Diversity of Pea-Associated F. proliferatum and F. verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis
Abstract
:1. Introduction
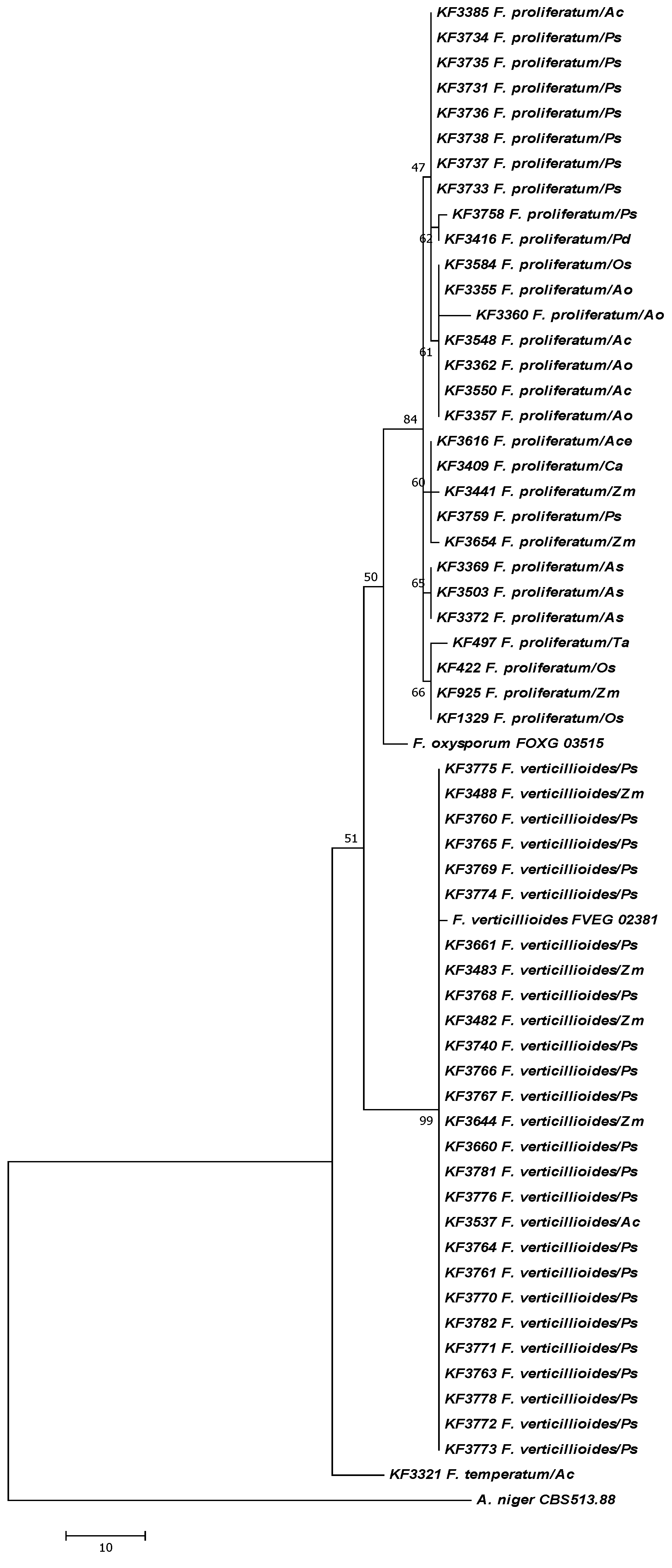
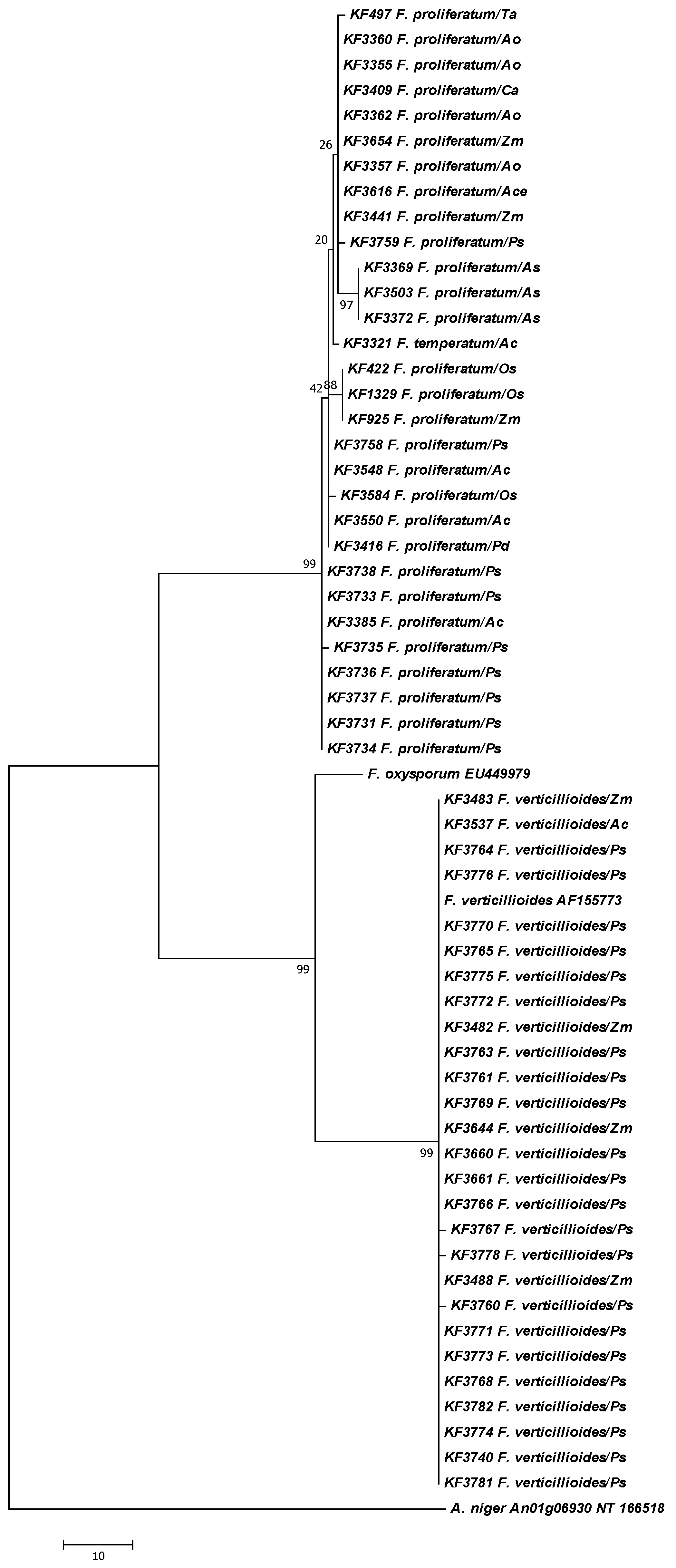
2. Results
| Strain | Species | Host/cultivar/locality | Year | Origin |
| KF 3758 | F. proliferatum | P. sativum/SOKOLIK/W | 2012 | Poland |
| KF 3759 | F. proliferatum | P. sativum/TARCHALSKA/W | 2012 | Poland |
| KF 3735 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3736 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3737 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3738 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3731 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3733 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3734 | F. proliferatum | P. sativum/TURNIA/R | 2012 | Poland |
| KF 3763 | F. verticillioides | P. sativum/WIATO/W | 2012 | Poland |
| KF 3764 | F. verticillioides | P. sativum/WIATO/W | 2012 | Poland |
| KF 3765 | F. verticillioides | P. sativum/WIATO/W | 2012 | Poland |
| KF 3661 | F. verticillioides | P. sativum/EUREKA/R | 2012 | Poland |
| KF 3740 | F. verticillioides | P. sativum/EUREKA/R | 2012 | Poland |
| KF 3660 | F. verticillioides | P. sativum/EUREKA/R | 2012 | Poland |
| KF 3766 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3767 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3768 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3769 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3770 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3771 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3772 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3773 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3774 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3775 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3776 | F. verticillioides | P. sativum/EZOP/W | 2012 | Poland |
| KF 3778 | F. verticillioides | P. sativum/TARCHALSKA/W | 2012 | Poland |
| KF 3760 | F. verticillioides | P. sativum/TURNIA/W | 2012 | Poland |
| KF 3761 | F. verticillioides | P. sativum/TURNIA/W | 2012 | Poland |
| KF 3781 | F. verticillioides | P. sativum/TURNIA/W | 2012 | Poland |
| KF 3782 | F. verticillioides | P. sativum/TURNIA/W | 2012 | Poland |
| Strain | Species | Collection strains | Year | Origin |
| CBS 513.88 | A. niger | NT_166526.1 | ||
| FOXG_03515 | F. oxysporum | |||
| FVEG_02381 | F. verticillioides | Z. mays | ||
| KF 497 | F. proliferatum | T. aestivum | 1987 | Portugal |
| KF 925 | F. proliferatum | Z. mays | 1986 | Poland |
| KF 3441 | F. proliferatum | Z. mays | 2006 | Poland |
| KF 3654 | F. proliferatum | Z. mays | 2011 | Poland |
| KF 3616 | F. proliferatum | A. cepa | 2011 | Poland |
| KF 3385 | F. proliferatum | A. comosus | 2009 | Vietnam |
| KF 3548 | F. proliferatum | A. comosus | 2011 | Ecuador |
| KF 3550 | F. proliferatum | A. comosus | 2011 | Ecuador |
| KF 3355 | F. proliferatum | A. officinalis | 2009 | Poland |
| KF 3357 | F. proliferatum | A. officinalis | 2009 | Poland |
| KF 3362 | F. proliferatum | A. officinalis | 2009 | Poland |
| KF 3360 | F. proliferatum | A. officinalis | 2009 | Poland |
| KF 3369 | F. proliferatum | A. sativum | 2009 | Poland |
| KF 3372 | F. proliferatum | A. sativum | 2009 | Poland |
| KF 3503 | F. proliferatum | A. sativum | 2010 | Poland |
| KF 3409 | F. proliferatum | Cambria | 2010 | |
| KF 422 | F. proliferatum | O. sativa | 1973 | Taiwan |
| KF 1329 | F. proliferatum | O. sativa | Japan | |
| KF 3584 | F. proliferatum | O. sativa | 2011 | Thailand |
| KF 3416 | F. proliferatum | P. dactylifera | 2010 | Tunisia |
| KF 3321 | F. temperatum | A. comosus | 2008 | Costa Rica |
| KF 3488 | F. verticillioides | Z. mays | 2010 | Poland |
| KF 3482 | F. verticillioides | Z. mays | 2010 | Poland |
| KF 3483 | F. verticillioides | Z. mays | 2010 | Poland |
| KF 3644 | F. verticillioides | Z. mays | 2010 | Poland |
| KF 3537 | F. verticillioides | A. comosus | 2010 | Costa Rica |
| Sample | FB1 | FB2 | FB3 |
|---|---|---|---|
| EUREKA_I (W) | 1.12 | 0.05 | 0.01 |
| EUREKA _II (W) | 1.34 | 0.07 | 0.02 |
| EUREKA _III (W) | 0.79 | 0.05 | 0.01 |
| EUREKA _IV (W) | 0.81 | 0.04 | 0.01 |
| Mean ± SD | 1.02 ± 0.26 | 0.05 ± 0.01 | 0.01 ± 0.01 |
| EUREKA_I (R) | 1.11 | 0.12 | 0.05 |
| EUREKA_II (R) | 0.72 | 0,03 | 0.00 |
| EUREKA III (R) | 0.45 | 0.12 | 0.08 |
| EUREKA_IV (R) | 0.63 | 0.04 | 0.01 |
| Mean ± SD | 0.73 ± 0.28 | 0.08 ± 0.05 | 0.04 ± 0.04 |
| TURNIA_I (W) | 0.85 | 0.11 | 0.04 |
| TURNIA _II (W) | 0.81 | 0.09 | 0.05 |
| TURNIA _III (W) | 0.92 | 0.07 | 0.01 |
| TURNIA _IV (W) | 0.91 | 0.08 | 0.02 |
| Mean ± SD | 0.87 ± 0.05 | 0.09 ± 0.02 | 0.03 ± 0.02 |
| TURNIA_I (R) | 0.55 | 0.06 | 0.02 |
| TURNIA_II (R) | 0.61 | 0.07 | 0.01 |
| TURNIA_III (R) | 1.27 | 0.14 | 0.08 |
| TURNIA_IV (R) | 1.48 | 0.15 | 0.09 |
| Mean ± SD | 0.98 ± 0.47 | 0.11 ± 0.05 | 0.05 ± 0.04 |
| Strain | Species | Host/cultivar/locality | FB1 | FB2 | FB3 |
|---|---|---|---|---|---|
| KF 3779 | F. verticillioides | P. sativum/TURNIA/W | 155.92 ± 12.31 | 0.60 ± 0.02 | 5.44 ± 1.13 |
| KF 3780 | F. verticillioides | P. sativum/TURNIA/W | 183.55 ± 14.03 | 0.98 ± 0.03 | 16.75 ± 2.52 |
| KF 3760 | F. verticillioides | P. sativum/TURNIA/W | 317.24 ± 16.32 | 34.00 ± 3.15 | 7.01 ± 1.14 |
| KF 3781 | F. verticillioides | P. sativum/TURNIA/W | 80.39 ± 7.44 | 0.49 ± 0.15 | 2.66 ± 0.09 |
| KF 3761 | F. verticillioides | P. sativum/TURNIA/W | 202.29 ± 10.18 | 95.61 ± 8.53 | 37.44 ± 4.79 |
| KF 3782 | F. verticillioides | P. sativum/TURNIA/W | 111.65 ± 11.59 | 52.35 ± 4.17 | 27.62 ± 5.56 |
| KF 3731 | F. proliferatum | P. sativum/TURNIA/R | 111.07 ± 9.47 | 49.61 ± 5.22 | 43.58 ± 6.85 |
| KF 3732 | F. proliferatum | P. sativum/TURNIA/R | 958.51 ± 21.06 | 271.22 ± 10.84 | 104.30 ± 9.48 |
| KF 3733 | F. proliferatum | P. sativum/TURNIA/R | 121.65 ± 10.11 | 51.99 ± 6.63 | 37.71 ± 5.54 |
| KF 3734 | F. proliferatum | P. sativum/TURNIA/R | 49.36 ± 5.33 | 29.26 ± 4.12 | 30.85 ± 3.52 |
| KF 3735 | F. proliferatum | P. sativum/TURNIA/R | 845.56 ± 42.67 | 227.99 ± 10.47 | 112.65 ± 9.65 |
| KF 3736 | F. proliferatum | P. sativum/TURNIA/R | 476.53 ± 25.13 | 227.78 ± 11.58 | 111.39 ± 8.83 |
| KF 3737 | F. proliferatum | P. sativum/TURNIA/R | 106.09 ± 9.88 | 49.72 ± 5.39 | 43.84 ± 5.28 |
| KF 3738 | F. proliferatum | P. sativum/TURNIA/R | 648.30 ± 21.56 | 180.68 ± 15.41 | 101.13 ± 4.69 |
| KF 3660 | F. verticillioides | P. sativum/EUREKA/R | 78.74 ± 7.40 | 24.20 ± 3.30 | 4.68 ± 0.08 |
| KF 3740 | F. verticillioides | P. sativum/EUREKA/R | 216.50 ±15.84 | 89.95 ± 7.17 | 43.33 ± 6.16 |
| KF 3661 | F. verticillioides | P. sativum/EUREKA/R | 1861.52 ± 54.23 | 108.16 ± 7.49 | 40.61 ± 5.58 |
| KF 3758 | F. proliferatum | P. sativum/EZOP/W | 212.02 ± 12.36 | 63.50 ± 8.82 | 44.01 ± 6.85 |
| KF 3416 | F. proliferatum | P. dactylifera | 46.34 ± 5.41 | 26.11 ± 5.41 | 6.53 ± 0.08 |
| KF 3357 | F. proliferatum | A. officinalis | 1536.00 ± 52.33 | 657.09 ± 80.35 | 123.72 ± 8.71 |
| KF 3654 | F. proliferatum | Z. mays | 1578.04 ± 41.25 | 529.96 ± 69.15 | 91.34 ± 9.13 |
| KF 3584 | F. proliferatum | O. sativa | 201.20 ± 7.69 | 53.60 ± 7.74 | 33.00 ± 4.28 |
| KF 3409 | F. proliferatum | Cambria sp. | 668.72 ± 18.47 | 170.07 ± 25.13 | 70.19 ± 6.71 |
| KF 3503 | F. proliferatum | A. sativum | 1186.87 ± 87.42 | 185.54 ± 18.53 | 66.24 ± 5.93 |
| KF 3537 | F. verticillioides | A. comosus | 59.65 ± 6.06 | 19.37 ± 2.47 | 5.86 ± 0.98 |
| KF 3644 | F. verticillioides | Z. mays | 6.12 ± 1.52 | 0.37 ± 0.05 | 0.29 ± 0.03 |
| KF 3488 | F. verticillioides | Z. mays | 39.77 ± 4.15 | 0.96 ± 0.09 | 0.06 ± 0.01 |
| KF 3482 | F. verticillioides | Z. mays | 273.38 ± 14.39 | 60.35 ± 1.11 | 0.88 ± 0.04 |
| KF 3483 | F. verticillioides | Z. mays | 14.17 ± 2.08 | 0.04 ± 0.01 | 0.00 ± 0.00 |
3. Discussion
4. Experimental Section
4.1. Seed Samples and Purification of Fungal Strains
4.2. Fusarium Species Identification
4.3. Molecular Analyses: DNA Extraction, Primers and PCR Conditions
4.4. DNA Sequencing, Analysis and Phylogeny Reconstruction
4.5. Fumonisin Quantification
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Urbano, G.; Aranda, P.; Gómez-Villalva, E.; Frejnagel, S.; Porres, J.M.; Frías, J.; Vidal-Valverde, C.N.; López-Jurado, M. Nutritional evaluation of Pea (Pisum sativum L.) protein diets after mild hydrothermal treatment and with and without added phytase. J. Agric. Food Chem. 2003, 51, 2415–2420. [Google Scholar] [CrossRef]
- Marcinkowska, J. Fungi occurrence on seeds of field pea. Acta Mycol. 2008, 43, 77–89. [Google Scholar]
- Ozgonen, H.; Gulcu, M. Determination of mycoflora of pea (Pisum sativum) seeds and the effects of Rhizobium leguminosorum on fungal pathogens of peas. Afr. J. Biotechnol. 2011, 10, 6235–6240. [Google Scholar]
- Ali, M.; Nitschke, L.; Krause, M.; Cameron, B. Selection of pea lines for resistance to pathotypes of Ascochyta pinodes, A. pisi and Phoma medicaginis var. pinodella. Aust. J. Agric. Res. 1978, 29, 841–849. [Google Scholar]
- Bretag, T.W.; Keane, P.J.; Price, T.V. The epidemiology and control of ascochyta blight in field peas: A review. Aust. J. Agric. Res. 2006, 57, 883–902. [Google Scholar] [CrossRef]
- Susuri, L.; Hagedorn, D.J.; Rand, R.E. Alternaria blight of pea. Plant. Dis. 1982, 66, 328–330. [Google Scholar] [CrossRef]
- Desjardins, A.E. Fusarium Mycotoxins: Chemistry, Genetics and Biology; Amer Phytopathological Society: St. Paul, MN, USA, 2006. [Google Scholar]
- Jurado, M.; Marin, P.; Callejas, C.; Moretti, A.; Vazquez, C.; Gonzalez-Jaen, M.T. Genetic variability and fumonisin production by Fusarium proliferatum. Food Microbiol. 2010, 27, 50–57. [Google Scholar]
- Rheeder, J.P.; Marasas, W.F.O.; Vismer, H.F. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002, 68, 2101–2105. [Google Scholar]
- Waśkiewicz, A.; Beszterda, M.; Golinski, P. Occurrence of fumonisins in food—An interdisciplinary approach to the problem. Food Control 2012, 26, 491–499. [Google Scholar]
- Marasas, W.F.O. Discovery and occurrence of the fumonisins: A historical perspective. Environ. Health Perspect. 2001, 109, 239–243. [Google Scholar]
- Ueno, Y.; Iijima, K.; Wang, S.D.; Sugiura, Y.; Sekijima, M.; Tanaka, T.; Chen, C.; Yu, S.Z. Fumonisins as a possible contributory risk factor for primary liver cancer: A 3-year study of corn harvested in Hainan, China by HPLC and ELISA. Food Chem. Toxicol. 1997, 35, 1143–1150. [Google Scholar]
- Missmer, S.A.; Suarez, L.; Felkner, M.; Wang, E.; Merrill, A.H., Jr.; Rothman, K.J.; Hendricks, K.A. Exposure to fumonisins and the occurrence of neural tube defects along the Texas-Mexico border. Environ. Health Perspect. 2006, 114, 237–241. [Google Scholar] [CrossRef]
- Fincham, J.E.; Marasas, W.F.O.; Taljaard, J.J.; Kriek, N.P.; Badenhorst, C.J.; Gelderblom, W.C.; Seier, J.V. Atherogenic effects in a non-human primate of Fusarium moniliforme cultures added to a carbohydrate diet. Atherosclerosis 1992, 94, 13–25. [Google Scholar]
- International Agency for Research on Cancer (IARC) Fumonisin B1, IARC Monographs on the Evaluation of the Carcinogenic Risks to Humans: Some Traditional Herbal Medicines, Some Mycotoxins, Naphthalene and Styrene; IARC: Lyon, France, 2002; Volume 82, pp. 301–366.
- Covarelli, L.; Stifano, S.; Beccari, G.; Raggi, L.; Lattanzio, V.M.T.; Albertini, E. Characterization of Fusarium verticillioides strains isolated from maize in Italy: Fumonisin production, pathogenicity and genetic variability. Food Microbiol. 2012, 31, 17–24. [Google Scholar] [CrossRef]
- Scauflaire, J.; Gourgue, M.; Munaut, F. Fusarium temperatum sp. nov. from maize, an emergent species closely related to Fusarium subglutinans. Mycologia 2011, 103, 586–597. [Google Scholar] [CrossRef]
- Torelli, E.; Firrao, G.; Bianchi, G.; Saccardo, F.; Locci, R. The influence of local factors on the prediction of fumonisin contamination in maize. J. Sci. Food Agric. 2012, 92, 1808–1814. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Wit, M.; Goliński, P.; Chełkowski, J.; Warzecha, R.; Ochodzki, P.; Wakuliński, W. Kinetics of fumonisin B1 formation in maize ears inoculated with Fusarium verticillioides. Food Addt. Contam. 2012, 29, 1752–1761. [Google Scholar] [CrossRef]
- Hsuan, H.M.; Salleh, B.; Zakaria, L. Molecular identification of Fusarium species in Gibberella fujikuroi species complex from rice, sugarcane and maize from Peninsular Malaysia. Int. J. Mol. Sci. 2011, 12, 6722–6732. [Google Scholar] [CrossRef]
- Sharma, R.; Thakur, R.P.; Senthilvel, S.; Nayak, S.; Reddy, S.V.; Rao, V.P.; Varshney, R.K. Identification and characterization of toxigenic Fusaria associated with sorghum grain mold complex in India. Mycopathologia 2011, 171, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Chehri, K.; Jahromi, S.T.; Reddy, K.R.N.; Abbasi, S.; Salleh, B. Occurrence of Fusarium spp. and fumonisins in stored wheat grains marketed in Iran. Toxins 2010, 2, 2816–2823. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Busman, M.; Proctor, R.H.; Stessman, R. Wheat kernel black point and fumonisin contamination by Fusarium proliferatum. Food Addit. Contam. 2007, 24, 1131–1137. [Google Scholar] [CrossRef]
- Palmero, D.; de Cara, M.; Nosir, W.; Gálvez, L.; Cruz, A.; Woodward, S.; González-Jaén, M.T.; Tello, J.C. Fusarium proliferatum isolated from garlic in Spain: Identification, toxigenic potential and pathogenicity on related Allium species. Phytopathol. Mediterr. 2012, 51, 207–218. [Google Scholar]
- Stankovic, S.; Levic, J.; Petrovic, T.; Logrieco, A.; Moretti, A. Pathogenicity and mycotoxin production by Fusarium proliferatum isolated from onion and garlic in Serbia. Eur. J. Plant Pathol. 2007, 118, 165–172. [Google Scholar] [CrossRef]
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. Genetic and phenotypic variation of Fusarium proliferatum isolates from different host species. J. Appl. Genet. 2011, 52, 487–96. [Google Scholar] [CrossRef]
- Tonti, S.; Prà, M.D.; Nipoti, P.; Prodi, A.; Alberti, I. First report of Fusarium proliferatum causing rot of stored garlic bulbs (Allium sativum L.) in Italy. J. Phytopathol. 2012, 160, 761–763. [Google Scholar] [CrossRef]
- Von Bargen, S.; Martinez, O.; Schadock, I.; Eisold, A.M.; Gossmann, M.; Buttner, C. Genetic variability of phytopathogenic Fusarium proliferatum associated with crown rot in Asparagus officinalis. J. Phytopathol. 2009, 157, 446–456. [Google Scholar] [CrossRef]
- Karolewski, Z.; Waśkiewicz, A.; Irzykowska, L.; Bocianowski, J.; Kostecki, M.; Goliński, P.; Knaflewski, M.; Weber, Z. Fungi presence and their mycotoxins distribution in asparagus spears. Polish J. Environ. Stud. 2011, 20, 911–919. [Google Scholar]
- Waśkiewicz, A.; Irzykowska, L.; Bocianowski, J.; Karolewski, Z.; Kostecki, M.; Weber, Z.; Goliński, P. Occurrence of Fusarium fungi and mycotoxins in marketable asparagus spears. Polish J. Environ. Stud. 2010, 49, 367–372. [Google Scholar]
- Waśkiewicz, A.; Irzykowska, L.; Karolewski, Z.; Bocianowski, J.; Kostecki, M.; Goliński, P.; Knaflewski, M.; Weber, Z. Fusarium spp. and mycotoxins present in asparagus spears. Cereal Res. Comm. 2008, 36, 405–407. [Google Scholar]
- Waśkiewicz, A.; Stępień, L. Mycotoxins biosynthesized by plant-derived Fusarium isolates. Arch. Ind. Hyg. Toxicol. 2012, 63, 479–488. [Google Scholar]
- Garcia, D.; Barros, G.; Chulze, S.; Ramos, A.J.; Sanchis, V.; Marín, S. Impact of cycling temperatures on Fusarium verticillioides and Fusarium graminearum growth and mycotoxins production in soybean. J. Sci. Food Agric. 2012, 92, 2952–2959. [Google Scholar]
- Punja, Z.K.; Wan, A.; Rahman, M.; Goswami, R.S.; Barasubiye, T.; Seifert, K.A.; Lévesque, C.A. Growth, population dynamics, and diversity of Fusarium equiseti in ginseng fields. Eur. J. Plant Pathol. 2008, 121, 173–184. [Google Scholar] [CrossRef]
- Reynoso, M.M.; Chulze, S.N.; Zeller, K.A.; Torres, A.M.; Leslie, J.F. Genetic structure of Fusarium verticillioides populations isolated from maize in Argentina. Eur. J. Plant. Pathol. 2009, 123, 207–215. [Google Scholar] [CrossRef]
- Rocha, L.O.; Reis, G.M.; da Silva, V.N.; Braghini, R.; Teixeira, M.M.; Corrêa, B. Molecular characterization and fumonisin production by Fusarium verticillioides isolated from corn grains of different geographic origins in Brazil. Int. J. Food Microbiol. 2011, 145, 9–21. [Google Scholar]
- Rossi, V.; Scandolara, A.; Battilani, P. Effect of environmental conditions on spore production by Fusarium verticillioides, the causal agent of maize ear rot. Eur. J. Plant. Pathol. 2009, 123, 159–169. [Google Scholar] [CrossRef]
- Kulik, T.; Pszczółkowska, A.; Łojko, M. Multilocus phylogenetics show high intraspecific variability within Fusarium avenaceum. Int. J. Mol. Sci. 2011, 12, 5626–5640. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Alexander, N.J.; Desjardins, A.E. Evidence that a secondary metabolite gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 2009, 74, 1128–1142. [Google Scholar] [CrossRef]
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. FUM cluster divergence in fumonisins-producing Fusarium species. Fungal Biol. 2011, 115, 112–123. [Google Scholar] [CrossRef]
- Stępień, Ł.; Gromadzka, K.; Chełkowski, J. Polymorphism of mycotoxin biosynthetic genes among Fusarium equiseti isolates from Italy and Poland. J. Appl. Genet. 2012, 53, 227–36. [Google Scholar] [CrossRef]
- Stępień, Ł.; Waśkiewicz, A. Beauvericin and enniatins biosynthesis by Fusarium species and its relation to the divergence of enniatin synthase gene. Toxins 2013. submitted for publication. [Google Scholar]
- Butchko, R.A.; Brown, D.W.; Busman, M.; Tudzynski, B.; Wiemann, P. Lae1 regulates expression of multiple secondary metabolite gene clusters in Fusarium verticillioides. Fungal Genet. Biol. 2012, 49, 602–612. [Google Scholar] [CrossRef]
- Geiser, D.M.; der mar Jimenez-Gasco, M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’Donnell, K. FUSARIUM-ID v.1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004, 110, 473–479. [Google Scholar] [CrossRef]
- Kristensen, R.; Torp, M.; Kosiak, B.; Holst-Jensen, A. Phylogeny and toxigenic potential is correlated in Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences. Mycol. Res. 2005, 109, 173–186. [Google Scholar] [CrossRef]
- Watanabe, M.; Yonezawa, T.; Lee, K.-I.; Kumagai, S.; Sugita-Konishi, Y.; Goto, K.; Hara-Kudo, Y. Molecular phylogeny of the higher and lower taxonomy of the Fusarium genus and differences in the evolutionary histories of multiple genes. BMC Evol. Biol. 2011, 11, 322. [Google Scholar] [CrossRef]
- Proctor, R.H.; Desjardins, A.E.; Plattner, R.D.; Hohn, T.M. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population. A. Fungal Genet. Biol. 1999, 27, 100–112. [Google Scholar] [CrossRef]
- Glenn, A.E.; Zitomer, N.C.; Zimeri, A.M.; Williams, L.D.; Riley, R.T.; Proctor, R.H. Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol. Plant Microbe Interact. 2008, 21, 87–97. [Google Scholar] [CrossRef]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium Species. An Illustrated Manual for Identification; The Pennsylvania State University Press: White Oak, PA, USA, 1983. [Google Scholar]
- Stępień, Ł.; Chełkowski, J.; Wenzel, G.; Mohler, V. Combined use of linked markers for genotyping the Pm1 locus in common wheat. Cell. Mol. Biol. Lett. 2004, 9, 819–827. [Google Scholar]
- Błaszczyk, L.; Tyrka, M.; Chełkowski, J. PstIAFLP based markers for leaf rust resistance genes in common wheat. J. Appl. Genet. 2005, 46, 357–364. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Waśkiewicz, A.; Stępień, Ł.; Wilman, K.; Kachlicki, P. Diversity of Pea-Associated F. proliferatum and F. verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis. Toxins 2013, 5, 488-503. https://doi.org/10.3390/toxins5030488
Waśkiewicz A, Stępień Ł, Wilman K, Kachlicki P. Diversity of Pea-Associated F. proliferatum and F. verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis. Toxins. 2013; 5(3):488-503. https://doi.org/10.3390/toxins5030488
Chicago/Turabian StyleWaśkiewicz, Agnieszka, Łukasz Stępień, Karolina Wilman, and Piotr Kachlicki. 2013. "Diversity of Pea-Associated F. proliferatum and F. verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis" Toxins 5, no. 3: 488-503. https://doi.org/10.3390/toxins5030488
APA StyleWaśkiewicz, A., Stępień, Ł., Wilman, K., & Kachlicki, P. (2013). Diversity of Pea-Associated F. proliferatum and F. verticillioides Populations Revealed by FUM1 Sequence Analysis and Fumonisin Biosynthesis. Toxins, 5(3), 488-503. https://doi.org/10.3390/toxins5030488






