Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles
Abstract
:1. Introduction
2. Evolution and Unique Domain Architecture of Cereal RIPs
3. Biological Activity of Cereal RIP Domains
3.1. Structure of Non-Cereal and Cereal RIP Domains
3.1.1. Structure and Active Site of Classical Non-Cereal RIPs
3.1.2. Structure and Active Site of Cereal RIPs
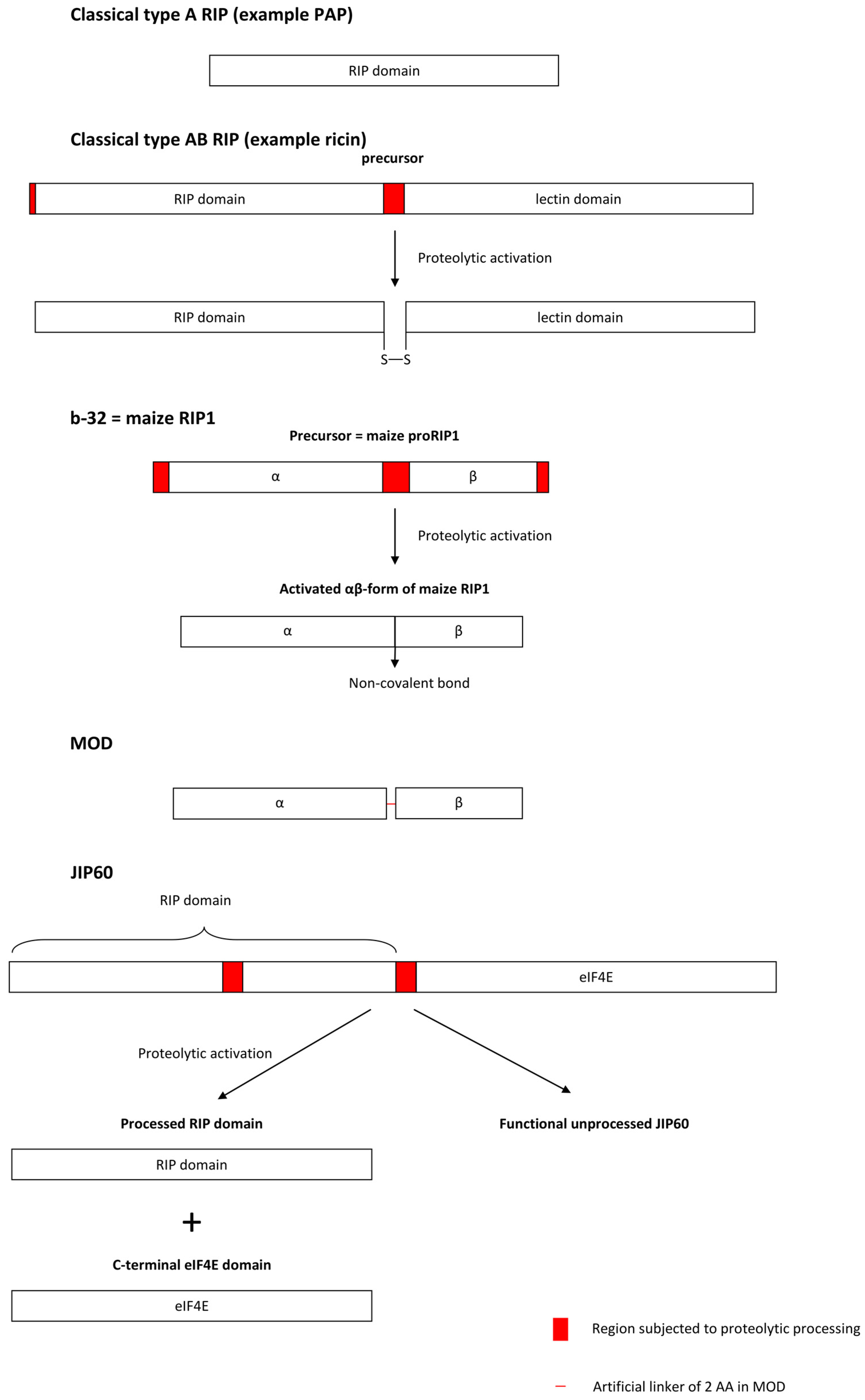
3.1.3. Proteolytic Activation Mechanisms of Cereal RIPs
3.1.4. “Switch Region” of Barley RIP
3.2. Interaction of RIPs with Ribosomes and Substrate Specificity
3.2.1. Interaction of Classical Non-Cereal RIPs with Ribosomes
3.2.2. Interaction of Cereal RIPs with Ribosomes
3.2.3. Substrate Specificity of Classical Non-Cereal RIPs
3.2.4. Substrate Specificity of Cytoplasmic RIPs from Cereals
4. Physiological Roles of Cereal RIPs
4.1. Role in Defense
4.1.1. Maize RIP1
4.1.2. Maize RIP2
4.1.3. Sorghum RIP
4.1.4. Barley RIPs
5. In planta Functions
5.1. Storage Function of Cereal Seed RIPs
5.2. Rice RIPs
5.3. Maize RIP2
5.4. Tritin
5.5. JIP60
6. Concluding Remarks
Acknowledgments
Conflict of Interest
References
- Peumans, W.J.; Van Damme, E.J.M. Evolution of plant ribosome-inactivating proteins. In Toxic Plant Proteins; Lord, J.M., Hartley, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 18, pp. 1–26. [Google Scholar]
- Peumans, W.; Shang, C.; Van Damme, E.J.M. Updated model of the molecular evolution of RIP genes. In Ribosome-Inactivating Proteins: Ricin and Related Proteins; Stirpe, F., Lappi, D., Eds.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2014; pp. 134–150. [Google Scholar]
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.; Polito, L. Ribosome-inactivating proteins from plants: A historical overview. Molecules 2016, 21, 1627:1–1627:19. [Google Scholar]
- Barbieri, L.; Stirpe, F. Ribosome-inactivating proteins from plants: Properties and possible uses. Cancer Surv. 1982, 1, 489–520. [Google Scholar] [CrossRef]
- Endo, Y.; Tsurugi, K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987, 262, 8128–8130. [Google Scholar] [PubMed]
- Ehrlich, P. Experimentelle Untersuchungen über Immunität. I. Ueber Ricin. Med. Wochenschr. 1891, 17, 976–979. [Google Scholar] [CrossRef]
- Hellin, H. Der giftige Eiweisskorper Abrin und seine Wirkung auf das Blut; University of Dorpat: Tartu, Estonia, 1891. [Google Scholar]
- Olsnes, S.; Pihl, A. Ricin—A potent inhibitor of protein synthesis. FEBS Lett. 1972, 20, 327–329. [Google Scholar] [CrossRef]
- Olsnes, S.; Pihl, A. Isolation and properties of abrin: A toxic protein inhibiting protein synthesis: Evidence for different biological functions of its two constituent-peptide chains. Eur. J. Biochem. 1973, 35, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Ferreras, J.M.; Barraco, A.; Ricci, P.; Stirpe, F. Some ribosome-inactivating proteins depurinate ribosomal RNA at multiple sites. Biochem. J. 1992, 286, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Bonora, E.; Gorini, P.; Bolognesi, A.; Stirpe, F. Polynucleotide:Adenosine glycosidase activity of ribosome-inactivating proteins: Effect on DNA, RNA and poly(A). Nucleic Acids Res. 1997, 25, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Gorini, P.; Valbonesi, P.; Castiglioni, P.; Stirpe, F. Unexpected activity of saporins. Nature 1994. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Righi, F.; Zuccheri, G.; Monti, F.; Gorini, P.; Samori, B.; Stirpe, F. Polynucleotide:adenosine glycosidase is the sole activity of ribosome-inactivating proteins on DNA. J. Biochem. 2000, 128, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Chambery, A.; Di Maro, A.; Mastroianni, A.; Parente, A.; Berisio, R. Crystallization and preliminary X-ray diffraction analysis of PD-L1, a highly glycosylated ribosome inactivating protein with DNase activity. Protein Pept. Lett. 2007, 14, 407–709. [Google Scholar] [CrossRef] [PubMed]
- Mock, J.W.Y.; Ng, T.B.; Wong, R.N.S.; Yao, Q.Z.; Yeung, H.W.; Fong, W.P. Demonstration of ribonuclease activity in the plant ribosome-inactivating proteins alpha- and beta-momorcharins. Life Sci. 1996, 59, 1853–1859. [Google Scholar] [CrossRef]
- Shih, N.R.; McDonald, K.A.; Jackman, A.P.; Girbés, T.; Iglesias, R. Bifunctional plant defence enzymes with chitinase and ribosome inactivating activities from Trichosanthes kirilowii cell cultures. Plant Sci. 1997, 130, 145–150. [Google Scholar]
- Chen, H.; Wang, Y.; Yan, M.G.; Yu, M.K.; Yao, Q.Z. The phosphatase activity of five ribosome-inactivating proteins. Biochem. J. 1996, 12, 125–129. [Google Scholar]
- Lombard, S.; Helmy, M.E.; Piéroni, G. Lipolytic activity of ricin from Ricinus sanguineus and Ricinus communis on neutral lipids. Biochem. J. 2001, 358, 773–81. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Chen, W.F.; Liu, W.Y.; Wang, G.H. Large-scale preparation of two new ribosome-inactivating proteins: Cinnamomin and camphorin from the seeds of Cinnamomum camphora. Protein Expr. Purif. 1997, 10, 27–31. [Google Scholar] [PubMed]
- Peumans, W.J.; Hao, Q.; Van Damme, E.J.M. Ribosome-inactivating proteins from plants: More than RNA N-glycosidases? FASEB J. 2001, 15, 1493–1506. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Pérez, Y.; De Torre, C.; Ferreras, J.M.; Antolín, P.; Jiménez, P.; Rojo, M.Á.; Méndez, E.; Girbés, T. Molecular characterization and systemic induction of single-chain ribosome-inactivating proteins (RIPs) in sugar beet (Beta vulgaris) leaves. J. Exp. Bot. 2005, 56, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Di Maro, A.; Ferreras, J.M. Biological activities of the antiviral protein BE27 from sugar beet (Beta vulgaris L.). Planta 2015, 241, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, R.; Citores, L.; Ragucci, S.; Russo, R.; Di Maro, A.; Ferreras, J.M. Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1256–1264. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Polito, L.; Bolognesi, A.; Ciani, M.; Pelosi, E.; Farini, V.; Jha, A.K.; Sharma, N.; Vivanco, J.M.; Chambery, A.; Parente, A.; Stirpe, F. Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta Gen. Subj. 2006, 1760, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Di Maro, A.; Severino, V.; Chambery, A.; Berisio, R. Crystal structure of PD-L1, a ribosome inactivating protein from Phytolacca dioica L. leaves with the property to induce DNA cleavage. Biopolym. Pept. Sci. Sect. 2009, 91, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Aceto, S.; Di Maro, A.; Conforto, B.; Siniscalco, G.G.; Parente, A.; Delli Bovi, P.; Gaudio, L. Nicking activity on pBR322 DNA of ribosome inactivating proteins from Phytolacca dioica L. leaves. Biol. Chem. 2005, 386, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Schrot, J.; Weng, A.; Melzig, M.F. Ribosome-inactivating and related proteins. Toxins (Basel) 2015, 7, 1556–1615. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.A.; Morgan, A.E.; Hey, T.D. Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize: Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kilodalton internal peptide segment. J. Biol. Chem. 1991, 266, 23422–23427. [Google Scholar] [PubMed]
- Reinbothe, S.; Reinbothe, C.; Lehmann, J.; Becker, W.; Apel, K.; Parthier, B. JIP60, a methyl jasmonate-induced ribosome-inactivating protein involved in plant stress reactions. Proc. Natl. Acad. Sci. USA 1994, 91, 7012–7016. [Google Scholar] [CrossRef] [PubMed]
- Stirpe, F.; Battelli, M.G. Ribosome-inactivating proteins: Progress and problems. Cell. Mol. Life Sci. 2006, 63, 1850–1866. [Google Scholar] [CrossRef] [PubMed]
- Battelli, M.G. Cytotoxicity and toxicity to animals and humans of ribosome-inactivating proteins. Mini Rev. Med. Chem. 2004, 4, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, P.; Gayoso, M.; Girbes, T. Non-toxic type 2 Ribosome-inactivating proteins. In Ribosome-Inactivating Proteins: Ricin and Related Proteins; Stirpe, F., Lappi, D., Eds.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2014; pp. 67–82. [Google Scholar]
- Polito, L.; Bortolotti, M.; Maiello, S.; Battelli, M.G.; Bolognesi, A. Plants producing ribosome-inactivating proteins in traditional medicine. Molecules 2016, 21, 1560:1–1560:27. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Tserng, K.Y.; Chen, C.C.; Lin, L.T.; Tung, T.C. Abrin and ricin: new anti-tumour substances. Nature 1970, 227, 292–293. [Google Scholar] [CrossRef] [PubMed]
- De Virgilio, M.; Lombardi, A.; Caliandro, R.; Fabbrini, M.S. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins (Basel) 2010, 2, 2699–2737. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, E.; Di Maro, A. A new age for biomedical applications of ribosome inactivating proteins (RIPs): From bioconjugate to nanoconstructs. J. Biomed. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, B.; Müller-Uri, F.; Cameron-Mills, V.; Gough, S.; Simpson, D.; Skriver, K.; Mundy, J. The barley 60 kDa jasmonate-induced protein (JIP60) is a novel ribosome-inactivating protein. Plant J. 1994, 6, 815–824. [Google Scholar] [PubMed]
- Jiang, S.Y.; Ramamoorthy, R.; Bhalla, R.; Luan, H.F.; Venkatesh, P.N.; Cai, M.; Ramachandran, S. Genome-wide survey of the RIP domain family in Oryza sativa and their expression profiles under various abiotic and biotic stresses. Plant Mol. Biol. 2008, 67, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Y.; Bhalla, R.; Ramamoorthy, R.; Luan, H.F.; Venkatesh, P.N.; Cai, M.; Ramachandran, S. Over-expression of OSRIP18 increases drought and salt tolerance in transgenic rice plants. Transgenic Res. 2012, 21, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Vargas, L.R.B.; Carlini, C.R. Insecticidal and antifungal activities of ribosome-inactivating proteins. In Ribosome-Inactivating Proteins: Ricin and Related Proteins; Stirpe, F., Lappi, D., Eds.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2014; pp. 212–222. [Google Scholar]
- Di, R.; Tumer, N.E. Pokeweed antiviral protein: Its cytotoxicity mechanism and applications in plant disease resistance. Toxins (Basel) 2015, 7, 755–772. [Google Scholar] [CrossRef] [PubMed]
- Krivdova, G.; Neller, K.; Parikh, B.A.; Hudak, K.A. Antiviral and antifungal properties of RIPs. In Ribosome-Inactivating Proteins: Ricin and Related Proteins; Stirpe, F., Lappi, D., Eds.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2014; pp. 198–211. [Google Scholar]
- Motto, M.; Lupotto, E. The genetics and properties of cereal ribosome-inactivating proteins. Mini Rev. Med. Chem. 2004, 4, 493–503. [Google Scholar] [PubMed]
- Balconi, C.; Lanzanova, C.; Motto, M. Ribosome-inactivating proteins in cereals. In Toxic Plant Proteins; Lord, J., Hartley, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 149–166. [Google Scholar]
- Lapadula, W.J.; Sánchez Puerta, M.V.; Juri Ayub, M. Revising the taxonomic distribution, origin and evolution of ribosome inactivating protein genes. PLoS ONE 2013, 8, e72825:1–e72825:8. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Peumans, W.J.; Van Damme, E.J.M. Occurrence and taxonomical distribution of ribosome-inactivating proteins belonging to the Ricin/Shiga toxin superfamily. In Ribosome-Inactivating Proteins: Ricin and Related Proteins; Stirpe, F., Lappi, D., Eds.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2014; pp. 11–27. [Google Scholar]
- Robertus, J.D.; Monzingo, A.F. The structure of ribosome inactivating proteins. Mini Rev. Med. Chem. 2004, 4, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Montfort, W.; Villafranca, J.E.; Monzingo, A.F.; Ernst, S.R.; Katzin, B.; Rutenber, E.; Xuong, N.H.; Hamlin, R.; Robertus, J.D. The three-dimensional structure of ricin at 2.8 A. J. Biol. Chem. 1987, 262, 5398–5403. [Google Scholar] [PubMed]
- Shi, W.W.; Mak, A.N.S.; Wong, K.B.; Shaw, P.C. Structures and ribosomal interaction of ribosome-inactivating proteins. Molecules 2016, 21, 1588:1–1588:13. [Google Scholar] [CrossRef] [PubMed]
- Robertus, J.D.; Monzingo, A.F. The structure and action of ribosome-inactivating proteins. In Ribosome-Inactivating Proteins: Ricin and Related Proteins; Stirpe, F., Lappi, D., Eds.; John Wiley & Sons, Inc.: Somerset, NJ, USA, 2014; pp. 111–133. [Google Scholar]
- Tahirov, T.H.; Lu, T.H.; Liaw, Y.C.; Chen, Y.L.; Lin, J.Y. Crystal structure of abrin-a at 2.14 A. J. Mol. Biol. 1995, 250, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Katzin, B.J.; Collins, E.J.; Robertus, J.D. Structure of ricin A-chain at 2.5 A. Proteins Struct. Funct. Bioinform. 1991, 10, 251–259. [Google Scholar]
- Mishra, V.; Bilgrami, S.; Sharma, R.S.; Kaur, P.; Yadav, S.; Krauspenhaar, R.; Betzel, C.; Voelter, W.; Babu, C.R.; Singh, T.P. Crystal structure of Himalayan mistletoe ribosome-inactivating protein reveals the presence of a natural inhibitor and a new functionally active sugar-binding site. J. Biol. Chem. 2005, 280, 20712–20721. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Robertus, J.D. Analysis of several key active site residues of ricin a chain by mutagenesis and x-ray crystallography. Protein Eng. Des. Sel. 1992, 5, 775–779. [Google Scholar] [CrossRef]
- Ho, M.-C.; Sturm, M.B.; Almo, S.C.; Schramm, V.L. Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 20276–20281. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Mlsna, D.; Monzingo, A.F.; Ready, M.P.; Frankel, A.; Robertus, J.D. Structure of a ricin mutant showing rescue of activity by a noncatalytic residue. Biochemistry 1992, 31, 3294–3296. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.B.; Ke, Y.B.; Dong, Y.C.; Li, X.B.; Guo, Y.W.; Yeung, H.W.; Shaw, P.C. Structure/function relationship study of Gln156, Glu160 and Glu189 in the active site of trichosanthin. Eur. J. Biochem. 1994, 221, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Mak, A.N.S.; Wong, Y.T.; An, Y.J.; Cha, S.S.; Sze, K.H.; Au, S.W.N.; Wong, K.B.; Shaw, P.C. Structure-function study of maize ribosome-inactivating protein: Implications for the internal inactivation region and the sole glutamate in the active site. Nucleic Acids Res. 2007, 35, 6259–6267. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.G.; Kim, M.K.; Kim, B.W.; Suh, S.W.; Song, H.K. Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Hartings, H.; Lazzaroni, N.; Marsan, P.A.; Aragay, A.; Thompson, R.; Salamini, F.; Di Fonzo, N.; Palau, J.; Motto, M. The b-32 protein from maize endosperm: Characterization of genomic sequences encoding two alternative central domains. Plant Mol. Biol. 1990, 14, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Di Fonzo, N.; Manzocchi, L.; Salamini, F.; Soave, C. Purification and properties of an endospermic protein of maize associated with the Opaque-2 and Opaque-6 genes. Planta 1986, 167, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Lohmer, S.; Maddaloni, M.; Motto, M.; Hartings, H.; Salamini, F.; Thompson, R.D. The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO 1991, 10, 617–624. [Google Scholar]
- Bass, H.W.; Webster, C.; OBrian, G.R.; Roberts, J.K.; Boston, R.S. A maize ribosome-inactivating protein is controlled by the transcriptional activator Opaque-2. Plant Cell Online 1992, 4, 225–234. [Google Scholar] [CrossRef]
- Hey, T.D.; Hartley, M.; Walsh, T.A. Maize ribosome-inactivating protein (b-32). Homologs in related species, effects on maize ribosomes, and modulation of activity by pro-peptide deletions. Plant Physiol. 1995, 107, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Krawetz, J.E.; Boston, R.S. Substrate specificity of a maize ribosome-inactivating protein differs across diverse taxa. Eur. J. Biochem. 2000, 267, 1966–1974. [Google Scholar] [CrossRef] [PubMed]
- Docherty, K.; Steiner, D.F. Post-translational proteolysis in polypeptide hormone biosynthesis. Annu. Rev. Physiol. 1982, 44, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Bass, H.W.; Krawetz, J.E.; Obrian, G.R.; Zinselmeier, C.; Habben, J.E.; Boston, R.S. Maize ribosome-inactivating proteins (RIPs) with distinct expression patterns have similar requirements for proenzyme activation. J. Exp. Bot. 2004, 55, 2219–2233. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Mak, A.N.S.; Shaw, P.C.; Sze, K.H. Solution structure of an active mutant of maize ribosome-inactivating protein (MOD) and its interaction with the ribosomal stalk protein P2. J. Mol. Biol. 2010, 395, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Chuang, W.P.; Herde, M.; Ray, S.; Castano-Duque, L.; Howe, G.A.; Luthe, D.S. Caterpillar attack triggers accumulation of the toxic maize protein RIP2. New Phytol. 2014, 201, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, S.; Pollmann, S.; Buhr, F.; Springer, A.; Reinbothe, C.; von Wettstein, D.; Reinbothe, S. JIP60-mediated, jasmonate- and senescence-induced molecular switch in translation toward stress and defense protein synthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 14181–14186. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Tsurugi, K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J. Biol. Chem. 1988, 263, 8735–8739. [Google Scholar] [PubMed]
- Hudak, K.A.; Dinman, J.D.; Tumer, N.E. Pokeweed antiviral protein accesses ribosomes by binding to L3. J. Biol. Chem. 1999, 274, 3859–3864. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.S.B.; Chu, L.O.; Lee, K.M.; Too, P.H.M.; Ma, K.W.; Sze, K.H.; Zhu, G.; Shaw, P.C.; Wong, K.B. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007, 35, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.J.; Poon, G.M.K.; Bolewska-Pedyczak, E.; Srikumar, T.; Jeram, S.M.; Raught, B.; Gariépy, J. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J. Mol. Biol. 2008, 378, 375–386. [Google Scholar] [PubMed]
- Chiou, J.C.; Li, X.P.; Remacha, M.; Ballesta, J.P.G.; Tumer, N.E. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin a chain in Saccharomyces cerevisiae. Mol. Microbiol. 2008, 70, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Hung, F.S.J.; Chan, D.S.B.; Shaw, P.C. Trichosanthin interacts with acidic ribosomal proteins P0 and P1 and mitotic checkpoint protein MAD2B. Eur. J. Biochem. 2001, 268, 2107–2112. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 2009, 461, 1234–1242. [Google Scholar] [PubMed]
- Bargis-Surgey, P.; Lavergne, J.P.; Gonzalo, P.; Vard, C.; Filhol-Cochet, O.; Reboud, J.P. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur. J. Biochem. 1999, 262, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Helgstrand, M.; Mandava, C.S.; Mulder, F.A.A.; Liljas, A.; Sanyal, S.; Akke, M. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 2007, 365, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Nomura, N.; Honda, T.; Baba, K.; Naganuma, T.; Tanzawa, T.; Arisaka, F.; Noda, M.; Uchiyama, S.; Tanaka, I.; Yao, M.; et al. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc. Natl. Acad. Sci. USA 2012, 109, 3748–3753. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.K.H.; Wong, E.C.K.; Lee, K.M.; Wong, K.B. Structures of eukaryotic ribosomal stalk proteins and its complex with trichosanthin, and their implications in recruiting ribosome-inactivating proteins to the ribosomes. Toxins (Basel) 2015, 7, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Diaconu, M.; Kothe, U.; Schlünzen, F.; Fischer, N.; Harms, J.M.; Tonevitsky, A.G.; Stark, H.; Rodnina, M.V.; Wahl, M.C. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTpase activation. Cell 2005, 121, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.R. Copies of proteins L7 and L12 and heterogeneity of the large subunit of Escherichia coli ribosome. J. Mol. Biol. 1975, 95, 1–8. [Google Scholar] [CrossRef]
- Lee, K.M.; Yu, C.W.H.; Chan, D.S.B.; Chiu, T.Y.H.; Zhu, G.; Sze, K.H.; Shaw, P.C.; Wong, K.B. Solution structure of the dimerization domain of ribosomal protein P2 provides insights for the structural organization of eukaryotic stalk. Nucleic Acids Res. 2010, 38, 5206–5216. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakagaki, M.; Nishi, Y.; Kobayashi, Y.; Hachimori, A.; Uchiumi, T. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 2002, 30, 2620–2627. [Google Scholar] [CrossRef] [PubMed]
- McCluskey, A.J.; Bolewska-Pedyczak, E.; Jarvik, N.; Chen, G.; Sidhu, S.S.; Gariépy, J. Charged and hydrophobic surfaces on the a chain of Shiga-Like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS ONE 2012, 7, e31191:1–e31191:11. [Google Scholar] [CrossRef] [PubMed]
- Too, P.H.M.; Ma, M.K.W.; Mak, A.N.S.; Wong, Y.T.; Tung, C.K.C.; Zhu, G.; Au, S.W.N.; Wong, K.B.; Shaw, P.C. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009, 37, 602–610. [Google Scholar] [PubMed]
- Shi, W.W.; Tang, Y.S.; Sze, S.Y.; Zhu, Z.N.; Wong, K.B.; Shaw, P.C. Crystal structure of ribosome-inactivating protein ricin a chain in complex with the C-terminal peptide of the ribosomal stalk protein P2. Toxins (Basel) 2016, 8, 296:1–296:12. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhu, Y.; Wang, C.; Niu, L.; Teng, M.; Li, X. Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin. Sci. Rep. 2016, 6, 37803:1–37803:10. [Google Scholar]
- Wong, Y.T.; Ng, Y.M.; Mak, A.N.S.; Sze, K.H.; Wong, K.B.; Shaw, P.C. Maize ribosome-inactivating protein uses Lys158-Lys161 to interact with ribosomal protein P2 and the strength of interaction is correlated to the biological activities. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lapadula, W.J.; Sanchez-Puerta, M.V.; Juri Ayub, M. Convergent evolution led ribosome inactivating proteins to interact with ribosomal stalk. Toxicon 2012, 59, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Sturm, M.B.; Schramm, V.L. Detecting ricin: Sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal. Chem. 2009, 81, 2847–2853. [Google Scholar] [CrossRef] [PubMed]
- Harley, S.M.; Beevers, H. Ricin inhibition of in vitro protein synthesis by plant ribosomes. Proc. Natl. Acad. Sci. USA 1982, 79, 5935–5938. [Google Scholar] [CrossRef] [PubMed]
- Hartley, M.R.; Chacidock, J.A.; Bonness, M.S. The structure and function of ribosome-inactivating proteins. Trends Plant Sci. 1996, 1, 254–260. [Google Scholar] [CrossRef]
- Prestle, J.; Schönfelder, M.; Adam, G.; Mundry, K.W. Type 1 ribosome-inactivating proteins depurinate plant 25S rRNA without species specificity. Nucleic Acids Res. 1992, 20, 3179–3182. [Google Scholar] [CrossRef] [PubMed]
- Hartley, M.R.; Lord, J.M. Structure, function and applications of ricin and related cytotoxic proteins. In Biosynthesis and Manipulation of Plant Products; Grierson, D., Ed.; Springer: Houten, The Netherlands, 1993; pp. 210–239. [Google Scholar]
- Neuhaus, J.M.; Rogers, J.C. Sorting of proteins to vacuoles in plant cells. In Protein Trafficking in Plant Cells; Soll, J., Ed.; Springer: Houten, The Netherlands, 1998; pp. 127–144. [Google Scholar]
- Vitale, A.; Raikhel, N.V. What do proteins need to reach different vacuoles? Trends Plant Sci. 1999, 4, 149–155. [Google Scholar] [PubMed]
- Wu, T.H.; Chow, L.P.; Lin, J.Y. Sechiumin, a ribosome-inactivating protein from the edible gourd, Sechium edule Swartz: Purification, characterization, molecular cloning and expression. Eur. J. Biochem. 1998, 255, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Ready, M.P.; Brown, D.T.; Robertus, J.D. Extracellular localization of pokeweed antiviral protein. Proc. Natl. Acad. Sci. USA 1986, 83, 5053–5056. [Google Scholar] [CrossRef] [PubMed]
- Leah, R.; Tommerup, H.; Svendsen, I.; Mundy, J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 1991, 266, 1564–1573. [Google Scholar] [PubMed]
- Habuka, N.; Kataoka, J.; Miyano, M.; Tsuge, H.; Ago, H.; Noma, M. Nucleotide sequence of a genomic gene encoding tritin, a ribosome-inactivating protein from Triticum aestivum. Plant Mol. Biol. 1993, 22, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.E.; Irvin, J.D. Depurination of plant ribosomes by pokeweed antiviral protein. FEBS Lett. 1990, 273, 144–146. [Google Scholar] [CrossRef]
- Massiah, A.J.; Hartley, M.R. Wheat ribosome-inactivating proteins: Seed and leaf forms with different specificities and cofactor requirements. Planta 1995, 197, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, J.; Habuka, N.; Miyano, M.; Masuta, C.; Koiwai, A. Adenine depurination and inactivation of plant ribosomes by an antiviral protein of Mirabilis jalapa (MAP). Plant Mol. Biol. 1992, 20, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Sawasaki, T.; Nishihara, M.; Endo, Y. RIP and RALyase cleave the sarcin/ricin domain, a critical domain for ribosome function, during senescence of wheat coleoptiles. Biochem. Biophys. Res. Commun. 2008, 370, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Dunaeva, M.; Goebel, C.; Wasternack, C.; Parthier, B.; Goerschen, E. The jasmonate-induced 60 kDa protein of barley exhibits N-glycosidase activity in vivo. FEBS Lett. 1999, 452, 263–266. [Google Scholar] [CrossRef]
- Soave, C.; Reggiani, R.; Di Fonzo, N.; Salamini, F. Clustering of genes for 20 kd zein subunits in the short arm of maize chromosome 7. Genetics 1981, 363–377. [Google Scholar]
- Loesch, P.J.; Foley, D.C.; Cox, D.F. Comparative resistance of opaque-2 and normal inbred lines of maize to ear-rotting pathogens. Crop Sci. 1976, 16, 841–842. [Google Scholar]
- Gupta, S.C.; Asnani, V.L.; Khare, B.P. Effect of the opaque-2 gene in maize (Zea mays L.) on the extent of infestation by Sitophilus oryzae L. J. Stored Prod. Res. 1970, 6, 191–194. [Google Scholar] [CrossRef]
- Maddaloni, M.; Forlani, F.; Balmas, V.; Donini, G.; Stasse, L.; Corazza, L.; Motto, M. Tolerance to the fungal pathogen Rhizoctonia solani AG4 of transgenic tobacco expressing the maize ribosome-inactivating protein b-32. Transgenic Res. 1997, 6, 393–402. [Google Scholar] [CrossRef]
- Nielsen, K.; Payne, G.A.; Boston, R.S. Maize ribosome-inactivating protein inhibits normal development of Aspergillus nidulans and Aspergillus flavus. Mol. Plant Microbe Interact. 2001, 14, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Duan, X.; Wu, R.; Seok, S.J.; Boston, R.S.; Jang, I.C.; Eun, M.Y.; Nahm, B.H. Molecular and genetic analysis of transgenic rice plants expressing the maize ribosome-inactivating protein b-32 gene and the herbicide resistance bar gene. Mol. Breed. 1999, 5, 85–94. [Google Scholar] [CrossRef]
- Balconi, C.; Lanzanova, C.; Conti, E.; Triulzi, T.; Forlani, F.; Cattaneo, M.; Lupotto, E. Fusarium head blight evaluation in wheat transgenic plants expressing the maize b-32 antifungal gene. Eur. J. Plant Pathol. 2007, 117, 129–140. [Google Scholar] [CrossRef]
- Lanzanova, C.; Giuffrida, M.G.; Motto, M.; Baro, C.; Donn, G.; Hartings, H.; Lupotto, E.; Careri, M.; Elviri, L.; Balconi, C. The Zea mays b-32 ribosome-inactivating protein efficiently inhibits growth of Fusarium verticillioides on leaf pieces in vitro. Eur. J. Plant Pathol. 2009, 124, 471–482. [Google Scholar] [CrossRef]
- Lanzanova, C.; Torri, A.; Motto, M.; Balconi, C. Characterization of the maize b-32 ribosome inactivating protein and its interaction with fungal pathogen development. Maydica 2011, 56, 1709:1–1709:11. [Google Scholar]
- Dowd, P.F.; Mehta, A.D.; Boston, R.S. Relative toxicity of the maize endosperm ribosome-inactivating protein to insects. J. Agric. Food Chem. 1998, 46, 3775–3779. [Google Scholar] [CrossRef]
- Dowd, P.F.; Zuo, W.N.; Gillikin, J.W.; Johnson, E.T.; Boston, R.S. Enhanced resistance to Helicoverpa zea in tobacco expressing an activated form of maize ribosome-inactivating protein. J. Agric. Food Chem. 2003, 51, 3568–3574. [Google Scholar] [CrossRef] [PubMed]
- Dowd, P.F.; Holmes, R.A.; Pinkerton, T.S.; Johnson, E.T.; Lagrimini, L.M.; Boston, R.S. Relative activity of a tobacco hybrid expressing high levels of a tobacco anionic peroxidase and maize ribosome-inactivating protein against Helicoverpa zea and Lasioderma serricorne. J. Agric. Food Chem. 2006, 54, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- Dowd, P.F.; Johnson, E.T.; Price, N.P. Enhanced pest resistance of maize leaves expressing monocot crop plant-derived ribosome-inactivating protein and agglutinin. J. Agric. Food Chem. 2012, 60, 10768–10775. [Google Scholar] [CrossRef] [PubMed]
- Bass, H.W.; Obrian, C.R.; Boston, R.S. Cloning and sequencing of a second ribosome-inactivating protein gene from maize. Plant Physiol. 1995, 107, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.D.; Willcox, M.C.; Williams, W.P.; Buckley, P.M. Quantitative trait loci conferring resistance to fall armyworm and southwestern corn borer leaf feeding damage. Crop Sci. 2005, 45, 2430–2434. [Google Scholar] [CrossRef]
- Vigers, A.J.; Roberts, W.K.; Selitrennikoff, C.P. A new family of plant antifungal proteins. Mol. Plant Microbe Interact. 1991, 4, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.R.; Chandrashekar, A.; Shetty, H.S. Proteins in developing sorghum endosperm that may be involved in resistance to grain moulds. J. Sci. Food Agric. 1992, 60, 275–282. [Google Scholar] [CrossRef]
- Darnetty; Leslie, J.F.; Muthukrishnan, S.; Swegle, M.; Vigers, A.J.; Selitrennikoff, C.P. Variability in antifungal proteins in the grains of maize, sorghum and wheat. Physiol. Plant. 1993, 88, 339–349. [Google Scholar] [CrossRef]
- Seetharaman, K.; Whitehead, E.; Keller, N.P.; Waniska, R.D.; Rooney, L.W. In vitro activity of sorgum seed antifungal proteins against grain mold pathogens. J. Agric. Food Chem. 1997, 45, 3666–3671. [Google Scholar] [CrossRef]
- Seetharaman, K.; Waniska, R.D.; Rooney, L.W. Physiological changes in sorghum antifungal proteins. J. Agric. Food Chem. 1996, 44, 2435–2441. [Google Scholar] [CrossRef]
- Rodríguez-Herrera, R.; Waniska, R.D.; Rooney, W.L. Antifungal proteins and grain mold resistance in sorghum with nonpigmented testa. J. Agric. Food Chem. 1999, 47, 4802–4806. [Google Scholar] [CrossRef] [PubMed]
- Asano, K.; Svensson, B.; Poulsen, F.M. Isolation and characterization of inhibitors of animal cell-free protein synthesis from barley seeds. Carlsb. Res. Commun 1984, 49, 619–626. [Google Scholar] [CrossRef]
- Oldach, K.H.; Becker, D.; Lörz, H. Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat. Mol. Plant Microbe Interact. 2001, 14, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Bieri, S.; Potrykus, I.; Fütterer, S. Expression of active barley seed ribosome-inactivating protein in transgenic wheat. Theor. Appl. Genet. 2000, 100, 755–763. [Google Scholar] [CrossRef]
- Bieri, S.; Potrykus, I.; Fütterer, J. Effects of combined expression of antifungal barley seed proteins in transgenic wheat on powdery mildew infection. Mol. Breed. 2003, 11, 37–48. [Google Scholar] [CrossRef]
- Chhikara, S.; Chaudhury, D.; Dhankher, O.P.; Jaiwal, P.K. Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae. Plant Cell Tissue Organ Cult. 2012, 108, 83–89. [Google Scholar] [CrossRef]
- M’hamdi, M.; Chikh-Rouhou, H.; Boughalleb, N.; de Galarreta, J.R. resistance to Rhizoctonia solani by combined expression of chitinase and ribosome inactivating protein in transgenic potatoes (Solanum tuberosum L). Span. J. Agric. Res. 2012, 10, 778–785. [Google Scholar] [CrossRef]
- Chopra, R.; Saini, R. Transformation of blackgram (Vigna mungo (L.) hepper) by barley chitinase and ribosome-inactivating protein genes towards improving resistance to Corynespora leaf spot fungal disease. Appl. Biochem. Biotechnol. 2014, 174, 2791–2800. [Google Scholar] [CrossRef] [PubMed]
- Motto, M.; Thompson, R.; Salamini, F. Genetic regulation of carbohydrate and protein accumulation in seeds. In Cellular and Molecular Biology of Plant Seed Development; Larkins, B.A., Vasil, I.K., Eds.; Springer: Houten, The Netherlands, 1997; pp. 479–522. [Google Scholar]
- Habben, J.E.; Kirleis, A.W.; Larkins, B.A. The origin of lysine-containing proteins in opaque-2 maize endosperm. Plant Mol. Biol. 1993, 23, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.J.; Wu, X.H.; Wang, T. The rice tapetum-specific gene RA39 encodes a type I ribosome-inactivating protein. Sex. Plant Reprod. 2002, 15, 205–212. [Google Scholar]
- Ozawa, A.; Sawasaki, T.; Takai, K.; Uchiumi, T.; Hori, H.; Endo, Y. RALyase; A terminator of elongation function of depurinated ribosomes. FEBS Lett. 2003, 555, 455–458. [Google Scholar] [CrossRef]
- Nam, H. The molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 1997, 8, 200–207. [Google Scholar] [CrossRef]
- Pennell, R.; Lamb, C. Programmed cell death in plants. Plant Cell 1997, 9, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Law, S.K.Y.; Wang, R.R.; Mak, A.N.S.; Wong, K.B.; Zheng, Y.T.; Shaw, P.C. A switch-on mechanism to activate maize ribosome-inactivating protein for targeting HIV-infected cells. Nucleic Acids Res. 2010, 38, 6803–6812. [Google Scholar] [CrossRef] [PubMed]
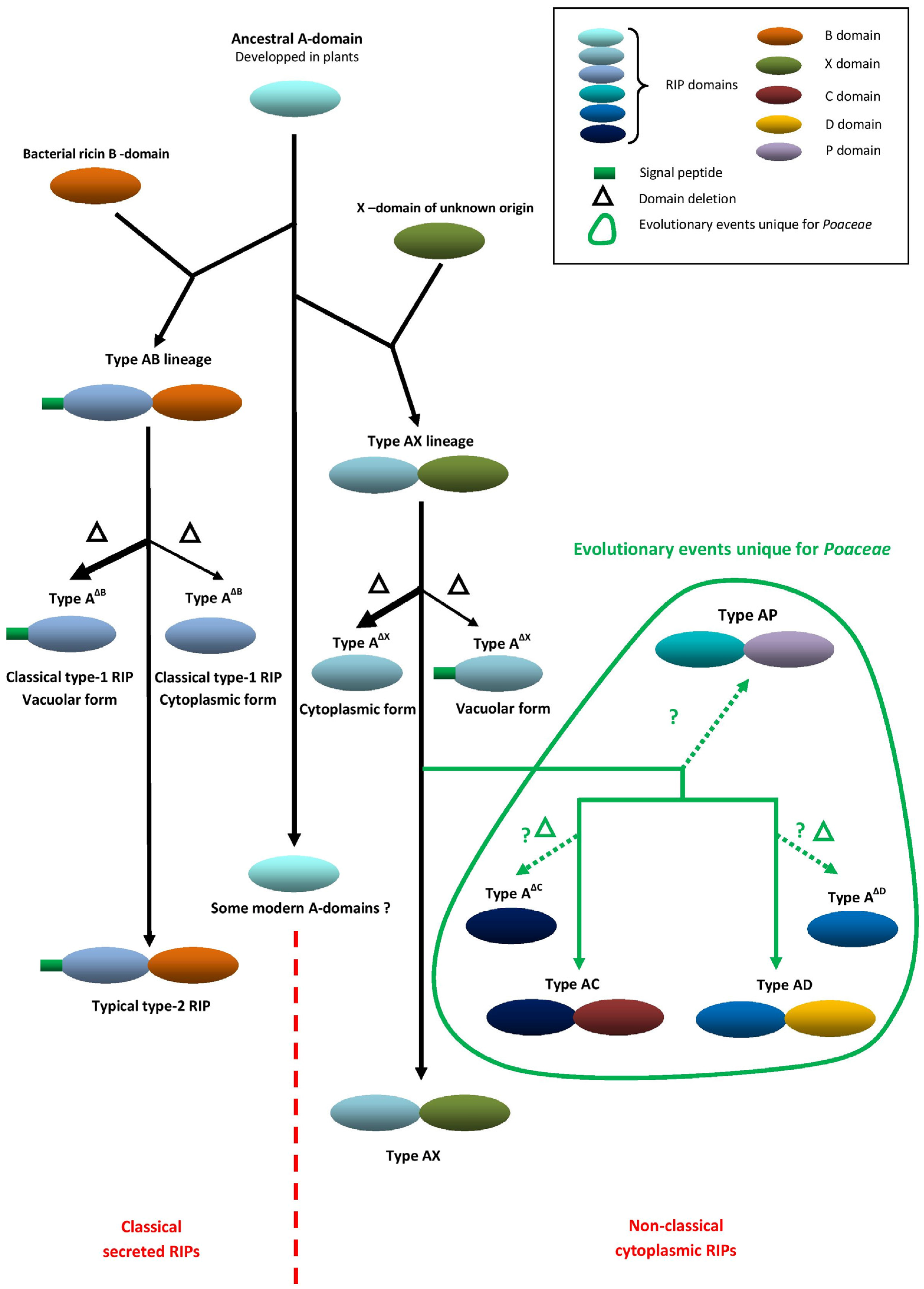

| Species | RIP Gene Architectures Reported |
|---|---|
| Oryza sativa (rice) | Au, AC, AP |
| Avena barbata (oat) | Au, AC |
| Hordeum vulgare (barley) | Au, AC |
| Triticum aestivum (wheat) | Au, AB, AP |
| Sorghum bicolor (sorghum) | Au, AB |
| Zea mays (maize) | Au, AB, AC, AD |
| RIP | Active Site Residues | ||||
|---|---|---|---|---|---|
| RTA | Y80 | Y123 | E177 | R180 | W211 |
| Maize RIP1 = b-32 | Y94 | Y130 | E207 | R210 | W241 |
| Barley bRIP1 | Y87 | Y118 | E175 | R178 | W213 |
| Species | RIP | Tissue | Role in Defense | In Planta Function |
|---|---|---|---|---|
| Zea mays | Maize RIP1 (= b-32) | Seeds | Antifungal, insecticidal | Storage function in seeds? |
| Maize RIP2 | Whole plant, except kernel | Expression upon herbivore attack, active against Spodoptera frugiperda | Involved in drought response? | |
| Sorghum bicolor | Sorghum RIP | Seeds | Antifungal protein | Not reported |
| Oryza sativa | OsRIP18 = RA39 | Tapetum | Not reported | Involved in drought and salt response. Involved in microspore maturation? |
| Other rice RIPs | Variable | Expression of several genes induced by Magnaporthe grisea or Xanthomonas oryzae | Expression of several genes is enhanced after abiotic stress | |
| Triticum aestivum | Tritin | Seed and leaf forms | Not reported | Involved in senescence |
| Hordeum vulgare | RIP30 and isoforms | Seeds | Antifungal protein | Not reported |
| JIP60 | Leaves | Re-organization of translational machinery in stress situations | Re-organization of translational machinery in stress situations |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaeytijd, J.D.; Damme, E.J.M.V. Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles. Toxins 2017, 9, 123. https://doi.org/10.3390/toxins9040123
Zaeytijd JD, Damme EJMV. Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles. Toxins. 2017; 9(4):123. https://doi.org/10.3390/toxins9040123
Chicago/Turabian StyleZaeytijd, Jeroen De, and Els J. M. Van Damme. 2017. "Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles" Toxins 9, no. 4: 123. https://doi.org/10.3390/toxins9040123
APA StyleZaeytijd, J. D., & Damme, E. J. M. V. (2017). Extensive Evolution of Cereal Ribosome-Inactivating Proteins Translates into Unique Structural Features, Activation Mechanisms, and Physiological Roles. Toxins, 9(4), 123. https://doi.org/10.3390/toxins9040123







