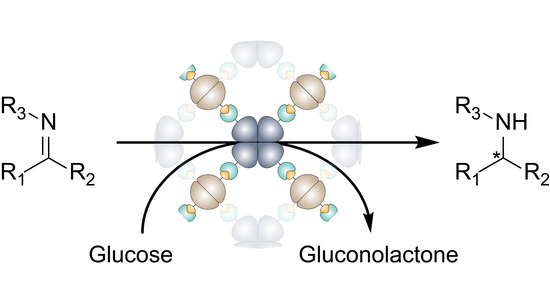
Imine Reductase Based All-Enzyme Hydrogel with Intrinsic Cofactor Regeneration for Flow Biocatalysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Constructions
- KSR227-for: TGAGATCCGGCTGCTAACAAAGCCC
- KSR65-rev: ATGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAGGGG
2.2. Proteinexpression and Purifcation
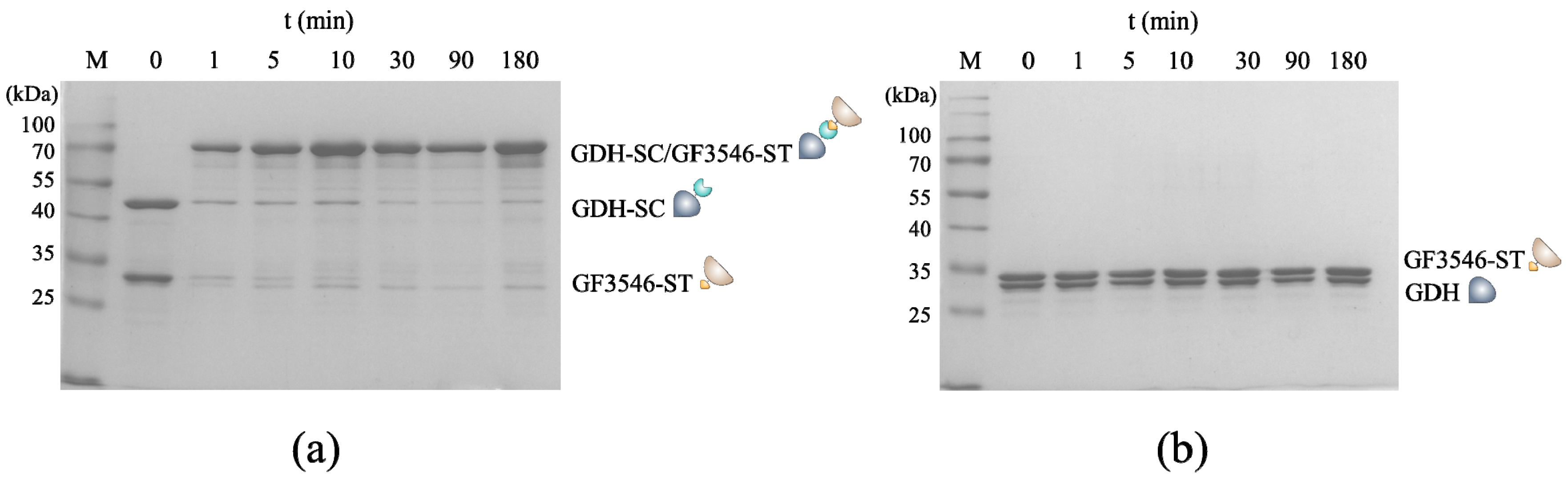
2.3. SDS Polyacrylamide Gel Electrophoresis Analysis
2.4. Dynamic Light Scattering Measurements
2.5. Microrheology Measurements
2.6. Determination of Enzyme Activity
- A:
- 97:3 (n-heptane:isopropanol) 0.5 mL/min 30 °C;
- B:
- 93:7 (n-heptane:isopropanol) 0.5 mL/min 30 °C.
2.7. Racemic Reduction of 6,7-Dimethoxy-1-methyl-3,4-dihydroisoquinoline 3
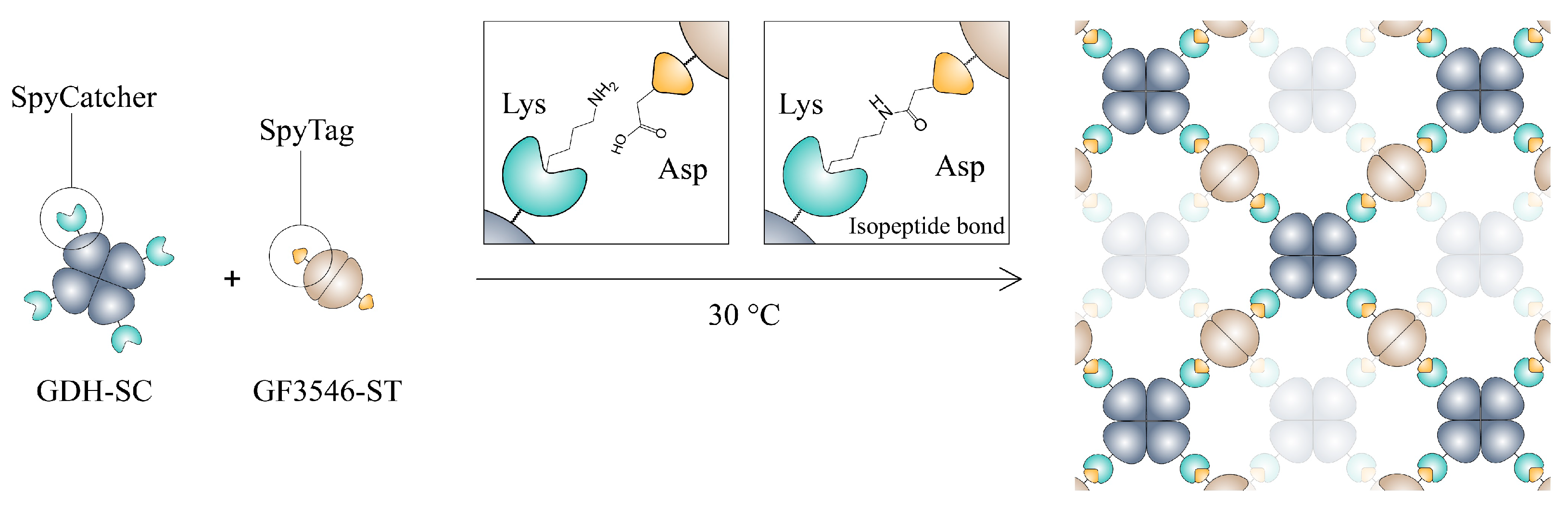
2.8. Hydrogel Preparation
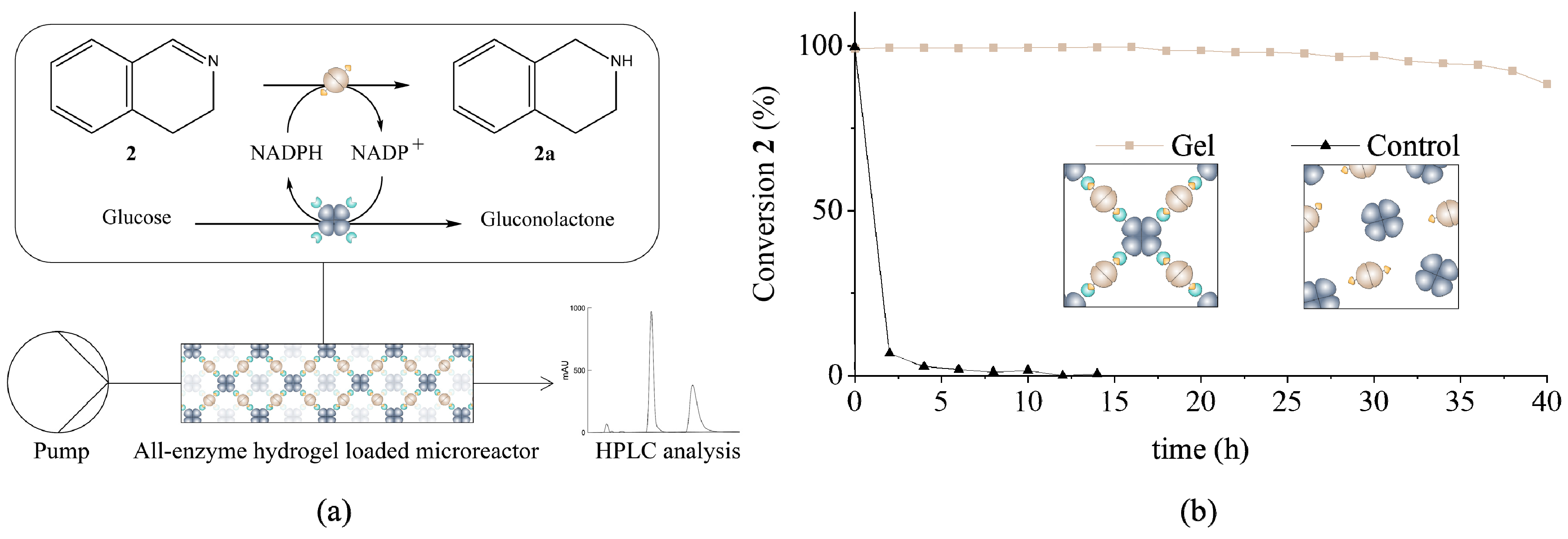
2.9. Microfluidic Setup and Analysis of the Hydrogels under Continuous Flow
3. Results and Discussion
3.1. Construction and Characterization of the All-Enzyme Hydrogels
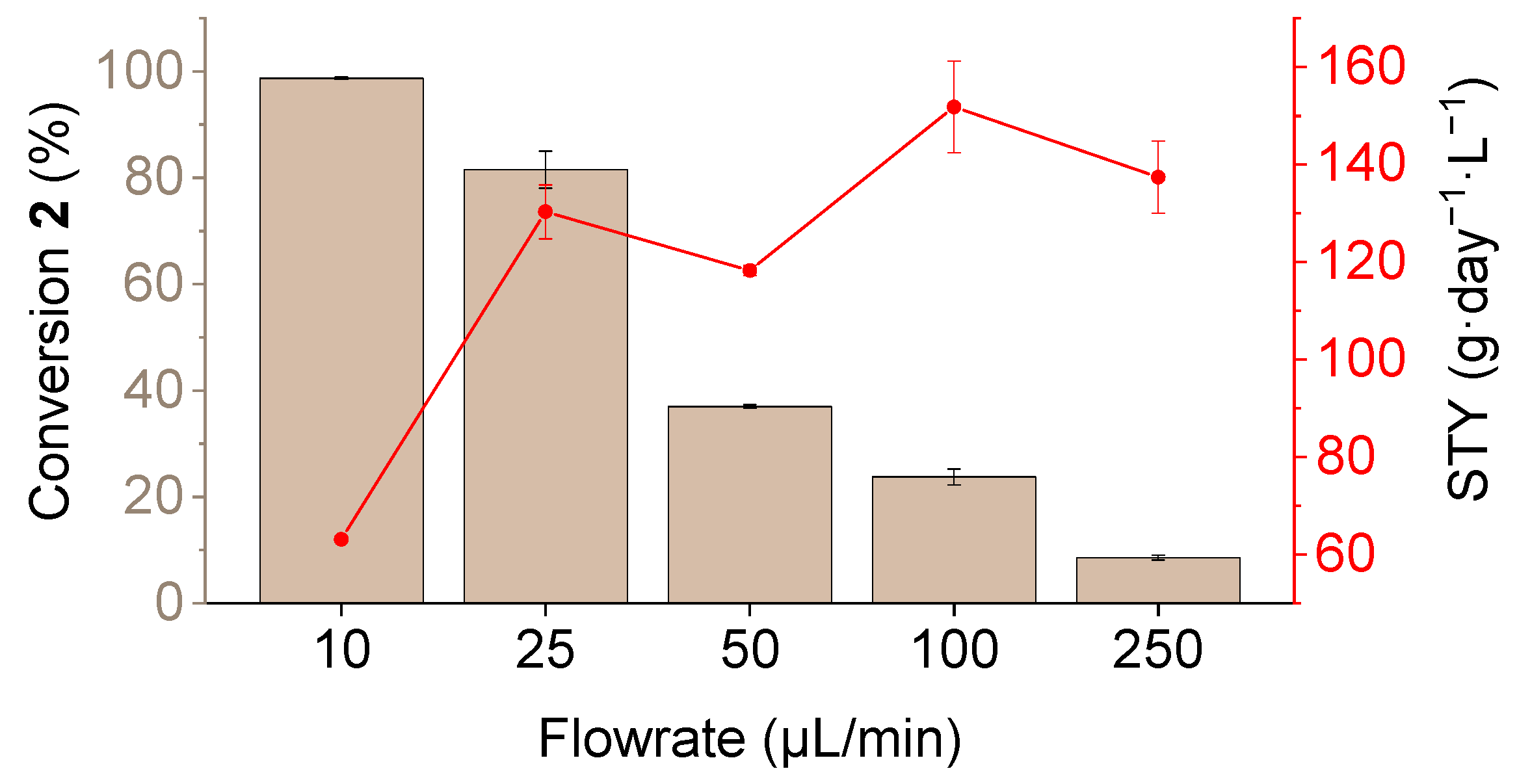
3.2. Determination of Catalytic Activity and Microfluidic Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef]
- Wheeldon, I.; Minteer, S.D.; Banta, S.; Barton, S.C.; Atanassov, P.; Sigman, M. Substrate channelling as an approach to cascade reactions. Nat. Chem. 2016, 8, 299–309. [Google Scholar] [CrossRef]
- Kuchler, A.; Yoshimoto, M.; Luginbuhl, S.; Mavelli, F.; Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol. 2016, 11, 409–420. [Google Scholar] [CrossRef] [PubMed]
- France, S.P.; Hepworth, L.J.; Turner, N.J.; Flitsch, S.L. Constructing biocatalytic cascades: In Vitro and in vivo approaches to de novo multi-enzyme pathways. ACS Catal. 2017, 7, 710–724. [Google Scholar] [CrossRef]
- Rabe, K.S.; Muller, J.; Skoupi, M.; Niemeyer, C.M. Cascades in compartments: En route to machine-assisted biotechnology. Angew. Chem. Int. Ed. 2017, 56, 13574–13589. [Google Scholar] [CrossRef]
- Britton, J.; Majumdar, S.; Weiss, G.A. Continuous flow biocatalysis. Chem. Soc. Rev. 2018, 47, 5891–5918. [Google Scholar] [CrossRef]
- Thompson, M.P.; Peñafiel, I.; Cosgrove, S.C.; Turner, N.J. Biocatalysis using immobilized enzymes in continuous flow for the synthesis of fine chemicals. Org. Process Res. Dev. 2018, 23, 9–18. [Google Scholar] [CrossRef]
- Schmid-Dannert, C.; Lopez-Gallego, F. Advances and opportunities for the design of self-sufficient and spatially organized cell-free biocatalytic systems. Curr. Opin. Chem. Biol. 2018, 49, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Gandomkar, S.; Żądło-Dobrowolska, A.; Kroutil, W. Extending designed linear biocatalytic cascades for organic synthesis. ChemCatChem 2019, 11, 225–243. [Google Scholar] [CrossRef]
- Mason, B.P.; Price, K.E.; Steinbacher, J.L.; Bogdan, A.R.; McQuade, D.T. Greener approaches to organic synthesis using microreactor technology. Chem. Rev. 2007, 107, 2300–2318. [Google Scholar] [CrossRef]
- Wohlgemuth, R.; Plazl, I.; Znidarsic-Plazl, P.; Gernaey, K.V.; Woodley, J.M. Microscale technology and biocatalytic processes: Opportunities and challenges for synthesis. Trends Biotechnol. 2015, 33, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Herr, A.E. Protein immobilization techniques for microfluidic assays. Biomicrofluidics 2013, 7, 041501. [Google Scholar] [CrossRef]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. A general overview of support materials for enzyme immobilization: Characteristics, properties, practical utility. Catalysts. 2018, 8, 92. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Cross-linked enzyme aggregates as industrial biocatalysts. Org. Process Res. Dev. 2011, 15, 213–223. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Peschke, T.; Bitterwolf, P.; Gallus, S.; Hu, Y.; Oelschlaeger, C.; Willenbacher, N.; Rabe, K.S.; Niemeyer, C.M. Self-assembling all-enzyme hydrogels for flow biocatalysis. Angew. Chem. Int. Ed. Engl. 2018, 57, 17028–17032. [Google Scholar] [CrossRef]
- Bitterwolf, P.; Gallus, S.; Peschke, T.; Mittmann, E.; Oelschlaeger, C.; Willenbacher, N.; Rabe, K.S.; Niemeyer, C.M. Valency engineering of monomeric enzymes for self-assembling biocatalytic hydrogels. Chem. Sci. 2019, 10, 9752–9757. [Google Scholar] [CrossRef]
- Mitsukura, K.; Kuramoto, T.; Yoshida, T.; Kimoto, N.; Yamamoto, H.; Nagasawa, T. A NADPH-dependent (S)-imine reductase (SIR) from Streptomyces sp GF3546 for asymmetric synthesis of optically active amines: Purification, characterization, gene cloning, and expression. Appl. Microbiol. Biotechnol. 2013, 97, 8079–8086. [Google Scholar] [CrossRef]
- Mangas-Sanchez, J.; France, S.P.; Montgomery, S.L.; Aleku, G.A.; Man, H.; Sharma, M.; Ramsden, J.I.; Grogan, G.; Turner, N.J. Imine reductases (IREDs). Curr. Opin. Chem. Biol. 2017, 37, 19–25. [Google Scholar] [CrossRef]
- Lenz, M.; Borlinghaus, N.; Weinmann, L.; Nestl, B.M. Recent advances in imine reductase-catalyzed reactions. World J. Microb. Biot. 2017, 33, 199. [Google Scholar] [CrossRef] [PubMed]
- Hestericova, M.; Correro, M.R.; Lenz, M.; Corvini, P.F.X.; Shahgaldian, P.; Ward, T.R. Immobilization of an artificial imine reductase within silica nanoparticles improves its performance. Chem. Commun. 2016, 52, 9462–9465. [Google Scholar] [CrossRef] [PubMed]
- Gand, M.; Thole, C.; Muller, H.; Brundiek, H.; Bashiri, G.; Hohne, M. A NADH-accepting imine reductase variant: Immobilization and cofactor regeneration by oxidative deamination. J. Biotechnol. 2016, 230, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Hilt, W.; Pfleiderer, G.; Fortnagel, P. Glucose dehydrogenase from Bacillus subtilis expressed in Escherichia coli. I: Purification, characterization and comparison with glucose dehydrogenase from Bacillus megaterium. Biochim. Biophys. Acta 1991, 1076, 298–304. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Oelschlaeger, C.; Willenbacher, N. Visualization of micro-scale inhomogeneities in acrylic thickener solutions: A multiple particle tracking study. Polymer 2015, 58, 170–179. [Google Scholar] [CrossRef]
| Substrate | Specific Activity (μmolsubstrate·(μmolenzyme·min)−1) | Ee (%) |
|---|---|---|
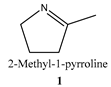 | 2.4 a ± 0.4 | n. d. |
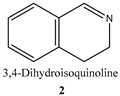 | 83.3 a ± 8.2 | - |
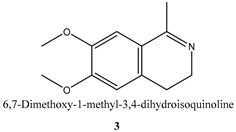 | 2.3 ± 0.3 b | >99 b |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bitterwolf, P.; Ott, F.; Rabe, K.S.; Niemeyer, C.M. Imine Reductase Based All-Enzyme Hydrogel with Intrinsic Cofactor Regeneration for Flow Biocatalysis. Micromachines 2019, 10, 783. https://doi.org/10.3390/mi10110783
Bitterwolf P, Ott F, Rabe KS, Niemeyer CM. Imine Reductase Based All-Enzyme Hydrogel with Intrinsic Cofactor Regeneration for Flow Biocatalysis. Micromachines. 2019; 10(11):783. https://doi.org/10.3390/mi10110783
Chicago/Turabian StyleBitterwolf, Patrick, Felix Ott, Kersten S. Rabe, and Christof M. Niemeyer. 2019. "Imine Reductase Based All-Enzyme Hydrogel with Intrinsic Cofactor Regeneration for Flow Biocatalysis" Micromachines 10, no. 11: 783. https://doi.org/10.3390/mi10110783
APA StyleBitterwolf, P., Ott, F., Rabe, K. S., & Niemeyer, C. M. (2019). Imine Reductase Based All-Enzyme Hydrogel with Intrinsic Cofactor Regeneration for Flow Biocatalysis. Micromachines, 10(11), 783. https://doi.org/10.3390/mi10110783