The Potential Mechanisms of Ochratoxin A in Prostate Cancer Development: An Integrated Study Combining Network Toxicology, Machine Learning, and Molecular Docking
Abstract
1. Introduction
2. Results
2.1. Toxicity Model Computation of Ochratoxin A
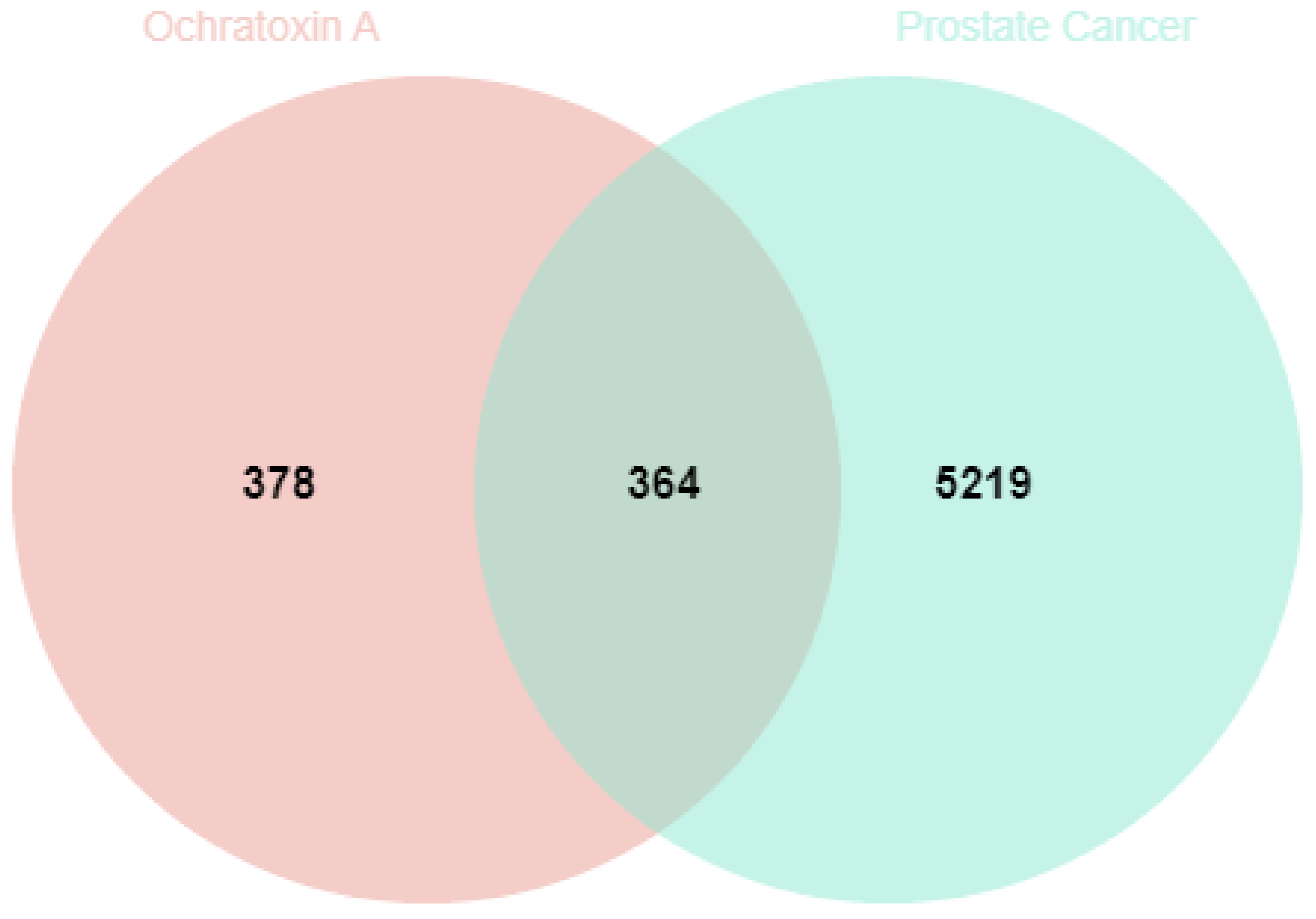
2.2. Targets of Ochratoxin A in Prostate Cancer
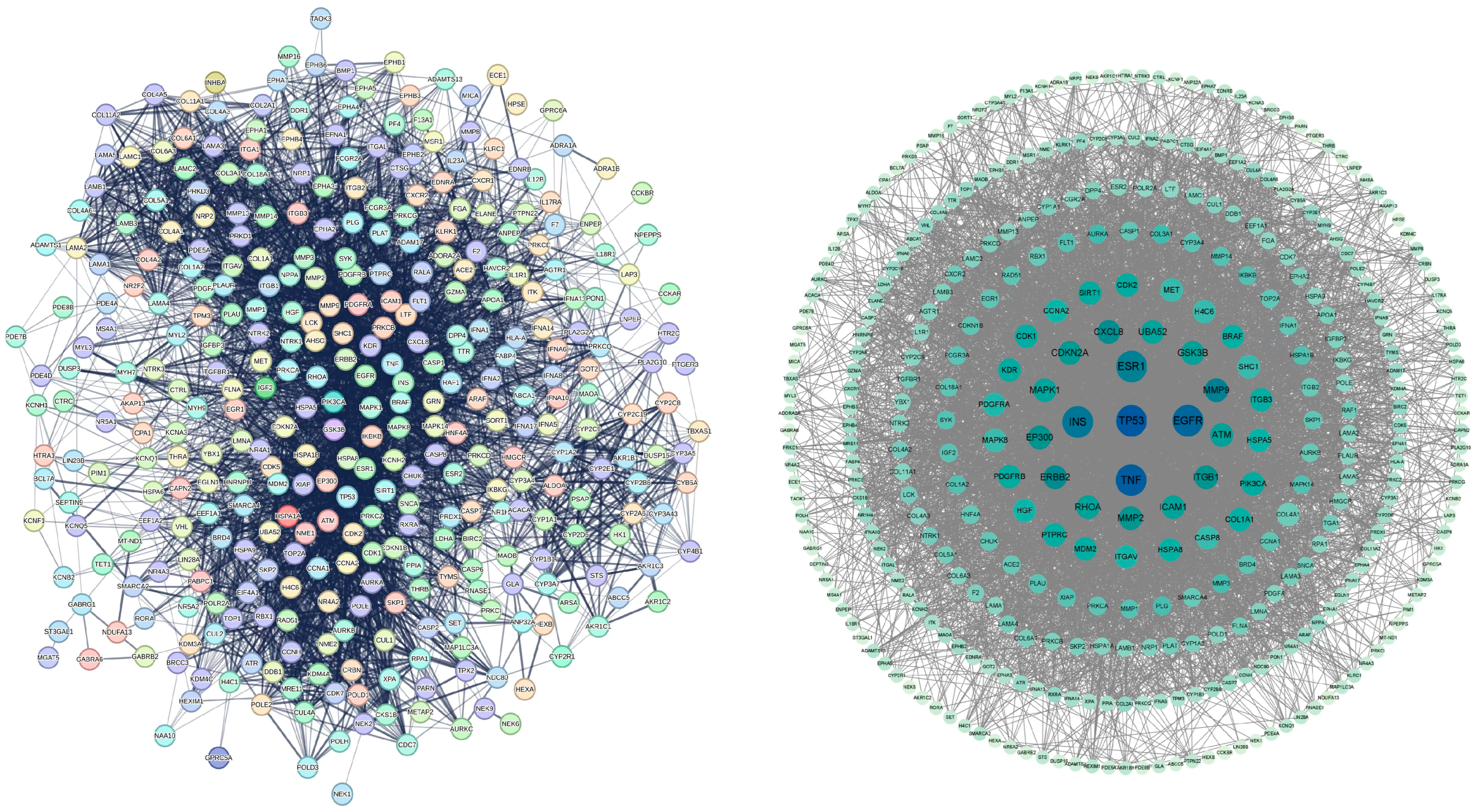
2.3. Protein–Protein Interaction Network of Common Targets
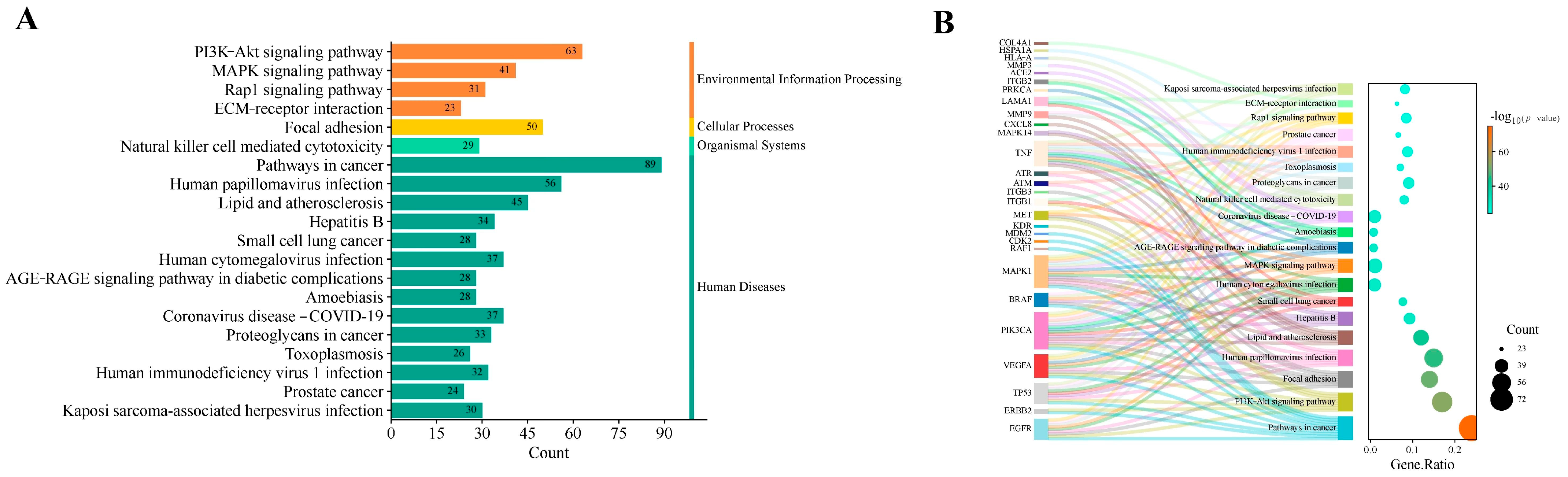
2.4. GO Functional and KEGG Pathway Enrichment Analyses
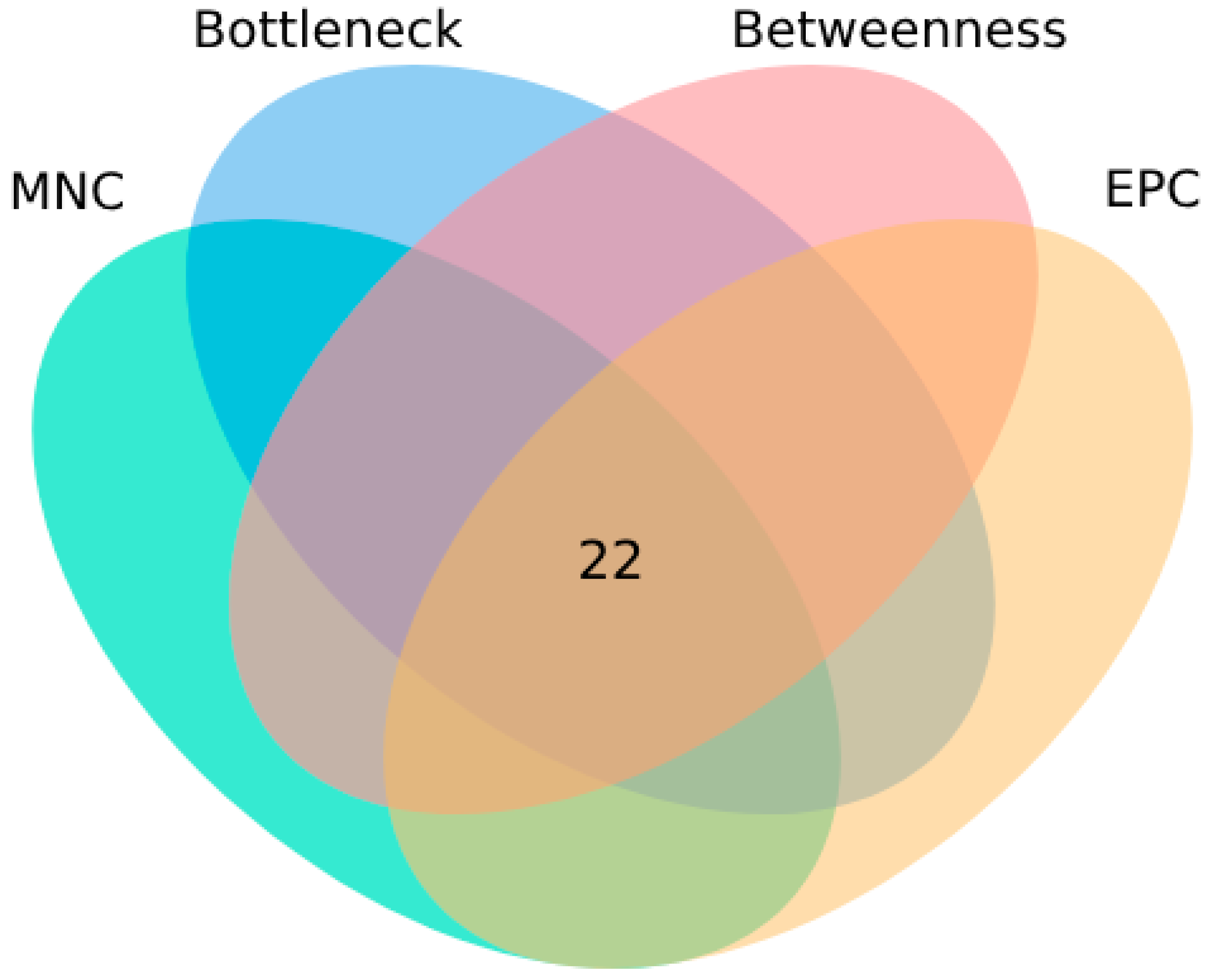
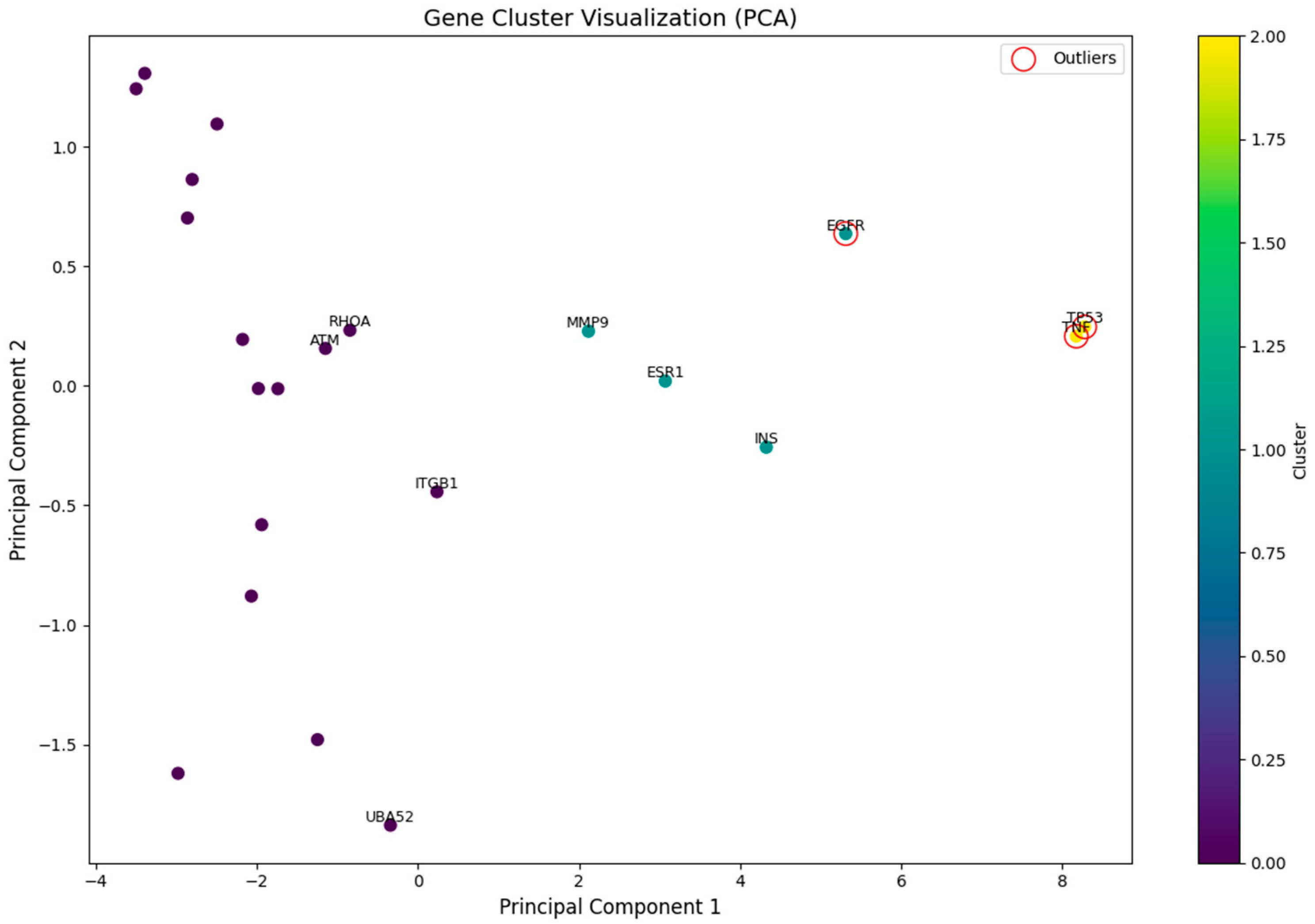
2.5. Identification of Potential Core Targets
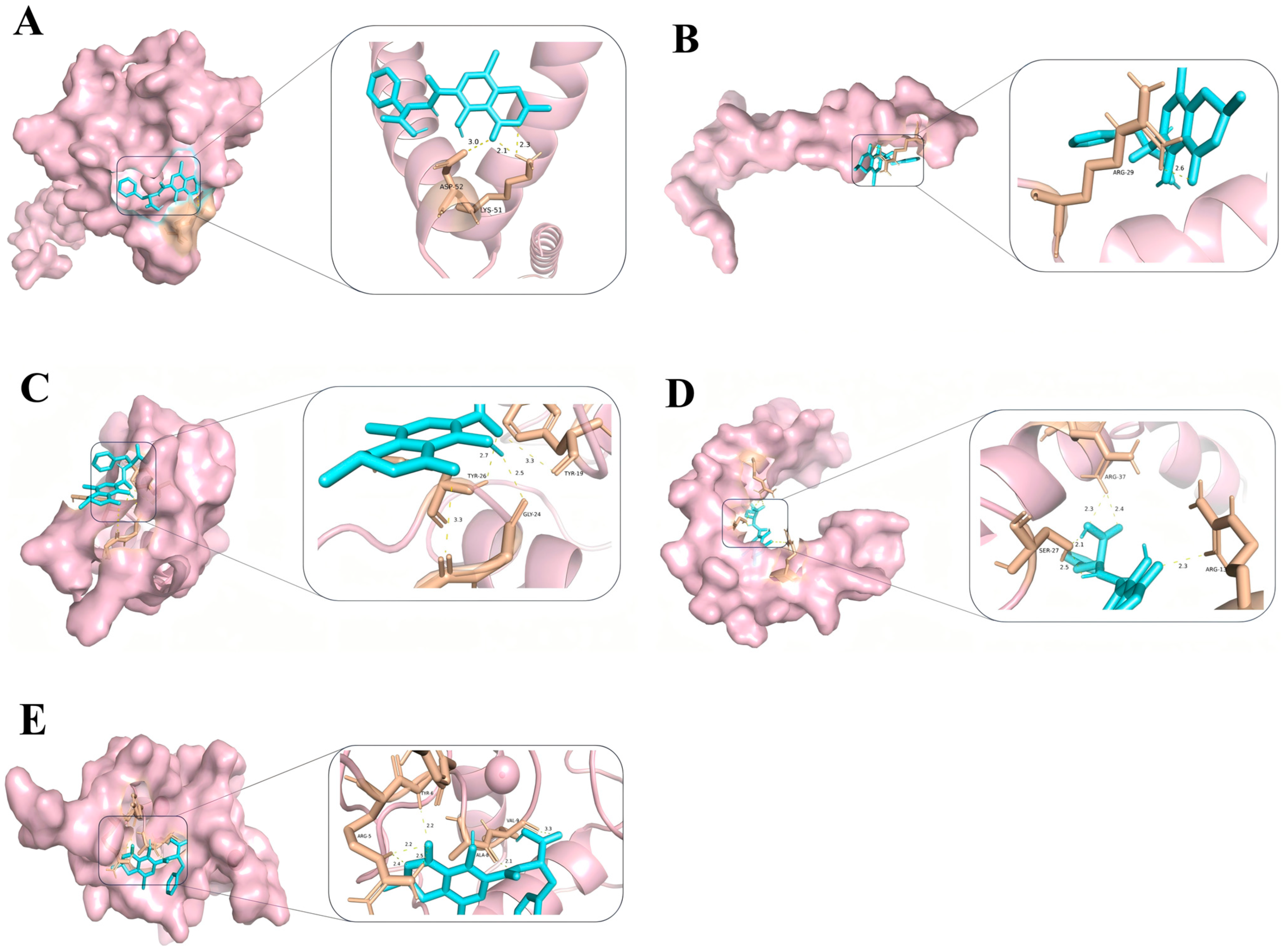
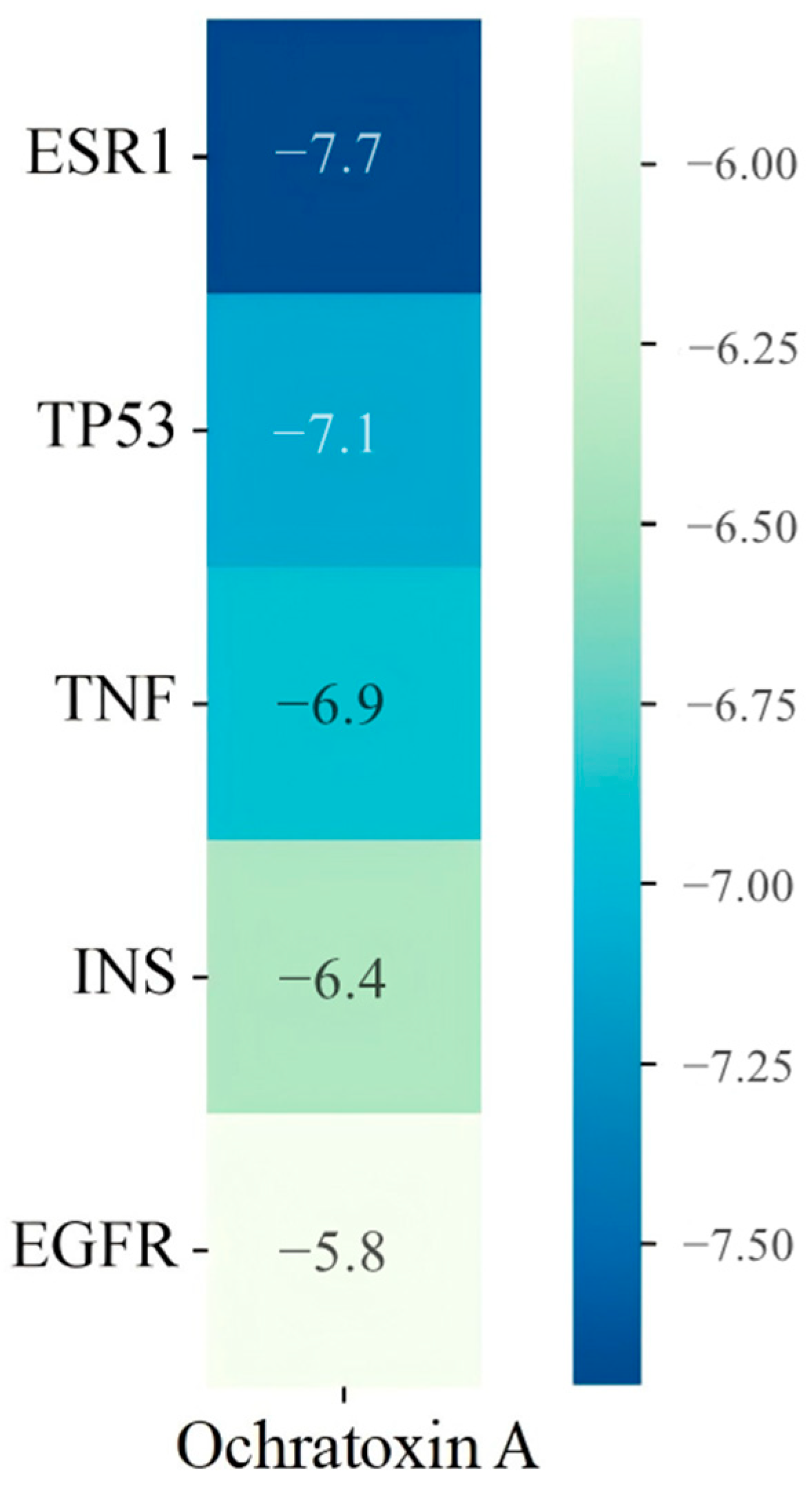
2.6. Molecular Docking Results of Core Targets of OTA in PCa
3. Discussion
3.1. Key Targets and Molecular Interactions
3.2. KEGG Pathway Analysis: Significance for Prostate Cancer
3.3. GO Functional Analysis: Cellular and Molecular Effects
3.4. Implications for Prostate Cancer Subtypes
4. Conclusions
5. Methods
5.1. Prediction of Ochratoxin A Toxicity
5.2. Target Screening of Ochratoxin A
5.3. Target Screening of Prostate Cancer
5.4. Common Targets
5.5. Protein–Protein Interaction Analysis
5.6. Gene Ontology (GO) Functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses
5.7. Machine Learning Workflow and Identification of Potential Core Targets
5.7.1. Feature Engineering
5.7.2. Model Selection and Hyper-Parameters
- (1)
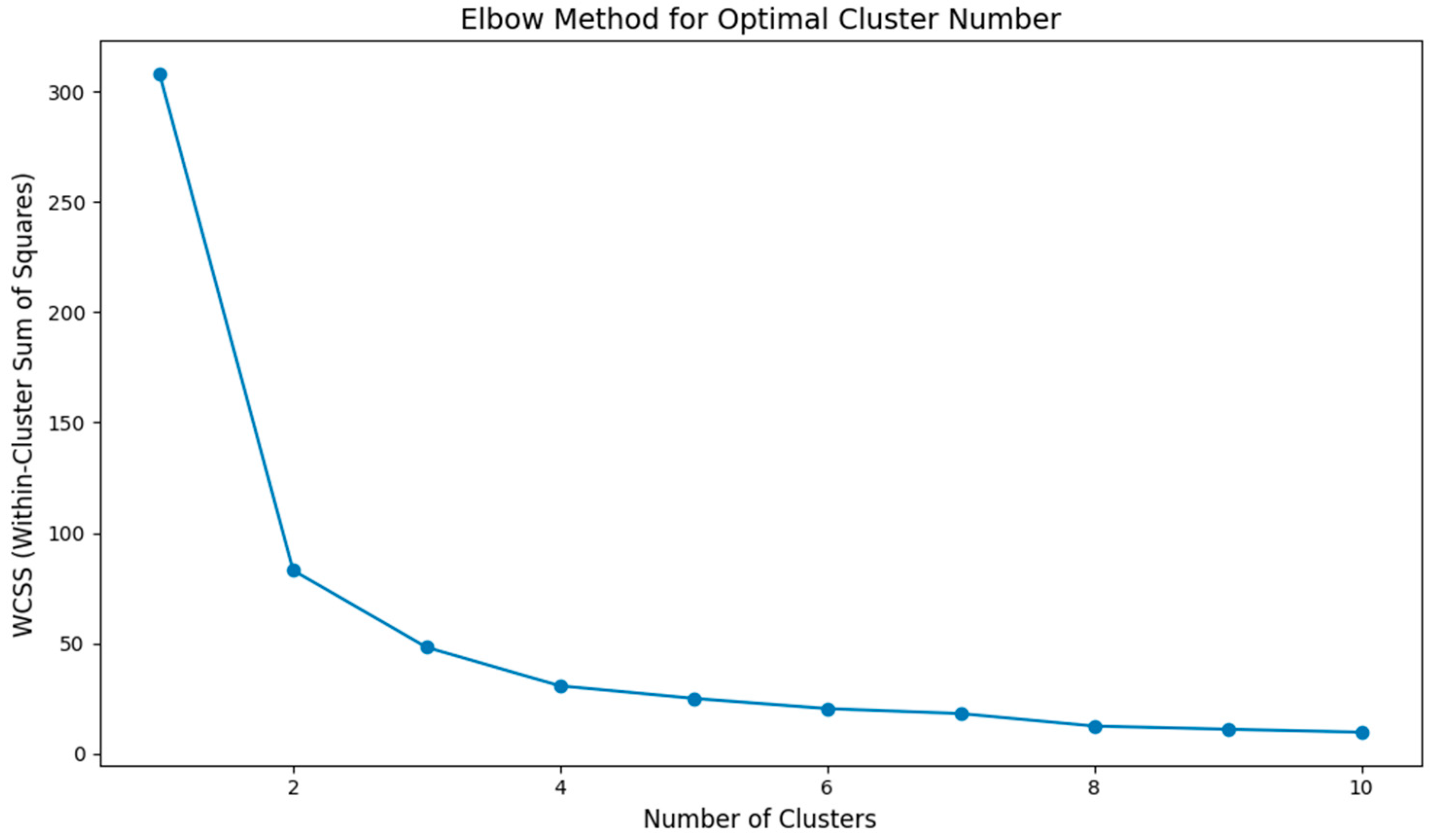
- K-means clustering: Euclidean distance, scikit-learn KMeans, max_iter = 300, and tol = 1 × 10−4. The optimal cluster number was determined using the elbow method: WCSS was computed for k = 1–10; the second-difference maximum indicated an elbow at k = 3 (Figure 7).
- (2)
- (3)
- PCA visualization: the first two principal components (n_components = 2) were retained.
5.7.3. Composite Score Construction
- (1)
- Centrality indices in which higher values indicate importance (11 indices, e.g., MNC, EPC, and Betweenness) were ranked in descending order;
- (2)
- Path-based indices in which lower values indicate importance (3 indices, e.g., AverageShortestPathLength and TopologicalCoefficient) were ranked in ascending order.
5.7.4. Integration and Core Gene Definition
- (1)
- IsolationForest anomaly flag = −1 (Is_outlier = 1);
- (2)
- Composite_rank ≤ 20.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited ‘FAO estimate’ of 25. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef]
- Marin, S.; Ramos, A.J.; Cano-Sancho, G.; Sanchis, V. Mycotoxins: Occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2013, 60, 218–237. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Scientific report on the 2018 European Union report on pesticide residues in food. EFSA J. 2020, 18, 6057. [Google Scholar] [CrossRef]
- Dahal, S.; Lee, H.J.; Gu, K.; Ryu, D. Heat Stability of Ochratoxin A in an Aqueous Buffered Model System. J. Food Prot. 2016, 79, 1748–1752. [Google Scholar] [CrossRef] [PubMed]
- Pavlović, M.; Plestina, R.; Krogh, P. Ochratoxin A contamination of foodstuffs in an area with Balkan (endemic) nephropathy. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. 1979, 87, 243–246. [Google Scholar] [CrossRef]
- Petkova-Bocharova, T.; Castegnaro, M. Ochratoxin A contamination of cereals in an area of high incidence of Balkan endemic nephropathy in Bulgaria. Food Addit. Contam. 1985, 2, 267–270. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. In Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins; World Health Organization: Geneva, Switzerland; International Agency for Research on Cancer: Lyon, France, 1993; Volume 56. [Google Scholar]
- Więckowska, M.; Cichon, N.; Szelenberger, R.; Gorniak, L.; Bijak, M. Ochratoxin A and Its Role in Cancer Development: A Comprehensive Review. Cancers 2024, 16, 3473. [Google Scholar] [CrossRef]
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate Cancer Review: Genetics, Diagnosis, Treatment Options, and Alternative Approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef]
- Oczkowski, M.; Dziendzikowska, K.; Pasternak-Winiarska, A.; Włodarek, D.; Gromadzka-Ostrowska, J. Dietary Factors and Prostate Cancer Development, Progression, and Reduction. Nutrients 2021, 13, 496. [Google Scholar] [CrossRef]
- Li, Q.; Dong, Z.; Lian, W.; Cui, J.; Wang, J.; Shen, H.; Liu, W.; Yang, J.; Zhang, X.; Cui, H. Ochratoxin A causes mitochondrial dysfunction, apoptotic and autophagic cell death and also induces mitochondrial biogenesis in human gastric epithelium cells. Arch. Toxicol. 2019, 93, 1141–1155. [Google Scholar] [CrossRef] [PubMed]
- Horvath, A.; Upham, B.L.; Ganev, V.; Trosko, J.E. Determination of the epigenetic effects of ochratoxin in a human kidney and a rat liver epithelial cell line. Toxicon 2002, 40, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Izco, M.; Vettorazzi, A.; de Toro, M.; Sáenz, Y.; Alvarez-Erviti, L. Oral Sub-chronic Ochratoxin A Exposure Induces Gut Microbiota Alterations in Mice. Toxins 2021, 13, 106. [Google Scholar] [CrossRef] [PubMed]
- Stoev, S.D. New Evidences about the Carcinogenic Effects of Ochratoxin A and Possible Prevention by Target Feed Additives. Toxins 2022, 14, 380. [Google Scholar] [CrossRef]
- Valls-Margarit, J.; Piñero, J.; Füzi, B.; Cerisier, N.; Taboureau, O.; Furlong, L.I. Assessing network-based methods in the context of system toxicology. Front. Pharmacol. 2023, 14, 1225697. [Google Scholar] [CrossRef]
- Al-Tashi, Q.; Saad, M.B.; Muneer, A.; Qureshi, R.; Mirjalili, S.; Sheshadri, A.; Le, X.; Vokes, N.I.; Zhang, J.; Wu, J. Machine Learning Models for the Identification of Prognostic and Predictive Cancer Biomarkers: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 7781. [Google Scholar] [CrossRef]
- Santos, L.H.S.; Ferreira, R.S.; Caffarena, E.R. Integrating Molecular Docking and Molecular Dynamics Simulations. Methods Mol. Biol. 2019, 2053, 13–34. [Google Scholar] [CrossRef]
- Neill, T.; Buraschi, S.; Kapoor, A.; Iozzo, R.V. Proteoglycan-driven Autophagy: A Nutrient-independent Mechanism to Control Intracellular Catabolism. J. Histochem. Cytochem. 2020, 68, 733–746. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Smith, R.D.; Engdahl, A.L.; Dunbar, J.B., Jr.; Carlson, H.A. Biophysical limits of protein-ligand binding. J. Chem. Inf. Model. 2012, 52, 2098–2106. [Google Scholar] [CrossRef]
- Kondo, K.; Honda, K.; Goshima, K.; Inoue, N.; Shinjo, D.; Tsutsumi, T.; Fushimi, K. Otologic disease trends in Japan post-COVID-19 outbreak: A retrospective time-series analysis. Auris Nasus Larynx 2024, 51, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, J.; Edmiston, J. Underreporting of non-study cigarette use by study participants confounds the interpretation of results from ambulatory clinical trial of reduced nicotine cigarettes. Harm Reduct. J. 2024, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Zhang, M. Author Correction: Spatio-temporal evolution and driving forces of habitat quality in Guizhou Province. Sci. Rep. 2023, 13, 8278. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S. Natural Products and Disease Prevention, Relief and Treatment. Nutrients 2022, 14, 2396. [Google Scholar] [CrossRef]
- Michaud, J.E.; Billups, K.L.; Partin, A.W. Testosterone and prostate cancer: An evidence-based review of pathogenesis and oncologic risk. Ther. Adv. Urol. 2015, 7, 378–387. [Google Scholar] [CrossRef]
- Zheng, Q.W.; Ding, X.F.; Cao, H.J.; Ni, Q.Z.; Zhu, B.; Ma, N.; Zhang, F.K.; Wang, Y.K.; Xu, S.; Chen, T.W.; et al. Ochratoxin A Induces Steatosis via PPARγ-CD36 Axis. Toxins 2021, 13, 802. [Google Scholar] [CrossRef]
- Paradells, S.; Rocamonde, B.; Llinares, C.; Herranz-Pérez, V.; Jimenez, M.; Garcia-Verdugo, J.M.; Zipancic, I.; Soria, J.M.; Garcia-Esparza, M.A. Neurotoxic effects of ochratoxin A on the subventricular zone of adult mouse brain. J. Appl. Toxicol. 2015, 35, 737–751. [Google Scholar] [CrossRef]
- Wang, Y.M.; Liu, Z.W.; Guo, J.B.; Wang, X.F.; Zhao, X.X.; Zheng, X. ESR1 Gene Polymorphisms and Prostate Cancer Risk: A HuGE Review and Meta-Analysis. PLoS ONE 2013, 8, e66999. [Google Scholar] [CrossRef]
- Olczak, M.; Orzechowska, M.J.; Bednarek, A.K.; Lipiński, M. The Transcriptomic Profiles of ESR1 and MMP3 Stratify the Risk of Biochemical Recurrence in Primary Prostate Cancer beyond Clinical Features. Int. J. Mol. Sci. 2013, 24, 8399. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, B. Extracellular matrix stiffness: Mechanisms in tumor progression and therapeutic potential in cancer. Exp. Hematol. Oncol. 2025, 14, 54. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, X.; Wang, W.; Pu, N.; Liu, L. Epithelial-mesenchymal transition orchestrates tumor microenvironment: Current perceptions and challenges. J. Transl. Med. 2025, 23, 386. [Google Scholar] [CrossRef]
- Hernández Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta. Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Ofner, H.; Kramer, G.; Shariat, S.F.; Hassler, M.R. TP53 Deficiency in the Natural History of Prostate Cancer. Cancers 2025, 17, 645. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Yang, L.V.; Abrams, S.L.; Steelman, L.S.; Follo, M.Y.; Cocco, L.; Ratti, S.; Martelli, A.M.; Augello, G.; Cervello, M. Effects of TP53 Mutations and miRs on Immune Responses in the Tumor Microenvironment Important in Pancreatic Cancer Progression. Cells 2022, 11, 2155. [Google Scholar] [CrossRef] [PubMed]
- Efe, G.; Rustgi, A.K.; Prives, C. p53 at the crossroads of tumor immunity. Nat. Cancer 2024, 5, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Reinberg, D. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014, 28, 723–734. [Google Scholar] [CrossRef]
- Macedo-Silva, C.; Miranda-Gonçalves, V.; Tavares, N.T.; Barros-Silva, D.; Lencart, J.; Lobo, J.; Oliveira, Â.; Correia, M.P.; Altucci, L.; Jerónimo, C. Epigenetic regulation of TP53 is involved in prostate cancer radioresistance and DNA damage response signaling. Signal Transduct. Target. Ther. 2023, 8, 395. [Google Scholar] [CrossRef]
- Ofer, P.; Heidegger, I.; Eder, I.E.; Schöpf, B.; Neuwirt, H.; Geley, S.; Klocker, H.; Massoner, P. Both IGF1R and INSR Knockdown Exert Antitumorigenic Effects in Prostate Cancer In Vitro and In Vivo. Mol. Endocrinol. 2015, 29, 1694–1707. [Google Scholar] [CrossRef]
- Heikkinen, T.; Küblbeck, J.; Rysä, J. Metabolic disruption by mycotoxins: Focus on metabolic endpoints steatosis, adipogenesis and glucose metabolism in vivo and in vitro. Arch. Toxicol. 2025, 99, 1749–1767. [Google Scholar] [CrossRef]
- Song, Y.; Hou, Z.; Zhu, L.; Chen, Y.; Li, J. Oxidative stress as a catalyst in prostate cancer progression: Unraveling molecular mechanisms and exploring therapeutic interventions. Discov. Oncol. 2025, 16, 457. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Zhang, P.; Chen, L.; Zhou, F.; He, Z.; Wang, G.; Luo, Y. NRP1 promotes prostate cancer progression via modulating EGFR-dependent AKT pathway activation. Cell Death Dis. 2023, 14, 159. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Hiroki, K.; Yamashita, Y. The role of epidermal growth factor receptor in cancer metastasis and microenvironment. BioMed Res. Int. 2013, 2013, 546318. [Google Scholar] [CrossRef] [PubMed]
- Uribe, M.L.; Marrocco, I.; Yarden, Y. EGFR in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance. Cancers 2021, 13, 2748. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Carotenuto, A.; De Feo, G.; Caponigro, F.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef]
- Day, K.C.; Lorenzatti Hiles, G.; Kozminsky, M.; Dawsey, S.J.; Paul, A.; Broses, L.J.; Shah, R.; Kunja, L.P.; Hall, C.; Palanisamy, N.; et al. HER2 and EGFR Overexpression Support Metastatic Progression of Prostate Cancer to Bone. Cancer Res. 2017, 77, 74–85. [Google Scholar] [CrossRef]
- Hashemi, M.; Taheriazam, A.; Daneii, P.; Hassanpour, A.; Kakavand, A.; Rezaei, S.; Hejazi, E.S.; Aboutalebi, M.; Gholamrezaie, H.; Saebfar, H.; et al. Targeting PI3K/Akt signaling in prostate cancer therapy. J. Cell Commun. Signal. 2023, 17, 423–443. [Google Scholar] [CrossRef]
- Bahar, M.E.; Kim, H.J.; Kim, D.R. Targeting the RAS/RAF/MAPK pathway for cancer therapy: From mechanism to clinical studies. Signal Transduct. Target. Ther. 2023, 8, 455. [Google Scholar] [CrossRef]
- Sharma, B.; Shekhar, H.; Sahu, A.; Kaur, D.; Haque, S.; Tuli, H.S.; Sharma, H.; Sharma, U. Epigenetic insights into prostate cancer: Exploring histone modifications and their therapeutic implications. Front. Oncol. 2025, 15, 1570193. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- He, X.; Lee, B.; Jiang, Y. Extracellular matrix in cancer progression and therapy. Med Rev. 2022, 2, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Elango, J.; Hou, C.; Bao, B.; Wang, S.; Maté Sánchez de Val, J.E.; Wenhui, W. The Molecular Interaction of Collagen with Cell Receptors for Biological Function. Polymers 2022, 14, 876. [Google Scholar] [CrossRef] [PubMed]
- Carver, B.S.; Chapinski, C.; Wongvipat, J.; Hieronymus, H.; Chen, Y.; Chandarlapaty, S.; Arora, V.K.; Le, C.; Koutcher, J.; Scher, H.; et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011, 19, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
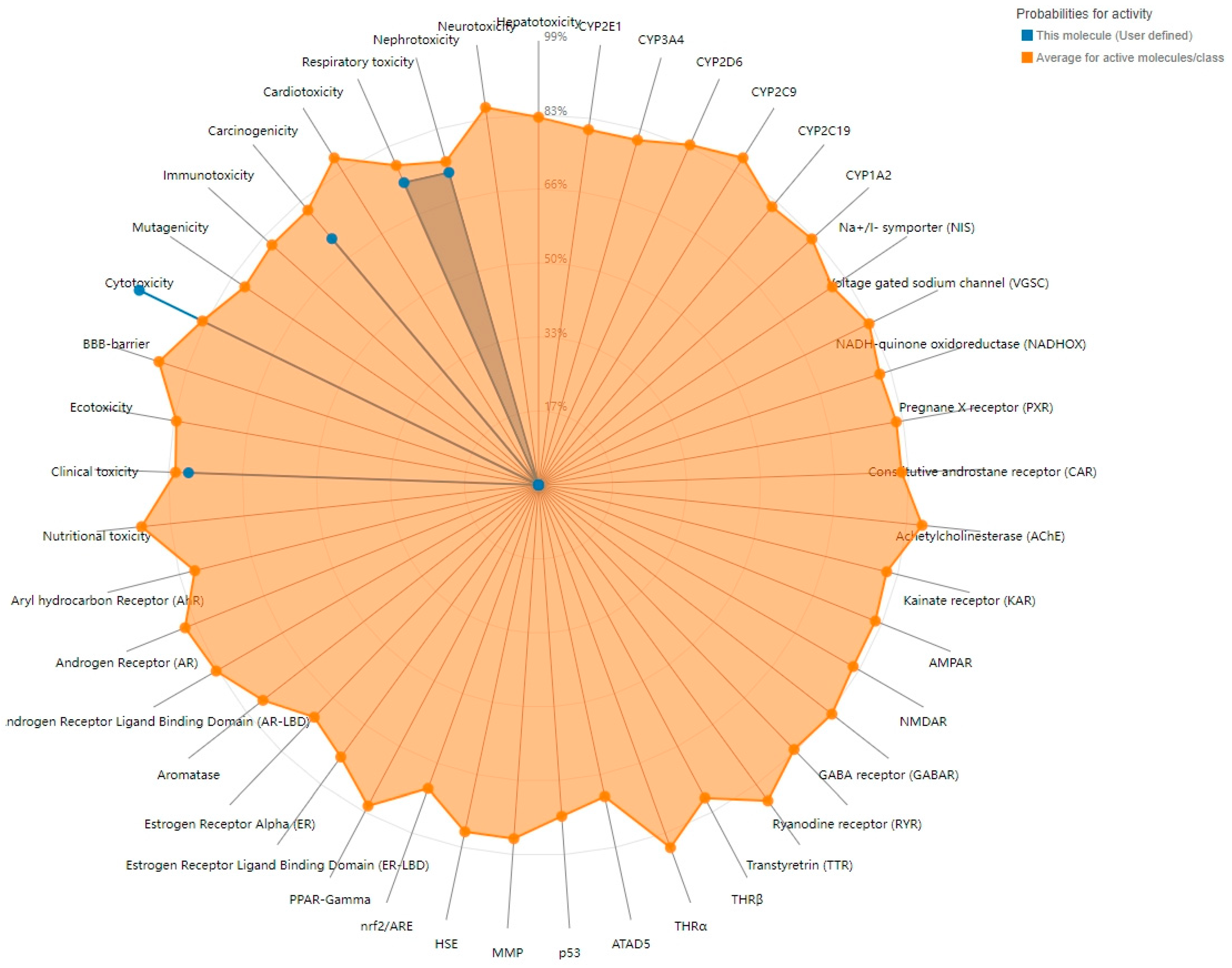

| Target | Prediction | Probability |
|---|---|---|
| Cytotoxicity | Active | 0.99 |
| Clinical toxicity | Active | 0.78 |
| Respiratory toxicity | Active | 0.73 |
| Nephrotoxicity | Active | 0.72 |
| Carcinogenicity | Active | 0.71 |
| Immunotoxicity | Inactive | 0.97 |
| Mutagenicity | Inactive | 0.92 |
| BBB-barrier | Inactive | 0.68 |
| Hepatotoxicity | Inactive | 0.65 |
| Cardiotoxicity | Inactive | 0.65 |
| Nutritional toxicity | Inactive | 0.64 |
| Ecotoxicity | Inactive | 0.60 |
| Neurotoxicity | Inactive | 0.54 |
| Gene | Composite Score |
|---|---|
| TP53 | 4.14 |
| TNF | 4.71 |
| INS | 6.14 |
| EGFR | 6.18 |
| ESR1 | 6.86 |
| MMP9 | 7.53 |
| ITGB1 | 8.46 |
| RHOA | 9.25 |
| ATM | 9.89 |
| UBA52 | 10.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, H.; Shen, D.; Hu, X.; Yin, H.; Yan, Z. The Potential Mechanisms of Ochratoxin A in Prostate Cancer Development: An Integrated Study Combining Network Toxicology, Machine Learning, and Molecular Docking. Toxins 2025, 17, 388. https://doi.org/10.3390/toxins17080388
Cai H, Shen D, Hu X, Yin H, Yan Z. The Potential Mechanisms of Ochratoxin A in Prostate Cancer Development: An Integrated Study Combining Network Toxicology, Machine Learning, and Molecular Docking. Toxins. 2025; 17(8):388. https://doi.org/10.3390/toxins17080388
Chicago/Turabian StyleCai, Hong, Dandan Shen, Xiangjun Hu, Hongwei Yin, and Zhangren Yan. 2025. "The Potential Mechanisms of Ochratoxin A in Prostate Cancer Development: An Integrated Study Combining Network Toxicology, Machine Learning, and Molecular Docking" Toxins 17, no. 8: 388. https://doi.org/10.3390/toxins17080388
APA StyleCai, H., Shen, D., Hu, X., Yin, H., & Yan, Z. (2025). The Potential Mechanisms of Ochratoxin A in Prostate Cancer Development: An Integrated Study Combining Network Toxicology, Machine Learning, and Molecular Docking. Toxins, 17(8), 388. https://doi.org/10.3390/toxins17080388





