Micromanipulation System for Isolating a Single Cryptosporidium Oocyst
Abstract
1. Introduction
- Manipulation of biological micro-objects particularly smaller than 10 m by using contact micromanipulation methods are not common as it is hard to detect 3D position of the target object and align the end effector with the target object successfully for capturing purposes. In this work, the possibility of handling biological micro-objects as small as 5 m is demonstrated.
- Manipulating a micro-object in the same liquid environment is a very common way to demonstrate the capability of micromanipulation systems. However, picking up a micro-object (particularly as small as 5 m) from one environment and transferring it to another environment is an arduous task and scarce in the literature. We demonstrate this is possible and actually crucial for some complicated tasks, e.g., oocyst isolation for DNA sequencing.
- Many studies in the literature have attempted to mitigate adhesion forces during the release of the target object by developing complex mechanisms. In this work, we show the possibility of utilizing forces at micro-scale to capture the target object and to release it without using any complicated mechanism.
2. Materials and Methods
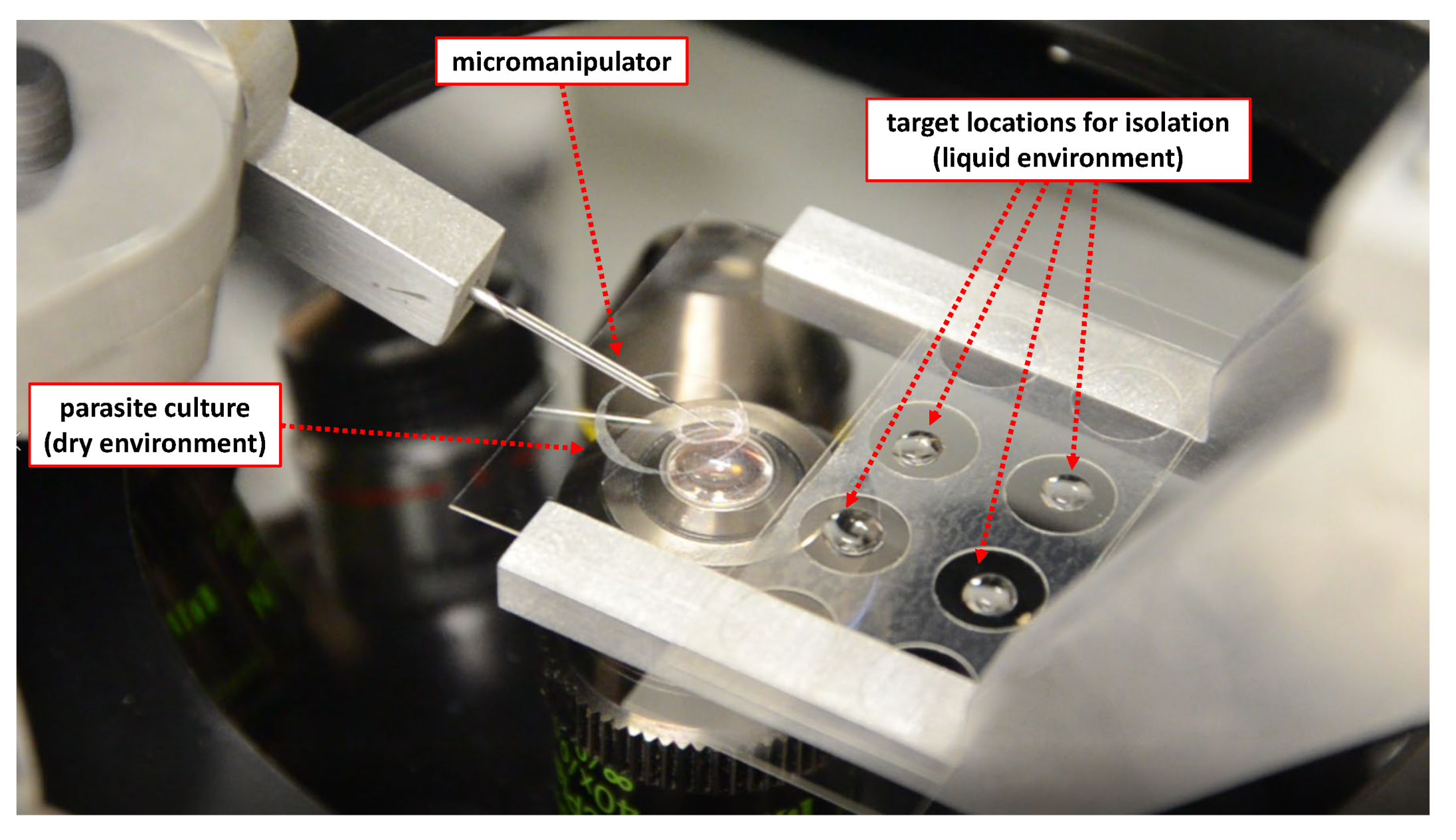
2.1. Overview
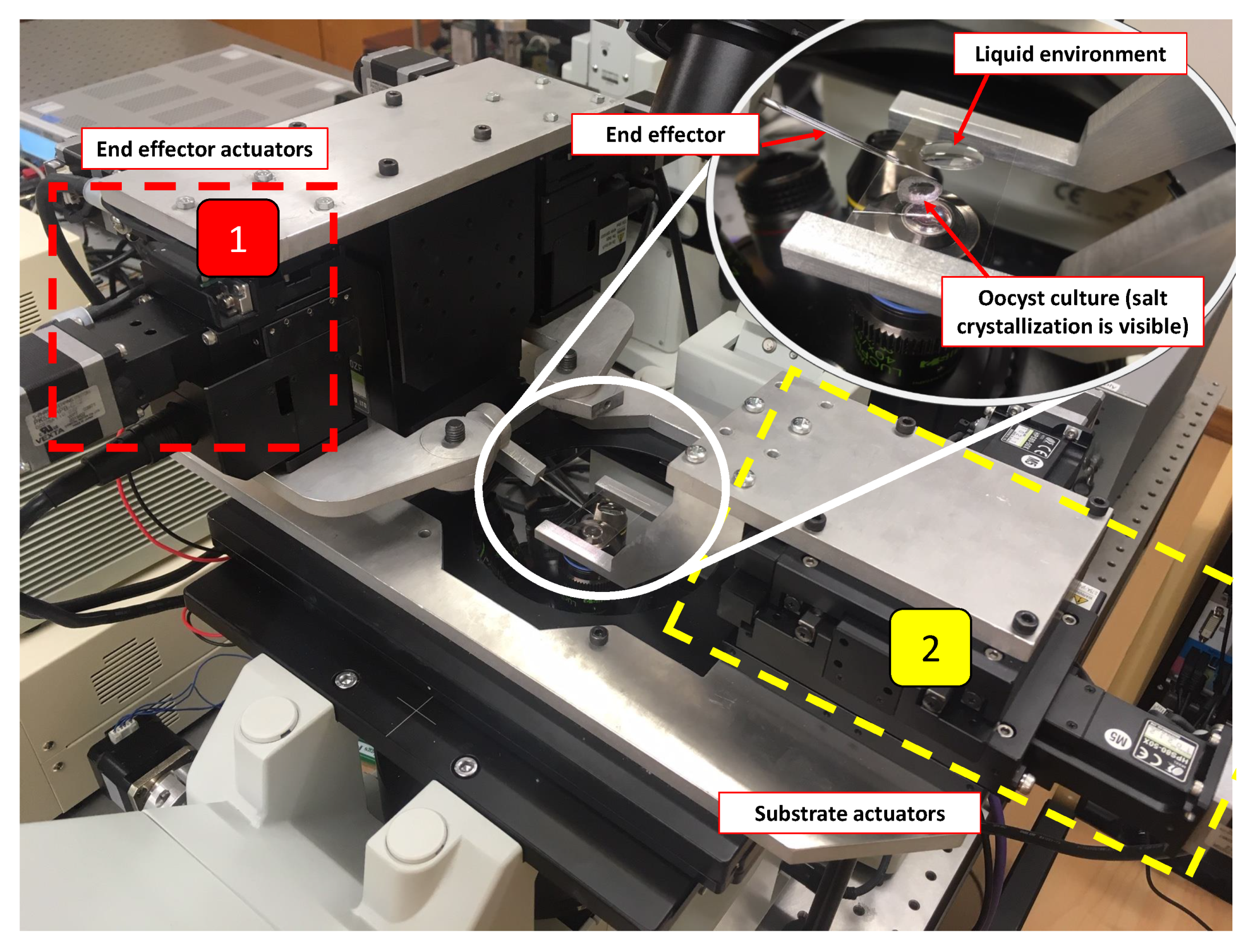
2.2. Physical System
2.3. Control System
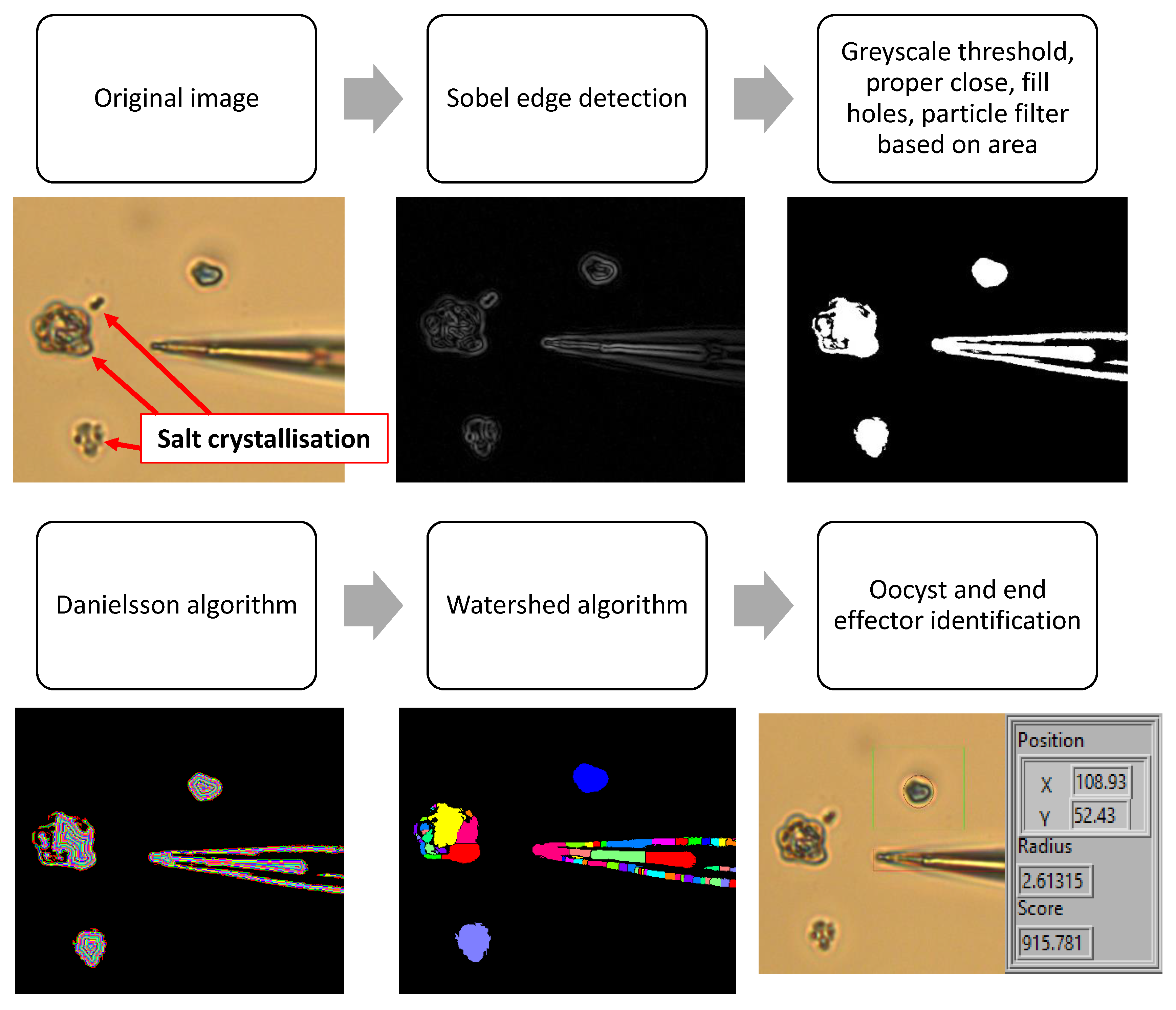
2.3.1. Object and End Effector Identification
2.3.2. 3D Location Method
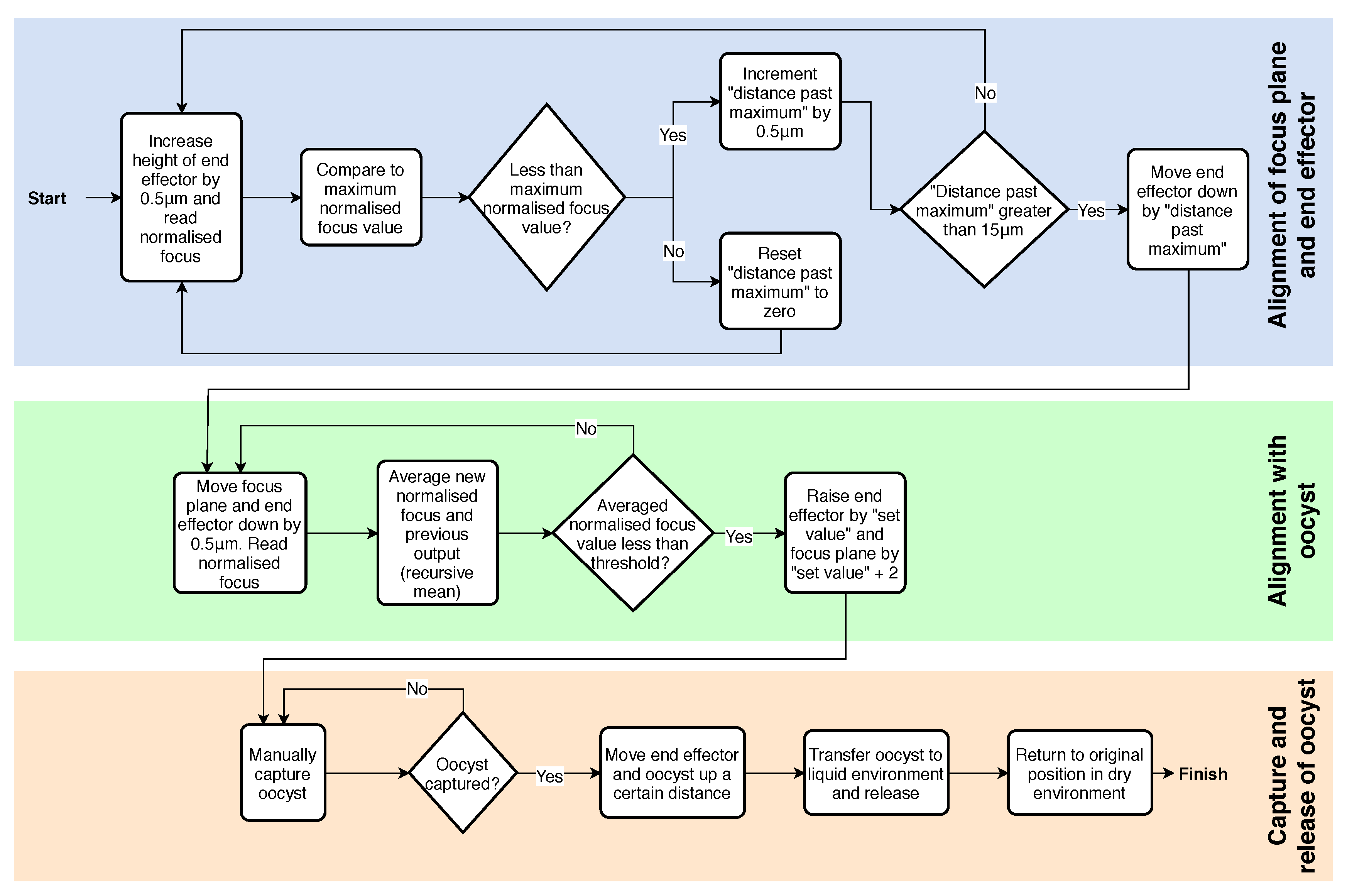
2.3.3. Control System Algorithm
2.3.4. Capture and Release Method
2.4. Statistical Analyses
3. Results
3.1. Preparing Cryptosporidium Sample
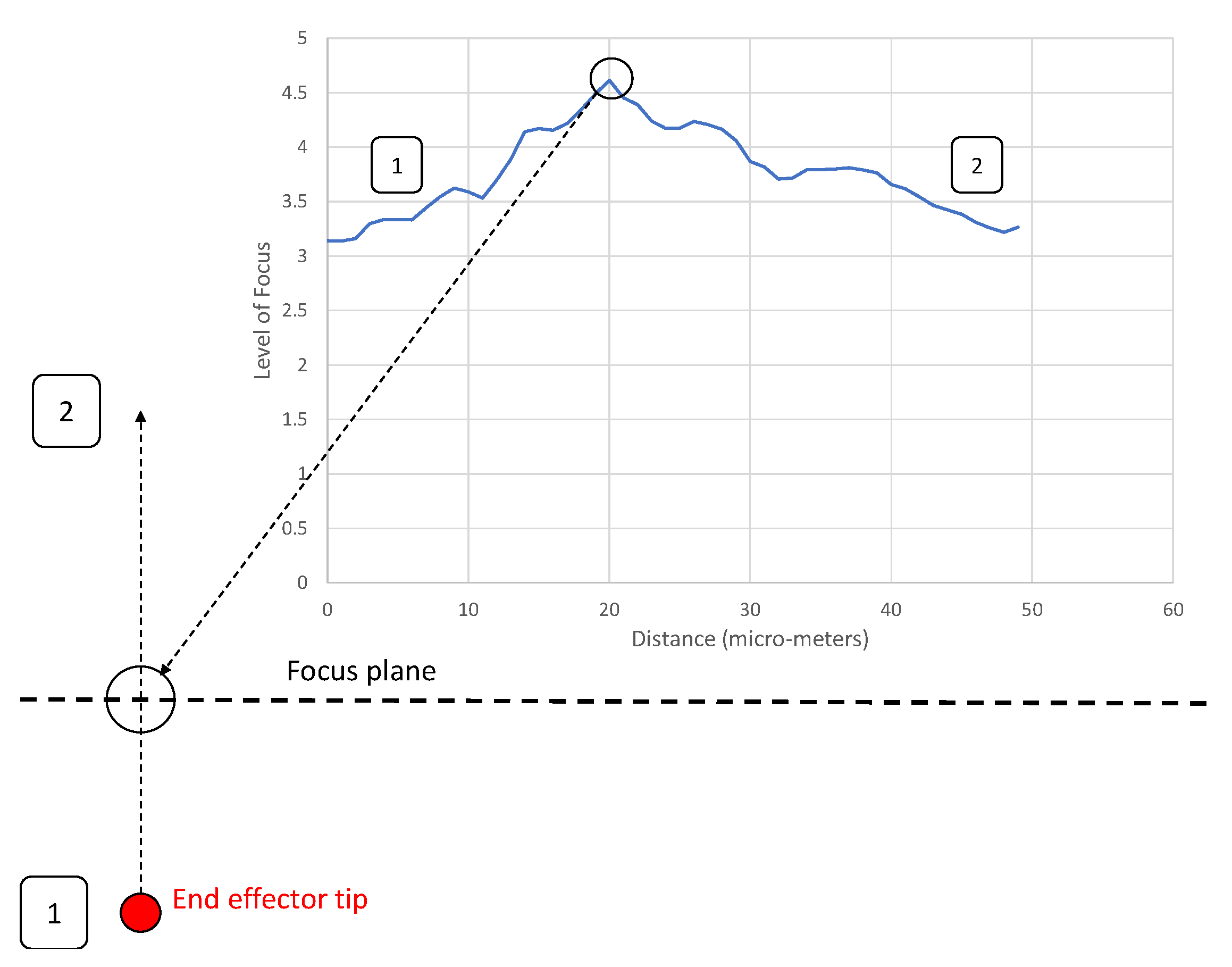
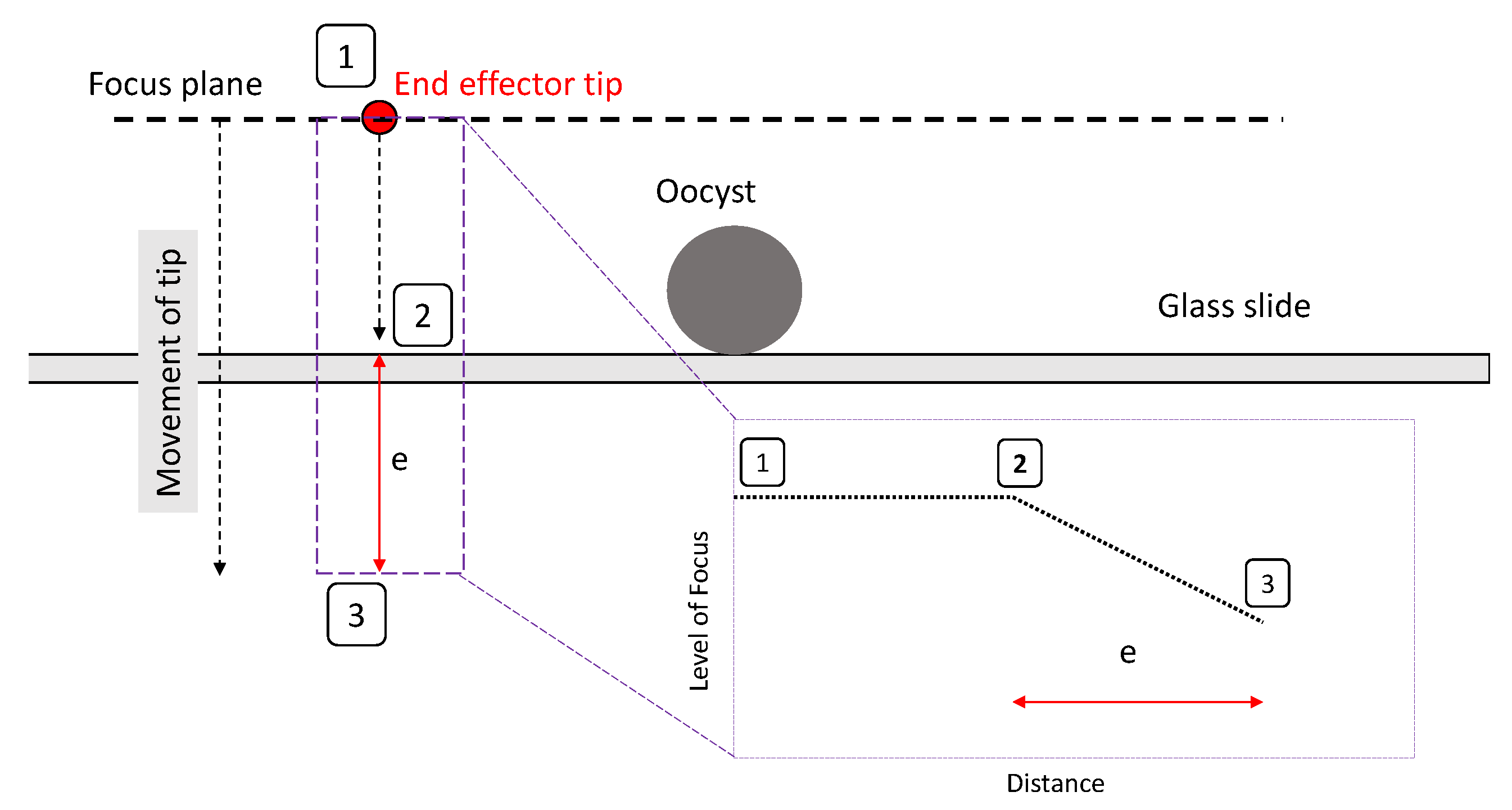
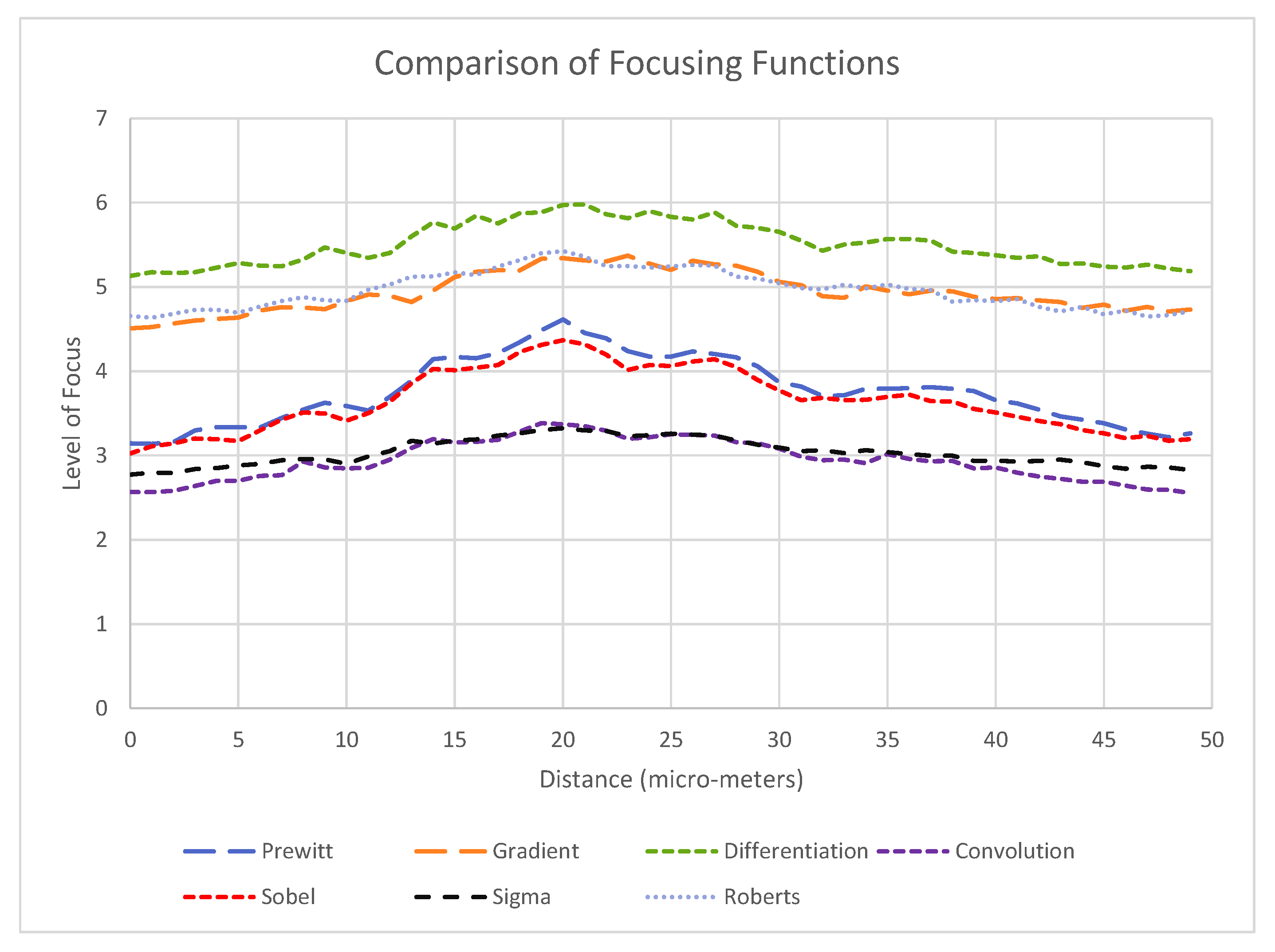
3.2. Alignment Experiments
- A clearly defined peak—it is important that the operator has one clear main peak, and that this peak is steep and narrow enough to find the center.
- An accurate peak location—to use the alignment method for micro-manipulation an accuracy of at least 2 m is required. Therefore, the peak must be in the same place each time.Given the typical oocyst diameter is 5 m and perfect alignment with the oocyst is not required (as our aim is to scrape an oocyst from the substrate), a 2 m accuracy is sufficient for the pick and place operation.
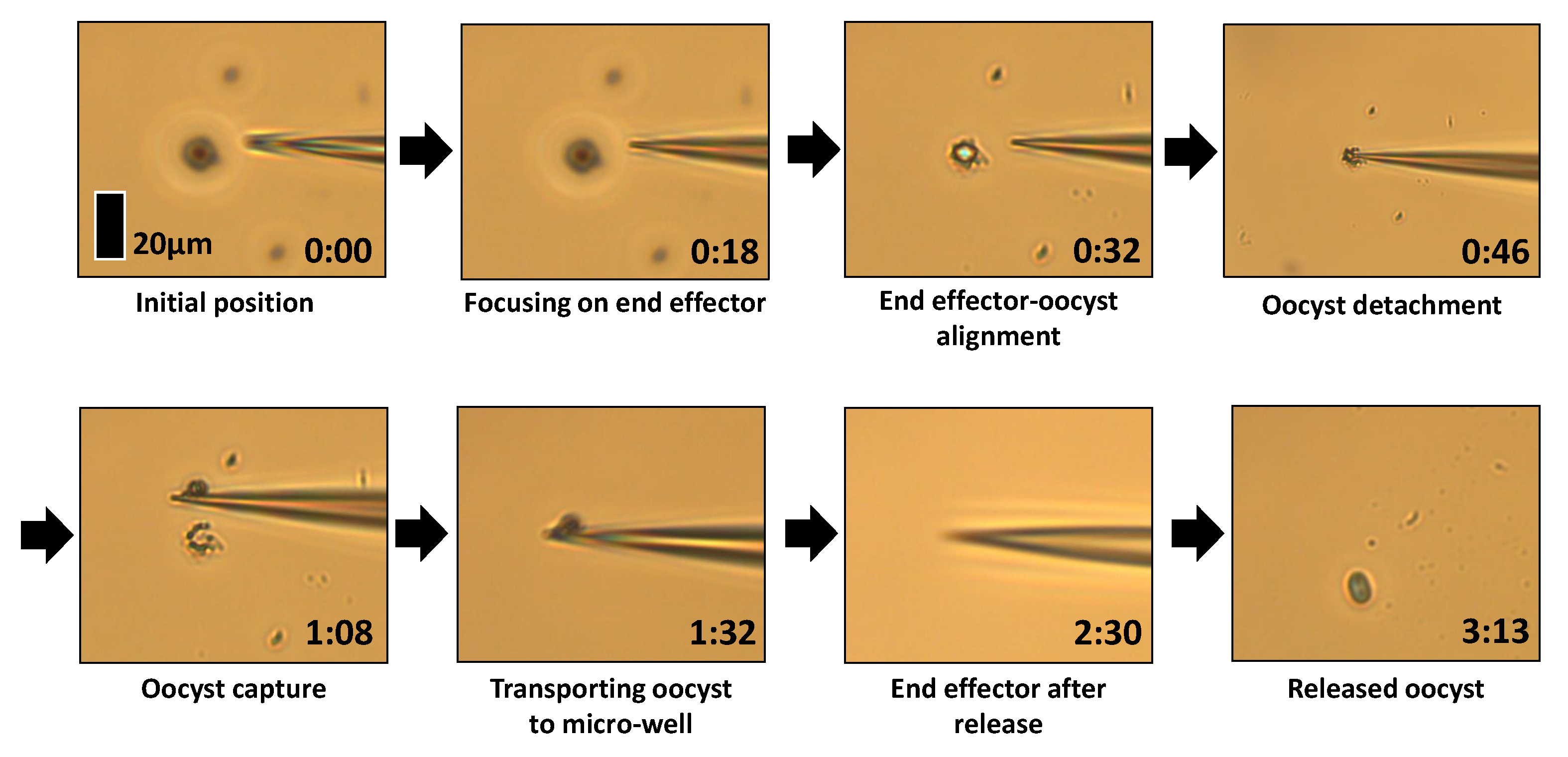
3.3. Pick and Place Reliability Experiments
- The automated system aligning the end effector and focus plane with the oocyst.
- The oocyst being manually picked from the dry substrate using the end effector.
- The end effector and oocyst automatically moving to the desired location, releasing the oocyst into an aqueous environment, and returning to the point it was initially picked from.
4. Discussion
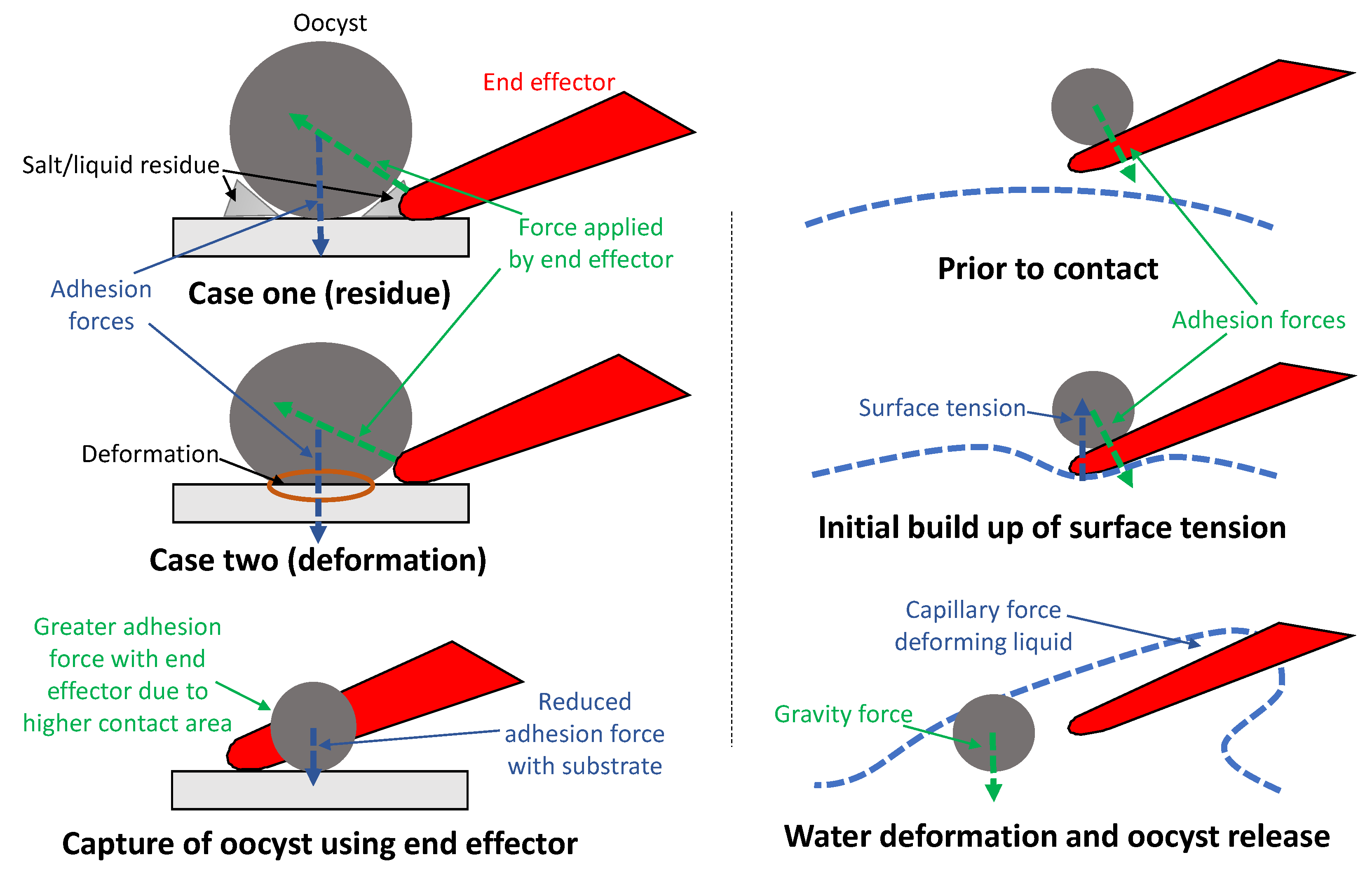
4.1. Alignment Method
4.2. Contact Adhesion
4.3. Comparison to Existing Systems
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SCA | Single cell analysis |
| FACS | Fluorescence activated cell sorters |
| ROI | Region of interest |
References
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for single-cell isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919. [Google Scholar] [CrossRef] [PubMed]
- Method of the Year 2013. Nat. Methods 2014, 11, 1. [CrossRef] [PubMed]
- Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.; Piat, E. Control of a particular micro-macro positioning system applied to cell micromanipulation. IEEE Trans. Autom. Sci. Eng. 2006, 3, 264–271. [Google Scholar] [CrossRef]
- Keloth, A.; Anderson, O.; Risbridger, D.; Paterson, L. Single Cell Isolation Using Optical Tweezers. Micromachines 2018, 9, 434. [Google Scholar]
- Ye, Z.; Diller, E.; Sitti, M. Micro-manipulation using rotational fluid flows induced by remote magnetic micro-manipulators. J. Appl. Phys. 2012, 112, 064912. [Google Scholar] [CrossRef]
- Oakey, J.; Allely, J.; Marr, D.W. Laminar-flow-based separations at the microscale. Biotechnol. Prog. 2002, 18, 1439–1442. [Google Scholar] [CrossRef]
- Aekbote, B.L.; Fekete, T.; Jacak, J.; Vizsnyiczai, G.; Ormos, P.; Kelemen, L. Surface-modified complex SU-8 microstructures for indirect optical manipulation of single cells. Biomed. Opt. Express 2016, 7, 45–56. [Google Scholar] [CrossRef]
- Brasko, C.; Smith, K.; Molnar, C.; Farago, N.; Hegedus, L.; Balind, A.; Balassa, T.; Szkalisity, A.; Sukosd, F.; Kocsis, K.; et al. Intelligent image-based in situ single-cell isolation. Nat. Commun. 2018, 9, 226. [Google Scholar] [CrossRef]
- Valihrach, L.; Androvic, P.; Kubista, M. Platforms for single-cell collection and analysis. Int. J. Mol. Sci. 2018, 19, 807. [Google Scholar] [CrossRef]
- Troell, K.; Hallstrom, B.; Divne, A.M.; Alsmark, C.; Arrighi, R.; Huss, M.; Beser, J.; Bertilsson, S. Cryptosporidium as a testbed for single cell genome characterization of unicellular eukaryotes. BMC Genom. 2016, 17, 471. [Google Scholar] [CrossRef]
- Savia, M.; Koivo, H.N. Contact micromanipulation—Survey of strategies. IEEE/ASME Trans. Mechatron. 2009, 14, 504–514. [Google Scholar] [CrossRef]
- Hériban, D.; Agnus, J.; Gauthier, M. MICRO-Manipulation of Silicate Micro-Sized Particles for Biological applications. IWMF’06. 2006. Number sur CD ROM. Available online: https://hal.archives-ouvertes.fr/hal-00334037/ (accessed on 1 October 2019).
- Xie, H.; Régnier, S. Three-dimensional automated micromanipulation using a nanotip gripper with multi-feedback. J. Micromech. Microeng. 2009, 19, 075009. [Google Scholar] [CrossRef]
- Chen, F.; Chen, X.; Xie, X.; Feng, X.; Yang, L. Full-field 3D measurement using multi-camera digital image correlation system. Opt. Lasers Eng. 2013, 51, 1044–1052. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Wang, L.; Yu, T. Mechanics of adhesion in MEMS—A review. J. Adhes. Sci. Technol. 2003, 17, 519–546. [Google Scholar] [CrossRef]
- Reid, A.J.; Talman, A.M.; Bennett, H.M.; Gomes, A.R.; Sanders, M.J.; Illingworth, C.J.; Billker, O.; Berriman, M.; Lawniczak, M.K. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. eLife 2018, 7, e33105. [Google Scholar] [CrossRef]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.I.; Morgan, B.; Trautwein, J.; et al. Inertial focusing for tumor antigen–dependent and–independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef]
- Burger, R.; Kurzbuch, D.; Gorkin, R.; Kijanka, G.; Glynn, M.; McDonagh, C.; Ducree, J. An integrated centrifugo-opto-microfluidic platform for arraying, analysis, identification and manipulation of individual cells. Lab Chip 2015, 15, 378–381. [Google Scholar] [CrossRef]
- Zhang, R.; Chu, J.; Wang, H.; Chen, Z. A multipurpose electrothermal microgripper for biological micro-manipulation. Microsyst. Technol. 2013, 19, 89–97. [Google Scholar] [CrossRef]
- DPDx—Laboratory Identification of Parasites of Public Health Concern. Available online: https://www.cdc.gov/dpdx/cryptosporidiosis/index.html (accessed on 1 October 2019).
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Robertson, L.; Campbell, A.; Smith, H. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 1992, 58, 3494–3500. [Google Scholar] [PubMed]
- Ministry of Health. Drinking-Water Standards for New Zealand 2005 (Revised 2018); Ministry of Health: Ajax, ON, USA, 2018.
- Danielsson, P.E. Euclidean distance mapping. Comput. Graph. Image Process. 1980, 14, 227–248. [Google Scholar] [CrossRef]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 6, 583–598. [Google Scholar] [CrossRef]
- Gauthier, M.; Alvo, S.; Dejeu, J.; Tamadazte, B.; Rougeot, P.; Régnier, S. Analysis and specificities of adhesive forces between microscale and nanoscale. IEEE Trans. Autom. Sci. Eng. 2013, 10, 562–570. [Google Scholar] [CrossRef]
- Lofroth, M.; Avci, E. An Auto-Focusing Approach for Micro Objects at Different Focal Planes. In Proceedings of the IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM), Auckland, New Zealand, 9–12 July 2018; pp. 974–979. [Google Scholar]
- Zhang, X.; Wu, H.; Ma, Y. A new auto-focus measure based on medium frequency discrete cosine transform filtering and discrete cosine transform. Appl. Comput. Harmon. Anal. 2016, 40, 430–437. [Google Scholar] [CrossRef]
- Sun, Y.; Duthaler, S.; Nelson, B.J. Autofocusing in computer microscopy: Selecting the optimal focus algorithm. Microsc. Res. Tech. 2004, 65, 139–149. [Google Scholar] [CrossRef]
- Chang, R.J.; Chien, Y.C. Polymer Microgripper with Autofocusing and Visual Tracking Operations to Grip Particle Moving in Liquid. Actuators 2018, 7, 27. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, X.; Leung, C.; Esfandiari, N.; Casper, R.F.; Sun, Y. Automated cell manipulation: Robotic ICSI. In Proceedings of the 2011 IEEE International Conference on Robotics and Automation, Shanghai, China, 9–13 May 2011; pp. 2540–2545. [Google Scholar]
- Nguyen, C.N.; Ohara, K.; Avci, E.; Takubo, T.; Mae, Y.; Arai, T. Automated micromanipulation for a microhand with all-in-focus imaging system. In Proceedings of the 2011 IEEE/RSJ International Conference on Intelligent Robots and Systems, San Francisco, CA, USA, 25–30 September 2011; pp. 427–432. [Google Scholar]
- Haliyo, D.S.; Dionnet, F.; Régnier, S. Controlled rolling of microobjects for autonomous manipulation. J. Micromechatron. 2004, 3, 75–102. [Google Scholar] [CrossRef]
- Lai, T.; Chen, R.; Huang, P. Temperature dependence of microscale adhesion force between solid surfaces using an AFM. J. Adhes. Sci. Technol. 2015, 29, 133–148. [Google Scholar] [CrossRef]
- Dejeu, J.; Bechelany, M.; Rougeot, P.; Philippe, L.; Gauthier, M. Adhesion control for micro-and nanomanipulation. ACS Nano 2011, 5, 4648–4657. [Google Scholar] [CrossRef]
- Malachowski, K.; Jamal, M.; Jin, Q.; Polat, B.; Morris, C.J.; Gracias, D.H. Self-folding single cell grippers. Nano Lett. 2014, 14, 4164–4170. [Google Scholar] [CrossRef] [PubMed]
- King, B.; Monis, P. Critical processes affecting Cryptosporidium oocyst survival in the environment. Parasitology 2007, 134, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Amar, C.; Pedraza-Díaz, S.; McLauchlin, J. Extraction and Genotyping of Cryptosporidium parvum DNA from Fecal Smears on Glass Slides Stained Conventionally for Direct Microscope Examination. J. Clin. Microbiol. 2001, 39, 401–403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, Y.; Nelson, B.J. Biological cell injection using an autonomous microrobotic system. Int. J. Robot. Res. 2002, 21, 861–868. [Google Scholar] [CrossRef]
- Chronis, N.; Lee, L.P. Electrothermally activated SU-8 microgripper for single cell manipulation in solution. J. Microelectromech. Syst. 2005, 14, 857–863. [Google Scholar] [CrossRef]
- Beyeler, F.; Neild, A.; Oberti, S.; Bell, D.J.; Sun, Y.; Dual, J.; Nelson, B.J. Monolithically fabricated microgripper with integrated force sensor for manipulating microobjects and biological cells aligned in an ultrasonic field. J. Microelectromech. Syst. 2007, 16, 7–15. [Google Scholar] [CrossRef]
- Avci, E.; Hattori, T.; Kamiyama, K.; Kojima, M.; Horade, M.; Mae, Y.; Arai, T. Piezo-actuated parallel mechanism for biological cell release at high speed. Biomed. Microdevices 2015, 17, 98. [Google Scholar] [CrossRef]

| Accuracy | Successes | Success Percentage (95% Confidence Interval) |
|---|---|---|
| <2 m | 27 | 90% (73%–98%) |
| <1 m | 15 | 50% (31%–69%) |
| Successes | Success Percentage (95% Confidence Interval) |
|---|---|
| 49/50 | 98% (89%–100%) |
| Task | Average Time (s) | Maximum Time (s) |
|---|---|---|
| Alignment (end effector and focus plane) | 21 ( = 2.6) | 25 |
| Alignment (end effector and oocyst) | 16 ( = 2.4) | 20 |
| Manual capture | 41 ( = 50.2) | 270 |
| Oocyst release and return to origin | 140 ( = 0) | 140 |
| Total | 218 ( = 50.7) | 455 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penny, H.; Hayman, D.T.S.; Avci, E. Micromanipulation System for Isolating a Single Cryptosporidium Oocyst. Micromachines 2020, 11, 3. https://doi.org/10.3390/mi11010003
Penny H, Hayman DTS, Avci E. Micromanipulation System for Isolating a Single Cryptosporidium Oocyst. Micromachines. 2020; 11(1):3. https://doi.org/10.3390/mi11010003
Chicago/Turabian StylePenny, Hamish, David T. S. Hayman, and Ebubekir Avci. 2020. "Micromanipulation System for Isolating a Single Cryptosporidium Oocyst" Micromachines 11, no. 1: 3. https://doi.org/10.3390/mi11010003
APA StylePenny, H., Hayman, D. T. S., & Avci, E. (2020). Micromanipulation System for Isolating a Single Cryptosporidium Oocyst. Micromachines, 11(1), 3. https://doi.org/10.3390/mi11010003






