Spread of Tst–Positive Staphylococcus aureus Strains Belonging to ST30 Clone among Patients and Healthcare Workers in Two Intensive Care Units
Abstract
:1. Introduction
2. Results
2.1. Colonization Results and Studied Isolates
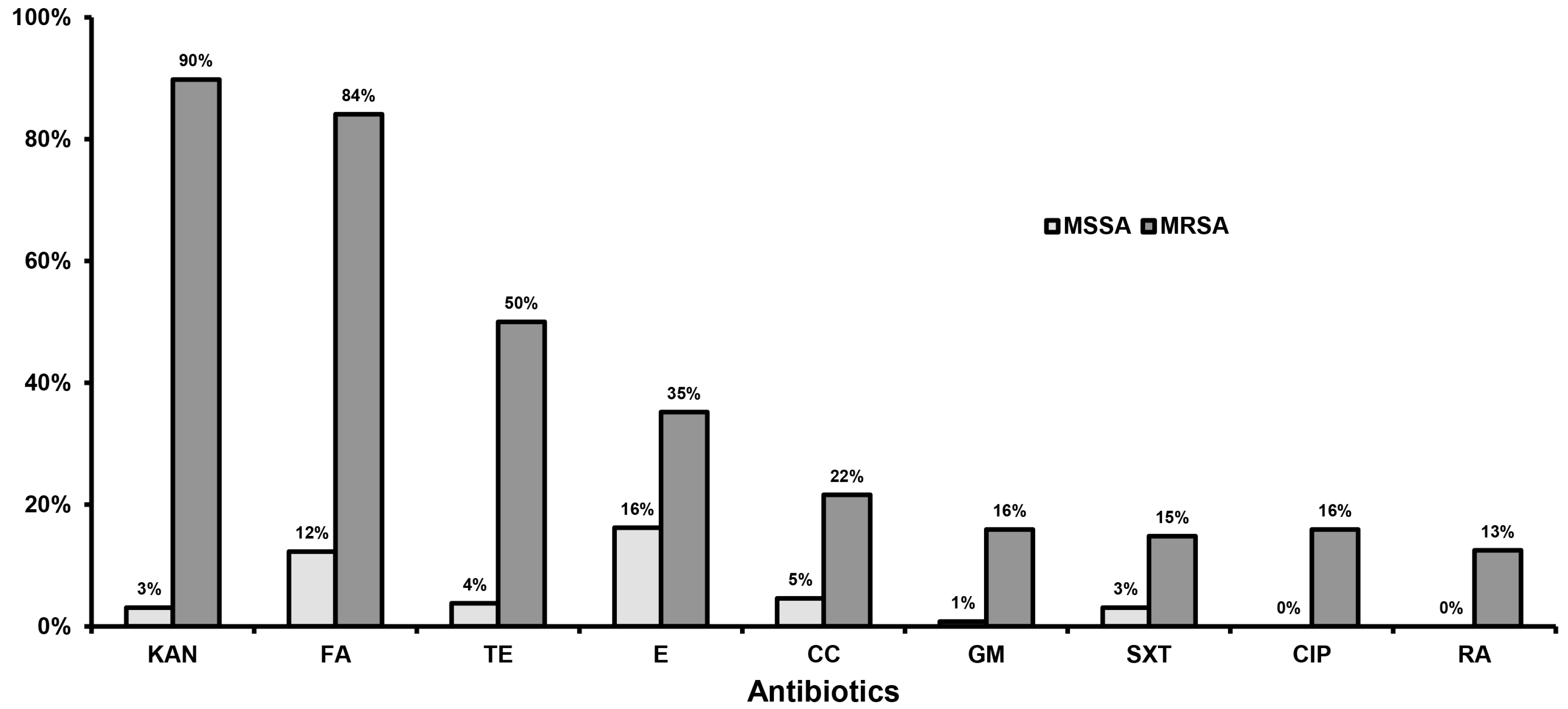
2.2. Susceptibility to Antistaphylococcal Agents
2.3. Toxin Genes Carriage
2.4. Clonal Identification
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Study Design and Hospital
5.2. Studied Groups
5.3. S. aureus Identification and Antibiotic Susceptibility Testing
5.4. Polymerase Chain Reaction Procedures
5.5. Clonal Identification
5.6. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- De Kraker, M.E.; Jarlier, V.; Monen, J.C.; Heuer, O.E.; van de Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in Europe: Trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infect. 2013, 19, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Drougka, E.; Foka, A.; Liakopoulos, A.; Doudoulakakis, A.; Jelastopulu, E.; Chini, V.; Spiliopoulou, A.; Levidiotou, S.; Panagea, T.; Vogiatzi, A.; et al. A 12-year survey of methicillin-resistant Staphylococcus aureus infections in Greece: ST80-IV epidemic? Clin. Microbiol. Infect. 2014, 20, O796–O803. [Google Scholar] [CrossRef] [PubMed]
- Robert, J.; Tristan, A.; Cavalié, L.; Decousser, J.W.; Bes, M.; Etienne, J.; Laurent, F.; ONERBA (Observatoire National de l’Epidémiologie de Résistance Bactérienne aux Antibiotiques). Panton-valentine leukocidin-positive and toxic shock syndrome toxin 1-positive methicillin-resistant Staphylococcus aureus: A French multicenter prospective study in 2008. Antimicrob. Agents Chemother. 2011, 55, 1734–1739. [Google Scholar] [CrossRef] [PubMed]
- Sdougkos, G.; Chini, V.; Papanastasiou, D.A.; Christodoulou, G.; Stamatakis, E.; Vris, A.; Christodoulidi, I.; Protopapadakis, G.; Spiliopoulou, I. Community-associated Staphylococcus aureus infections and nasal carriage among children: Molecular microbial data and clinical characteristics. Clin. Microbiol. Infect. 2008, 14, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Rolo, J.; Miragaia, M.; Turlej-Rogacka, A.; Empel, J.; Bouchami, O.; Faria, N.A.; Tavares, A.; Hryniewicz, W.; Fluit, A.C.; de Lencastre, H.; et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: Results from a multicenter study. PLoS ONE 2012, 7, e34768. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Liao, C.H.; Fang, C.T.; Chie, W.C.; Lai, M.S.; Lauderdale, T.L.; Chang, S.C. Incidence of and risk factors for community-associated methicillin-resistant Staphylococcus aureus acquired infection or colonization in intensive-care-unit patients. J. Clin. Microbiol. 2010, 48, 4439–4444. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.C.; Behrendt, M.D.; Melles, D.C.; Mollema, F.P.; de Groot, W.; Parlevliet, G.; Ott, A.; Horst-Kreft, D.; van Belkum, A.; Verbrugh, H.A. 5 years of experience implementing a methicillin-resistant Staphylococcus aureus search and destroy policy at the largest university medical center in the Netherlands. Infect. Control Hosp. Epidemiol. 2009, 30, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Mandsager, P.; Biddle, A.K.; Weber, D.J. Cost-effectiveness analysis of active surveillance screening for methicillin-resistant Staphylococcus aureus in an academic hospital setting. Infect. Control Hosp. Epidemiol. 2012, 33, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Argudín, M.A.; Argumosa, V.; Mendoza, M.C.; Guerra, B.; Rodicio, M.R. Population structure and exotoxin gene content of methicillin-susceptible Staphylococcus aureus from Spanish healthy carriers. Microb. Pathog. 2013, 54, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Kolonitsiou, F.; Papadimitriou-Olivgeris, M.; Spiliopoulou, A.; Stamouli, V.; Papakostas, V.; Apostolopoulou, E.; Panagiotopoulos, C.; Marangos, M.; Anastassiou, E.D.; Christofidou, M.; et al. Trends of Bloodstream Infections in a University Greek Hospital during a Three-Year Period: Incidence of Multidrug-Resistant Bacteria and Seasonality in Gram-negative Predominance. Pol. J. Microbiol. 2017, 66, 171–180. [Google Scholar] [PubMed]
- Layer, F.; Ghebremedhin, B.; König, W.; König, B. Heterogeneity of methicillin-susceptible Staphylococcus aureus strains at a German University Hospital implicates the circulating-strain pool as a potential source of emerging methicillin-resistant S. aureus clones. J. Clin. Microbiol. 2006, 44, 2179–2185. [Google Scholar] [CrossRef] [PubMed]
- Von Eiff, C.; Becker, K.; Machka, K.; Stammer, H.; Peters, G. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 2001, 344, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Dong, B.R.; Yang, M.; Chen, X.; Wu, T.; Liu, G.J. Linezolid versus vancomycin for skin and soft tissue infections. Cochrane Database Syst. Rev. 2016, 1, CD008056. [Google Scholar] [CrossRef]
- Papadimitriou-Olivgeris, M.; Kolonitsiou, F.; Zerva, L.; Lebessi, E.; Koutsia, C.; Drougka, E.; Sarrou, S.; Giormezis, N.; Vourli, S.; Doudoulakakis, A.; et al. Activity of vancomycin, linezolid, and daptomycin against staphylococci and enterococci isolated in 5 Greek hospitals during a 5-year period (2008–2012). Diagn. Microbiol. Infect. Dis. 2015, 83, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Peck, K.R.; Baek, J.Y.; Song, J.H.; Ko, K.S. Comparison of genotypes and enterotoxin genes between Staphylococcus aureus isolates from blood and nasal colonizers in a Korean hospital. J. Korean Med. Sci. 2009, 4, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Sauer, P.; Síla, J.; Stosová, T.; Vecerová, R.; Hejnar, P.; Vágnerová, I.; Kolár, M.; Raclavsky, V.; Petrzelová, J.; Lovecková, Y.; et al. Prevalence of genes encoding extracellular virulence factors among meticillin-resistant Staphylococcus aureus isolates from the University Hospital, Olomouc, Czech Republic. J. Med. Microbiol. 2008, 57, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Gbaguidi-Haore, H.; Thouverez, M.; Couetdic, G.; Cholley, P.; Talon, D.; Bertrand, X. Usefulness of antimicrobial resistance pattern for detecting PVL- or TSST-1-producing meticillin-resistant Staphylococcus aureus in a French university hospital. J. Med. Microbiol. 2009, 58, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- De Boeck, H.; Vandendriessche, S.; Hallin, M.; Batoko, B.; Alworonga, J.P.; Mapendo, B.; Van Geet, C.; Dauly, N.; Denis, O.; Jacobs, J. Staphylococcus aureus nasal carriage among healthcare workers in Kisangani, the Democratic Republic of the Congo. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gonzalez, C.; Alba, C.; Otero, J.R.; Sanz, F.; Chaves, F. Long persistence of methicillin-susceptible strains of Staphylococcus aureus causing sepsis in a neonatal intensive care unit. J. Clin. Microbiol. 2007, 45, 2301–2304. [Google Scholar] [CrossRef] [PubMed]
- Chini, V.; Dimitracopoulos, G.; Spiliopoulou, I. Occurrence of the enterotoxin gene cluster and the toxic shock syndrome toxin 1 gene among clinical isolates of methicillin-resistant Staphylococcus aureus is related to clonal type and agr group. J. Clin. Microbiol. 2006, 44, 1881–1883. [Google Scholar] [CrossRef] [PubMed]
- McGavin, M.J.; Arsic, B.; Nickerson, N.N. Evolutionary blueprint for host- and niche-adaptation in Staphylococcus aureus clonal complex CC30. Front. Cell. Infect. Microbiol. 2012, 2, 48. [Google Scholar] [CrossRef] [PubMed]
- Achermann, Y.; Seidl, K.; Kuster, S.P.; Leimer, N.; Durisch, N.; Ajdler-Schäffler, E.; Karrer, S.; Senn, G.; Holzmann-Bürgel, A.; Wolfensberger, A.; et al. Epidemiology of Methicillin-Susceptible Staphylococcus aureus in a Neonatology Ward. Infect. Control Hosp. Epidemiol. 2015, 36, 1305–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zheng, Y.; Mediavilla, J.R.; Chen, L.; Kreiswirth, B.N.; Song, Y.; Yang, R.; Du, H. Hospital Dissemination of tst-1-Positive Clonal Complex 5 (CC5) Methicillin-Resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2017, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sparling, J.; Chow, B.L.; Elsayed, S.; Hussain, Z.; Church, D.L.; Gregson, D.B.; Louie, T.; Conly, J.M. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004, 42, 4947–4955. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretations of MICs and Zone Diameters, version 3.1; EUCAST: Växjö, Sweden, 2013. [Google Scholar]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Nesme, X.; Etienne, J.; Vandenesch, F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 2002, 70, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Boutite, F.; Tristan, A.; Bes, M.; Etienne, J.; Vandenesch, F. Bacterial competition for human nasal cavity colonization: Role of Staphylococcal agr alleles. Appl. Environ. Microbiol. 2003, 69, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- MLST. Available online: http://saureus.mlst.net/misc/info.asp (accessed on 1 September 2017).
| Characteristics | Total (N = 143) | 1 Group A (N = 90) | 2 Group B (N = 53) | 3 p Value | ||
|---|---|---|---|---|---|---|
| Total (N = 90) | ICU Patients (N = 67) | ICU Personnel (N = 23) | ||||
| MRSA | 57 (39.9%) | 28 (31.1%) | 22 (32.8%) | 6 (26.1%) | 29 (54.7%) | 0.008 |
| PVL in total | 54 (37.8%) | 23 (25.6%) | 21 (31.3%) | 2 (8.7%) | 31 (58.5%) | <0.001 |
| tst in total | 20 (14%) | 19 (21.1%) | 14 (20.9%) | 5 (21.7%) | 1 (1.9%) | 0.002 |
| PVL in MSSA | 7 (8.1%) | 3 (4.8%) | 3 (6.7%) | 0 (0%) | 4 (16.7%) | 0.091 |
| tst in MSSA | 18 (20.1%) | 17 (27.4%) | 14 (31.1%) | 3 (17.6%) | 1 (4.2%) | 0.018 |
| PVL in MRSA | 47 (82.5%) | 20 (71.4%) | 18 (81.8%) | 2 (33.3%) | 27 (93.1%) | 0.041 |
| tst in MRSA | 2 (3.5%) | 2 (7.1%) | 0 (0%) | 2 (33.3%) | 0 (0%) | 0.237 |
| CA-MRSA in MRSA | 35 (61.4%) | 8 (28.6%) | 8 (36.4%) | 0 (0%) | 27 (93.1%) | <0.001 |
| PVL in CA-MRSA | 34 (97.1%) | 8 (100%) | 8 (100%) | 0 (0%) | 26 (96.3%) | >0.999 |
| PVL in HA-MRSA | 13 (59.1%) | 12 (60%) | 10 (71.4%) | 2 (33.3%) | 1 (50%) | 0.560 |
| tst in CA-MRSA | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | - |
| tst in HA-MRSA | 2 (9.1%) | 2 (10%) | 0 (0%) | 2 (33.3%) | 0 (0%) | >0.999 |
| 1 Clones | PFGE Type/agr Allele | CA-/HA-MRSA | 2 Group A (N = 90) | 3 Group B (N = 53) | ||
|---|---|---|---|---|---|---|
| ICU Patients (N = 67) | ICU Personnel (N = 23) | |||||
| PVL-positive MRSA (47) | ST80-IV (45) | C/3 (43), C/1 (1), C/NT (1) | 33/12 | 16 | 2 | 27 |
| ST72-IV (2) | B/1 (1), E/1 (1) | 0/2 | 2 | 0 | 0 | |
| PVL-positive MSSA (7) | ST14 (4) | Y/1 (2), A/1 (1), C/2 (1) | - | 3 | 0 | 1 |
| ST97 (1) | C/1 (1) | - | 0 | 0 | 1 | |
| ST101 (2) | C/2 (2) | - | 0 | 0 | 2 | |
| tst-positive MRSA (2) | ST30-IV (2) | A/3 (2) | 0/2 | 0 | 2 | 0 |
| tst-positive MSSA (18) | ST30 (10) | A/3 (9), A/4 (1) | - | 7 | 2 | 1 |
| ST2123 (6) | Y/1 (6) | - | 6 | 0 | 0 | |
| ST27 (1) | Y/2 (1) | - | 1 | 0 | 0 | |
| ST45 (1) | F/1 (1) | - | 0 | 1 | 0 | |
| Toxin negative MRSA (8) | SΤ239-III (4) | B/1 (3), B/NT (1) | 0/4 | 3 | 0 | 1 |
| ST225–II (2) | C/3 (2) | 0/2 | 1 | 1 | 0 | |
| ST30–IV (2) | A/3 (2) | 1/1 | 0 | 1 | 1 | |
| Toxin negative MSSA (61) | - | 16 PFGE/agr types | - | 28 | 14 | 19 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papadimitriou-Olivgeris, M.; Drougka, E.; Fligou, F.; Dodou, V.; Kolonitsiou, F.; Filos, K.S.; Anastassiou, E.D.; Petinaki, E.; Marangos, M.; Spiliopoulou, I. Spread of Tst–Positive Staphylococcus aureus Strains Belonging to ST30 Clone among Patients and Healthcare Workers in Two Intensive Care Units. Toxins 2017, 9, 270. https://doi.org/10.3390/toxins9090270
Papadimitriou-Olivgeris M, Drougka E, Fligou F, Dodou V, Kolonitsiou F, Filos KS, Anastassiou ED, Petinaki E, Marangos M, Spiliopoulou I. Spread of Tst–Positive Staphylococcus aureus Strains Belonging to ST30 Clone among Patients and Healthcare Workers in Two Intensive Care Units. Toxins. 2017; 9(9):270. https://doi.org/10.3390/toxins9090270
Chicago/Turabian StylePapadimitriou-Olivgeris, Matthaios, Eleanna Drougka, Fotini Fligou, Vasiliki Dodou, Fevronia Kolonitsiou, Kriton S. Filos, Evangelos D. Anastassiou, Efthimia Petinaki, Markos Marangos, and Iris Spiliopoulou. 2017. "Spread of Tst–Positive Staphylococcus aureus Strains Belonging to ST30 Clone among Patients and Healthcare Workers in Two Intensive Care Units" Toxins 9, no. 9: 270. https://doi.org/10.3390/toxins9090270
APA StylePapadimitriou-Olivgeris, M., Drougka, E., Fligou, F., Dodou, V., Kolonitsiou, F., Filos, K. S., Anastassiou, E. D., Petinaki, E., Marangos, M., & Spiliopoulou, I. (2017). Spread of Tst–Positive Staphylococcus aureus Strains Belonging to ST30 Clone among Patients and Healthcare Workers in Two Intensive Care Units. Toxins, 9(9), 270. https://doi.org/10.3390/toxins9090270







