Atomistic-Level Structural Insight into Vespa Venom (Ves a 1) and Lipid Membrane Through the View of Molecular Dynamics Simulation
Abstract
1. Introduction
2. Computational Methods
2.1. Parameterization
2.2. All-Atom Molecular Dynamics (MDs) Simulation
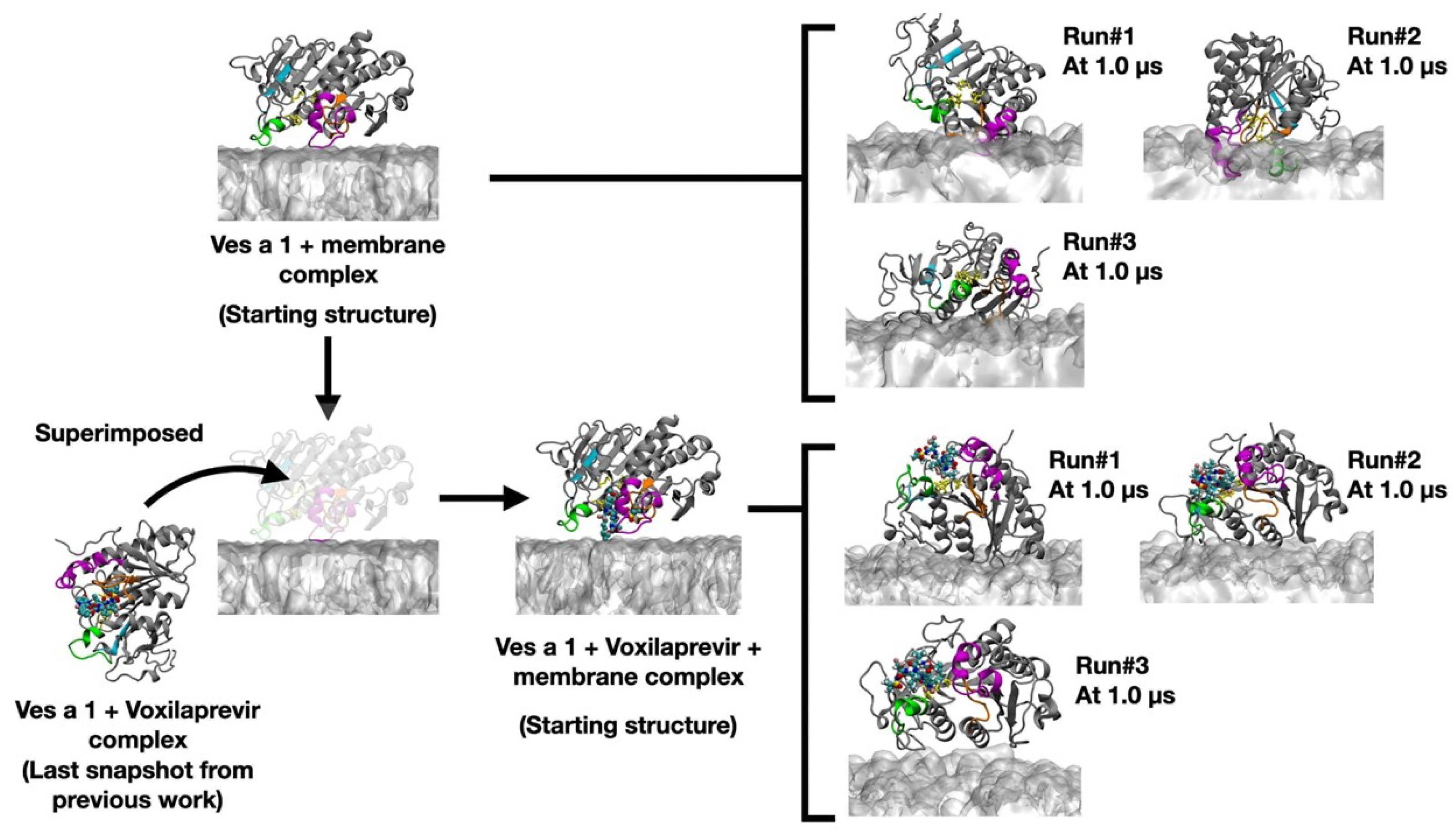
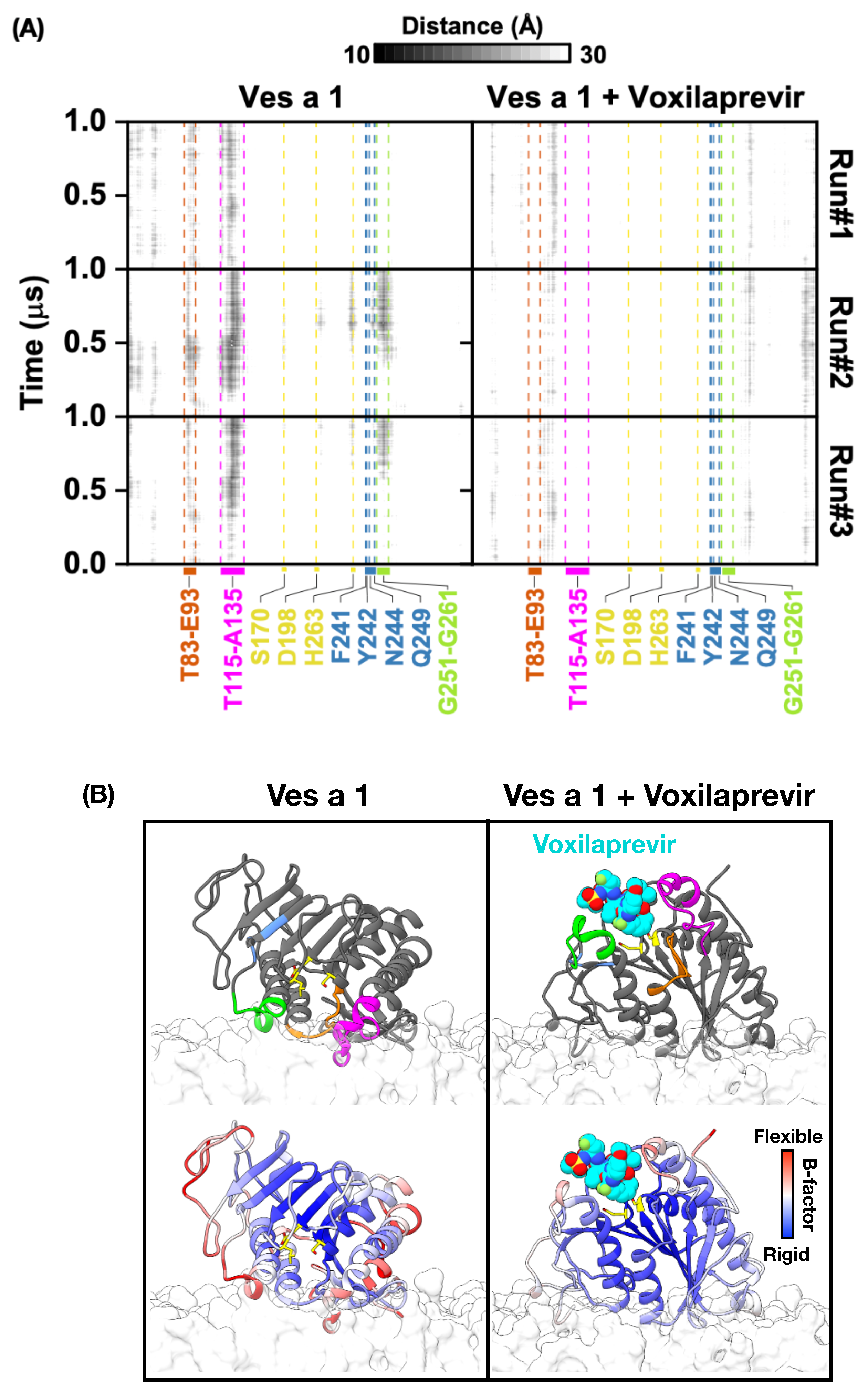
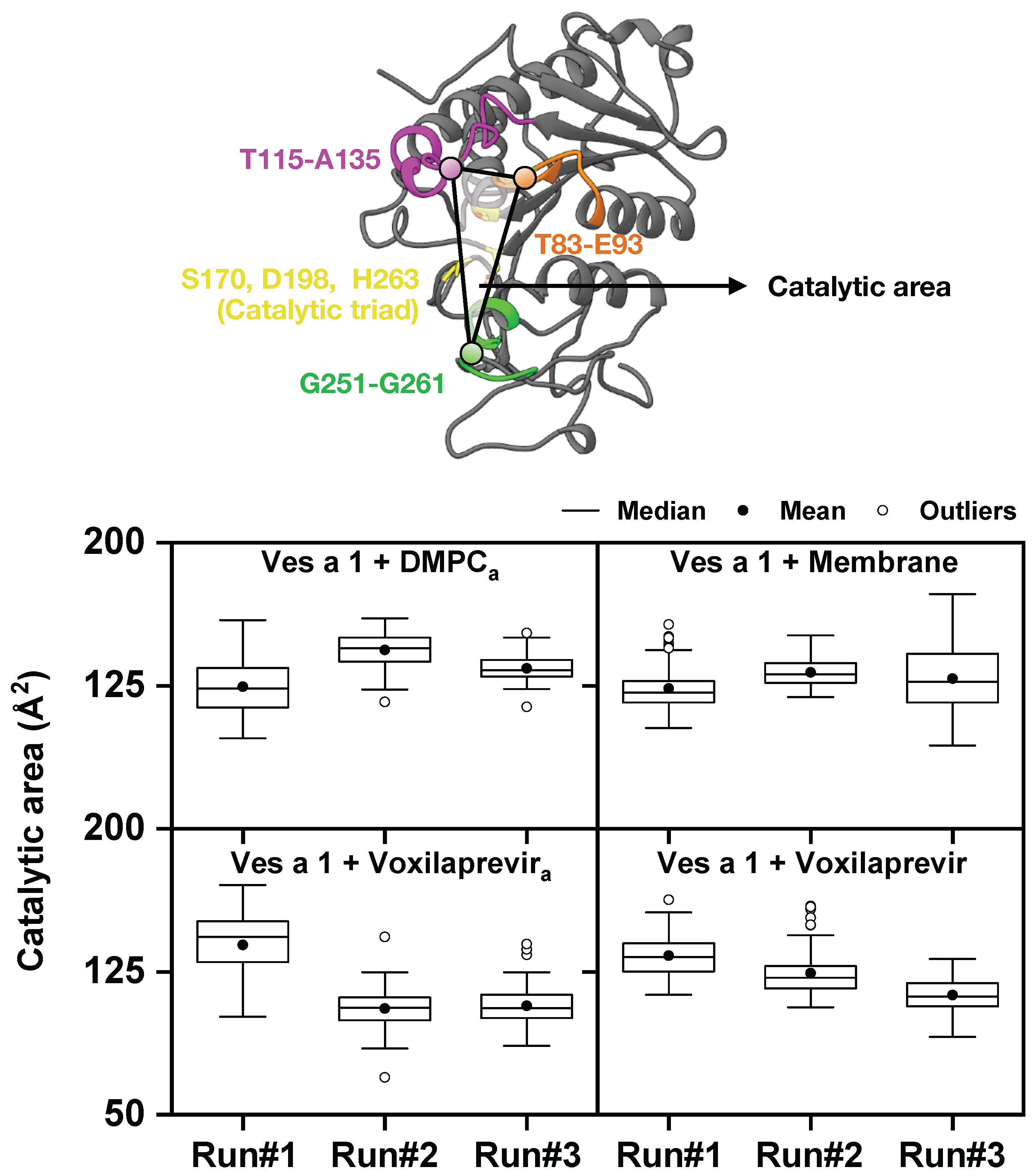
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- King, T.P.; Spangfort, M.D. Structure and biology of stinging insect venom allergens. Int. Arch. Allergy Immunol. 2000, 123, 99–106. [Google Scholar] [CrossRef]
- Sukprasert, S.; Rungsa, P.; Uawonggul, N.; Incamnoi, P.; Thammasirirak, S.; Daduang, J.; Daduang, S. Purification and structural characterisation of phospholipase A1 (Vespapase, Ves a 1) from Thai banded tiger wasp (Vespa affinis) venom. Toxicon 2013, 61, 151–164. [Google Scholar] [CrossRef]
- Hou, M.H.; Chuang, C.Y.; Ko, T.P.; Hu, N.J.; Chou, C.C.; Shih, Y.P.; Ho, C.L.; Wang, A.H.J. Crystal structure of vespid phospholipase A1 reveals insights into the mechanism for cause of membrane dysfunction. Insect Biochem. Mol. Biol. 2016, 68, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.D.; Santos, K.S.; de Souza, B.M.; Arcuri, H.A.; Cunha-Neto, E.; Castro, F.M.; Kalil, J.E.; Palma, M.S. Purification, sequencing and structural characterization of the phospholipase A1 from the venom of the social wasp Polybia paulista (Hymenoptera, Vespidae). Toxicon 2007, 50, 923–937. [Google Scholar] [CrossRef]
- King, T.P.; Jim, S.Y.; Wittkowski, K.M. Inflammatory role of two venom components of yellow jackets (Vespula vulgaris): A mast cell degranulating peptide mastoparan and phospholipase A1. Int. Arch. Allergy Immunol. 2003, 131, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xu, X.; Ma, D.; Zhang, K.; Lai, R. A phospholipase A1 platelet activator from the wasp venom of Vespa magnifica (Smith). Toxicon 2008, 51, 289–296. [Google Scholar] [CrossRef]
- Kularatne, S.; Gawarammana, I.; De Silva, P. Severe multi-organ dysfunction following multiple wasp (Vespa affinis) stings. Ceylon Med. J. 2011, 48, 146–147. [Google Scholar] [CrossRef]
- Kularatne, K.; Kannangare, T.; Jayasena, A.; Jayasekera, A.; Waduge, R.; Weerakoon, K.; Kularatne, S.A. Fatal acute pulmonary oedema and acute renal failure following multiple wasp/hornet (Vespa affinis) stings in Sri Lanka: Two case reports. J. Med. Case Rep. 2014, 8, 188. [Google Scholar] [CrossRef][Green Version]
- Yanagawa, Y.; Morita, K.; Sugiura, T.; Okada, Y. Cutaneous hemorrhage or necrosis findings after Vespa mandarinia (wasp) stings may predict the occurrence of multiple organ injury: A case report and review of literature. Clin. Toxicol. 2007, 45, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Ravikiran, S.; Manya, S.; Baliga, K.; Bhat, K.G. Acute Liver injury, Rhabdomyolysis and Acute Renal Failure in a Toddler due to Multiple Stings by Vespa affinis. J. Clin. Diagn. Res. 2019, 13, SD01–SD03. [Google Scholar] [CrossRef]
- de Oliveira, A.L.N.; Lacerda, M.T.; Ramos, M.J.; Fernandes, P.A. Viper venom phospholipase A2 database: The structural and functional anatomy of a primary toxin in envenomation. Toxins 2024, 16, 71. [Google Scholar] [CrossRef]
- Chou, C.C.; Hou, M.H. Crystallization and preliminary X-ray diffraction analysis of phospholipase A1 isolated from hornet (Vespa basalis) venom. Acta Crystallogr. Sect. Struct. Biol. Cryst. Commun. 2008, 64, 1118–1120. [Google Scholar] [CrossRef] [PubMed]
- Pattaranggoon, N.C.; Daduang, S.; Rungrotmongkol, T.; Teajaroen, W.; Tipmanee, V.; Hannongbua, S. Computational model for lipid binding regions in phospholipase (Ves a 1) from Vespa venom. Sci. Rep. 2023, 13, 10652. [Google Scholar] [CrossRef] [PubMed]
- Rungsa, P.; Peigneur, S.; Daduang, S.; Tytgat, J. Purification and biochemical characterization of VesT1s, a novel phospholipase A1 isoform isolated from the venom of the greater banded wasp Vespa tropica. Toxicon 2018, 148, 74–84. [Google Scholar] [CrossRef]
- Teajaroen, W.; Phimwapi, S.; Daduang, J.; Klaynongsruang, S.; Tipmanee, V.; Daduang, S. A role of newly found auxiliary site in phospholipase A1 from Thai banded tiger wasp (Vespa affinis) in its enzymatic enhancement: In silico homology modeling and molecular dynamics insights. Toxins 2020, 12, 510. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.W.; Haider, S.Z.; Nawaz, M.Z.; Irfan, M.; Al-Ghanim, K.A.; Sun, X.; Yuan, Q. Molecular simulations guided drugs repurposing to inhibit human GPx1 enzyme for cancer therapy. Bioorganic Chem. 2025, 157, 108279. [Google Scholar] [CrossRef]
- Jiménez, E.M.; Żołek, T.; Hernández Perez, P.G.; Miranda Ruvalcaba, R.; Nicolás-Vázquez, M.I.; Hernández-Rodríguez, M. Drug repurposing to inhibit histamine N-methyl transferase. Molecules 2023, 28, 576. [Google Scholar] [CrossRef]
- Del Prete, S.; Pagano, M. Enzyme inhibitors as multifaceted tools in medicine and agriculture. Molecules 2024, 29, 4314. [Google Scholar] [CrossRef]
- Mohanty, S.S.; Sahoo, C.R.; Padhy, R.N. Targeting some enzymes with repurposing approved pharmaceutical drugs for expeditious antiviral approaches against newer strains of COVID-19. AAPS PharmSciTech 2021, 22, 214. [Google Scholar] [CrossRef]
- Marrink, S.J.; Corradi, V.; Souza, P.C.; Ingolfsson, H.I.; Tieleman, D.P.; Sansom, M.S. Computational modeling of realistic cell membranes. Chem. Rev. 2019, 119, 6184–6226. [Google Scholar] [CrossRef]
- Róg, T.; Girych, M.; Bunker, A. Mechanistic understanding from molecular dynamics in pharmaceutical research 2: Lipid membrane in drug design. Pharmaceuticals 2021, 14, 1062. [Google Scholar] [CrossRef]
- Watkins, S.L. Current trends and changes in use of membrane molecular dynamics simulations within academia and the pharmaceutical industry. Membranes 2023, 13, 148. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Feng, S.; Park, S.; Choi, Y.K.; Im, W. CHARMM-GUI membrane builder: Past, current, and future developments and applications. J. Chem. Theory Comput. 2023, 19, 2161–2185. [Google Scholar] [CrossRef] [PubMed]
- Willems, N.; Lelimousin, M.; Koldsø, H.; Sansom, M.S. Interfacial activation of M37 lipase: A multi-scale simulation study. Biochim. Biophys. Acta (BBA)-Biomembr. 2017, 1859, 340–349. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Peeyatu, C.; Prompat, N.; Voravuthikunchai, S.P.; Roongsawang, N.; Sangkhathat, S.; Khongkow, P.; Saetang, J.; Tipmanee, V. Role of Non-Binding T63 Alteration in IL-18 Binding. Int. J. Mol. Sci. 2024, 25, 12992. [Google Scholar] [CrossRef] [PubMed]
- Naïm, M.; Bhat, S.; Rankin, K.N.; Dennis, S.; Chowdhury, S.F.; Siddiqi, I.; Drabik, P.; Sulea, T.; Bayly, C.I.; Jakalian, A.; et al. Solvated interaction energy (SIE) for scoring protein- ligand binding affinities. 1. Exploring the parameter space. J. Chem. Inf. Model. 2007, 47, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Sulea, T.; Purisima, E.O. The solvated interaction energy method for scoring binding affinities. In Computational Drug Discovery and Design; Springer: Berlin/Heidelberg, Germany, 2011; pp. 295–303. [Google Scholar]
- Somboon, T.; Mahalapbutr, P.; Sanachai, K.; Maitarad, P.; Lee, V.S.; Hannongbua, S.; Rungrotmongkol, T. Computational study on peptidomimetic inhibitors against SARS-CoV-2 main protease. J. Mol. Liq. 2021, 322, 114999. [Google Scholar] [CrossRef] [PubMed]


| Ligands | |||||
|---|---|---|---|---|---|
| Ves a 1 + Voxilaprevir | |||||
| Run#1 | −37.61 ± 4.89 | −1.16 ± 1.90 | 5.97 ± 2.09 | −7.00 ± 0.86 | −7.066 ± 0.51 |
| Run#2 | −44.48 ± 3.92 | −1.13 ± 2.41 | 6.49 ± 2.11 | −8.65 ± 0.65 | −7.89 ± 0.45 |
| Run#3 | −43.96 ± 4.84 | −1.40 ± 2.67 | 9.43 ± 3.00 | −8.89 ± 1.08 | −7.59 ± 0.57 |
| Ves a 1 + | |||||
| Run#1 | −56.20 ± 3.76 | −14.40 ± 2.78 | 19.37 ± 2.36 | −11.77 ± 0.50 | −9.49 ± 0.43 |
| Run#2 | −41.97 ± 6.09 | −3.80 ± 3.67 | 9.24 ± 2.86 | −9.80 ± 1.44 | −7.74 ± 0.64 |
| Run#3 | −53.36 ± 3.98 | −19.14 ± 4.78 | 22.13 ± 3.91 | −11.21 ± 0.70 | −9.34 ± 0.51 |
| Ves a 1 + | |||||
| Run#1 | −58.80 ± 3.60 | −84.00 ± 6.63 | 84.88 ± 4.98 | −11.45 ± 0.32 | −10.16 ± 0.43 |
| Run#2 | −57.96 ± 4.59 | −72.34 ± 11.32 | 75.11 ± 8.70 | −10.91 ± 0.63 | −9.81 ± 0.74 |
| Run#3 | −60.10 ± 3.03 | −83.61 ± 9.49 | 84.43 ± 8.21 | −11.52 ± 0.40 | −10.31 ± 0.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pattaranggoon, N.C.; Teajaroen, W.; Daduang, S.; Hannongbua, S.; Rungrotmongkol, T.; Tipmanee, V. Atomistic-Level Structural Insight into Vespa Venom (Ves a 1) and Lipid Membrane Through the View of Molecular Dynamics Simulation. Toxins 2025, 17, 387. https://doi.org/10.3390/toxins17080387
Pattaranggoon NC, Teajaroen W, Daduang S, Hannongbua S, Rungrotmongkol T, Tipmanee V. Atomistic-Level Structural Insight into Vespa Venom (Ves a 1) and Lipid Membrane Through the View of Molecular Dynamics Simulation. Toxins. 2025; 17(8):387. https://doi.org/10.3390/toxins17080387
Chicago/Turabian StylePattaranggoon, Nawanwat Chainuwong, Withan Teajaroen, Sakda Daduang, Supot Hannongbua, Thanyada Rungrotmongkol, and Varomyalin Tipmanee. 2025. "Atomistic-Level Structural Insight into Vespa Venom (Ves a 1) and Lipid Membrane Through the View of Molecular Dynamics Simulation" Toxins 17, no. 8: 387. https://doi.org/10.3390/toxins17080387
APA StylePattaranggoon, N. C., Teajaroen, W., Daduang, S., Hannongbua, S., Rungrotmongkol, T., & Tipmanee, V. (2025). Atomistic-Level Structural Insight into Vespa Venom (Ves a 1) and Lipid Membrane Through the View of Molecular Dynamics Simulation. Toxins, 17(8), 387. https://doi.org/10.3390/toxins17080387





