] |
| SERS | GO-AuNPs-antibody-magnetic bead | IgG | 31 fM | 0.1–10,000 pM | [41] |
| SERS | GO-AgNPs-antibody | PSA | 0.23 pg/mL | 0.5–500 pg/mL | [42] |
| SERS | GO-Aptamer-Au | ATP | 0.85 pM | 10 pM–10 nM | [43] |
| SERS | GO-Au-antibody-malachite green isothiocyanate | cTnI | 5 pg/mL | 0.01–1000 ng/mL | [44] |
| SERS | GO-AgNPs-ssDNA-cy3 | ssDNA | 10 fM | 10 fM–10 μM | [45] |
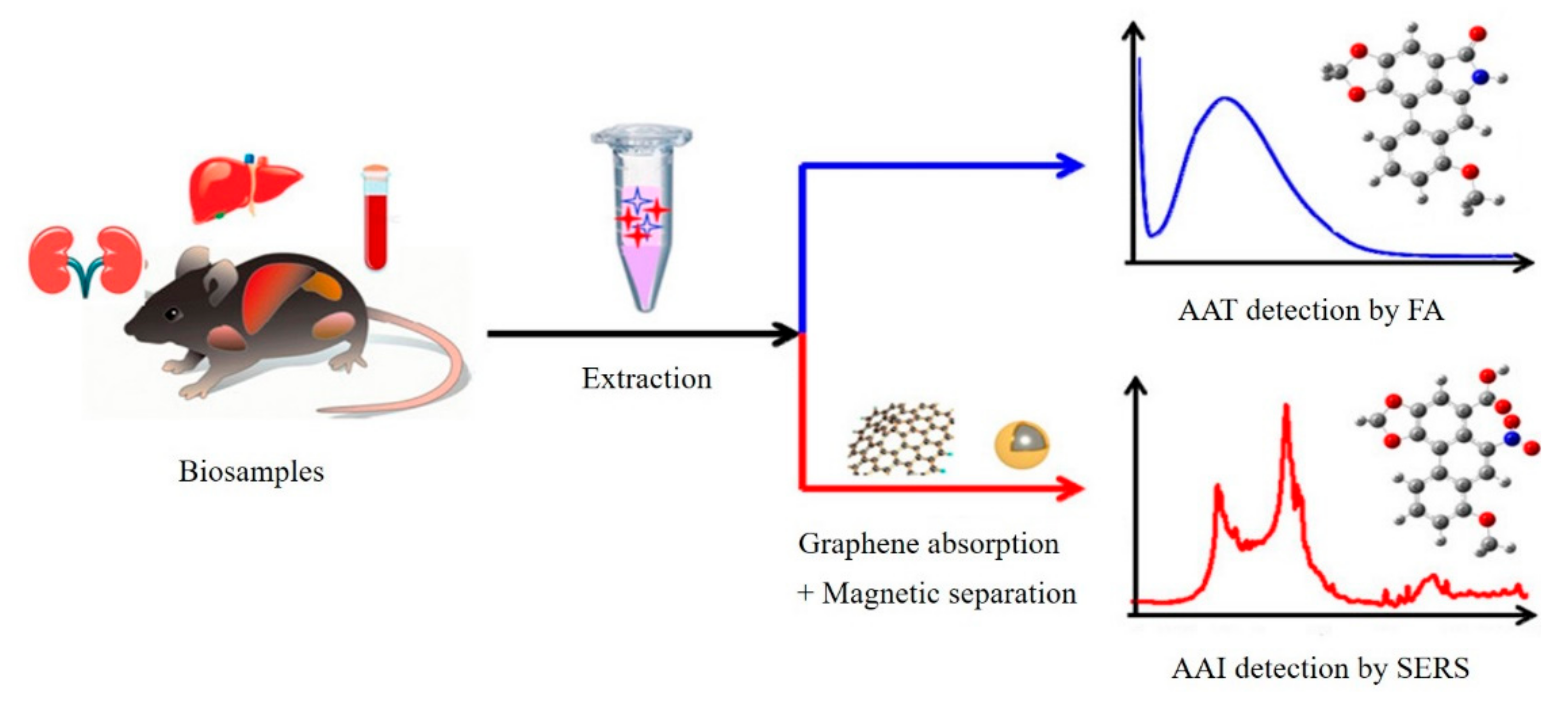
| SERS | Graphene-Fe3O4-AgNPs | Aristolochic acids | 100 ppb | 200 ppb–10 ppm | [46] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.; Xu, T.; Zhang, X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines 2020, 11, 60. https://doi.org/10.3390/mi11010060
Bai Y, Xu T, Zhang X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines. 2020; 11(1):60. https://doi.org/10.3390/mi11010060
Chicago/Turabian StyleBai, Yunlong, Tailin Xu, and Xueji Zhang. 2020. "Graphene-Based Biosensors for Detection of Biomarkers" Micromachines 11, no. 1: 60. https://doi.org/10.3390/mi11010060
APA StyleBai, Y., Xu, T., & Zhang, X. (2020). Graphene-Based Biosensors for Detection of Biomarkers. Micromachines, 11(1), 60. https://doi.org/10.3390/mi11010060







