Venom Proteomics of Trimeresurus gracilis, a Taiwan-Endemic Pitviper, and Comparison of Its Venom Proteome and VEGF and CRISP Sequences with Those of the Most Related Species
Abstract
1. Introduction
2. Results
2.1. Chromatographic and Electrophoretic Profiling of T. gracilis Venom
2.2. T. gracilis Venom Proteomic Analysis
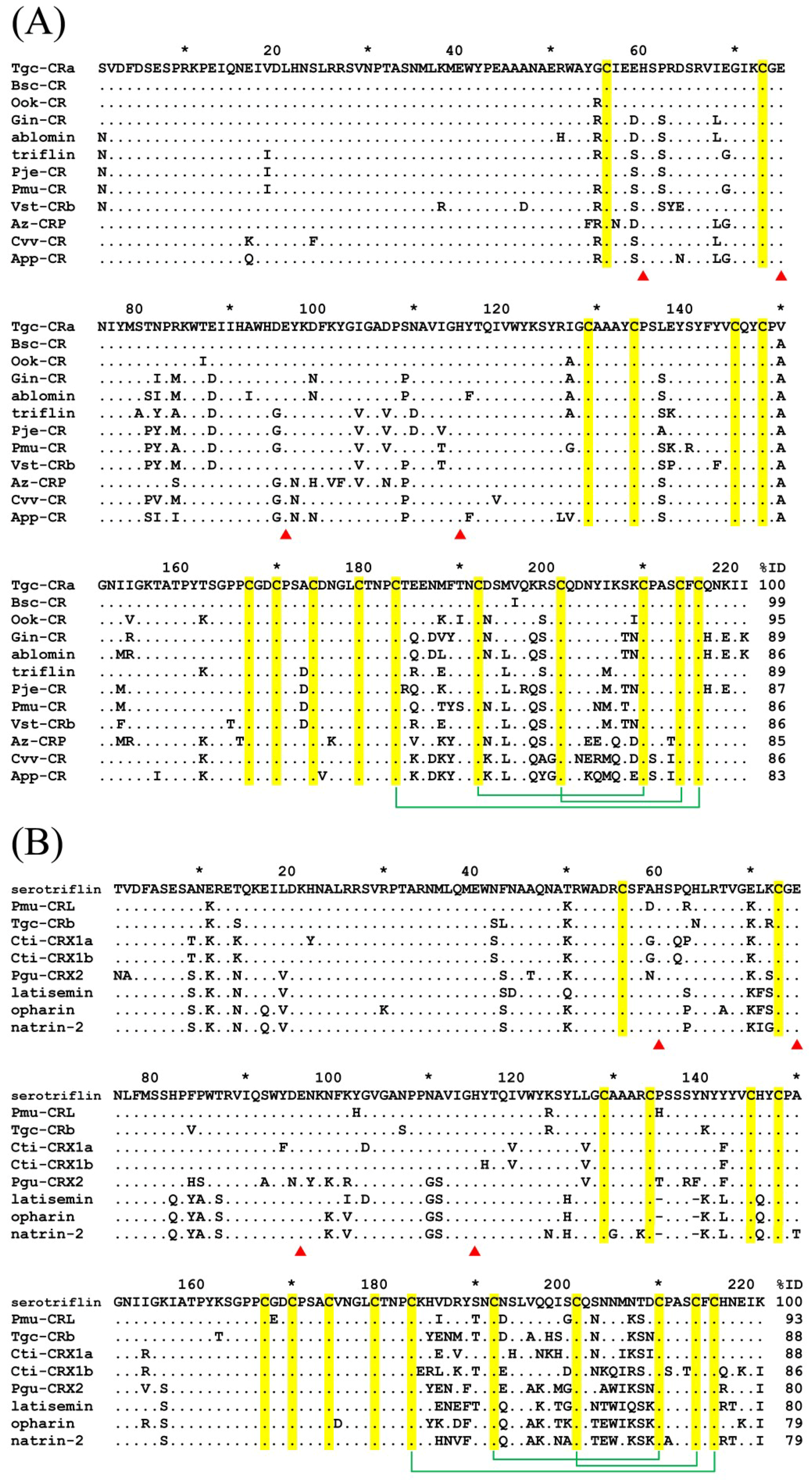
2.3. Two Cysteine-Rich Secretory Proteins (CRISPs) of T. gracilis Venom
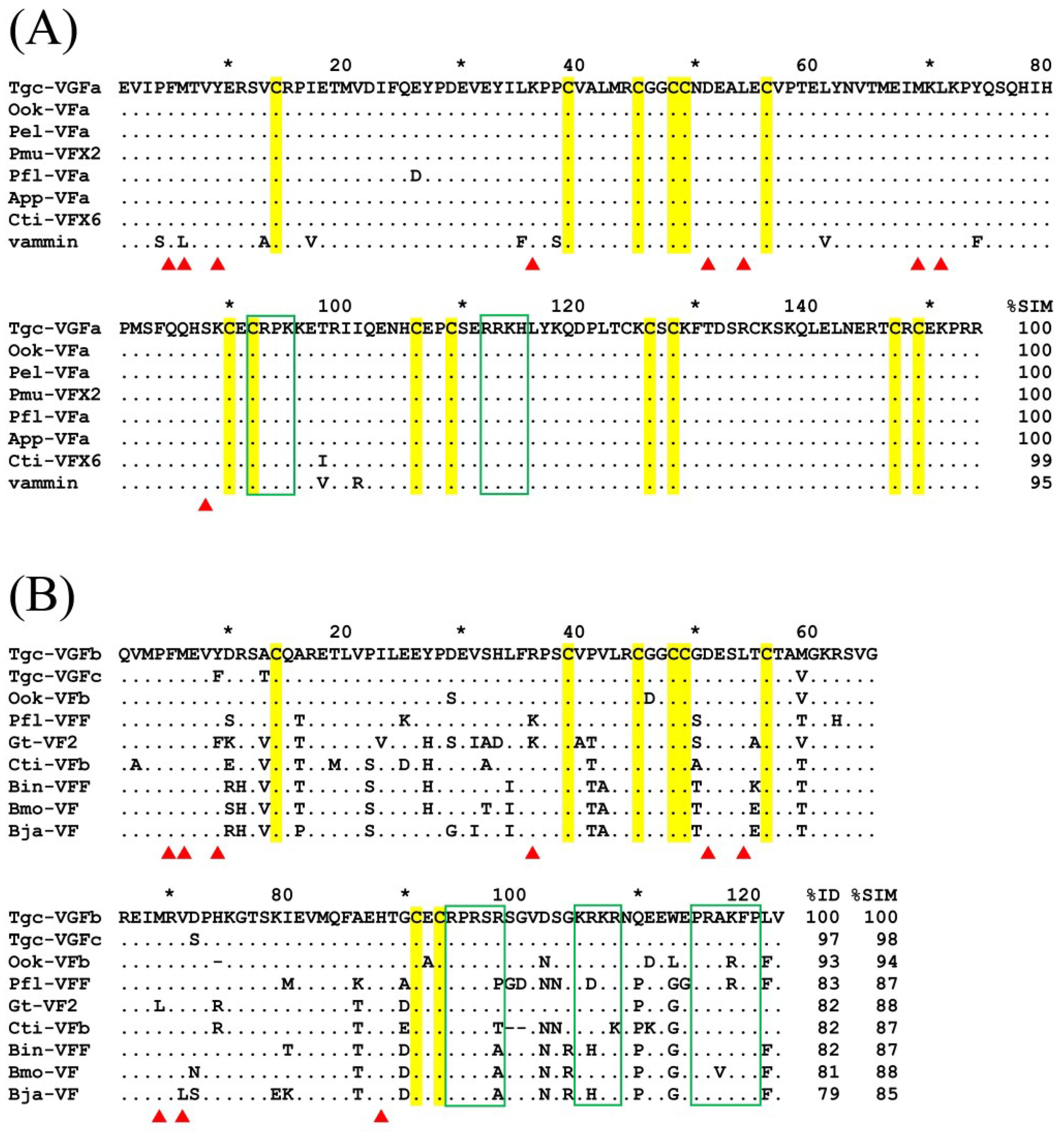
2.4. Three VEGFs Are Expressed in the T. gracilis Venom Gland
3. Discussion
3.1. T. gracilis Venom Proteome
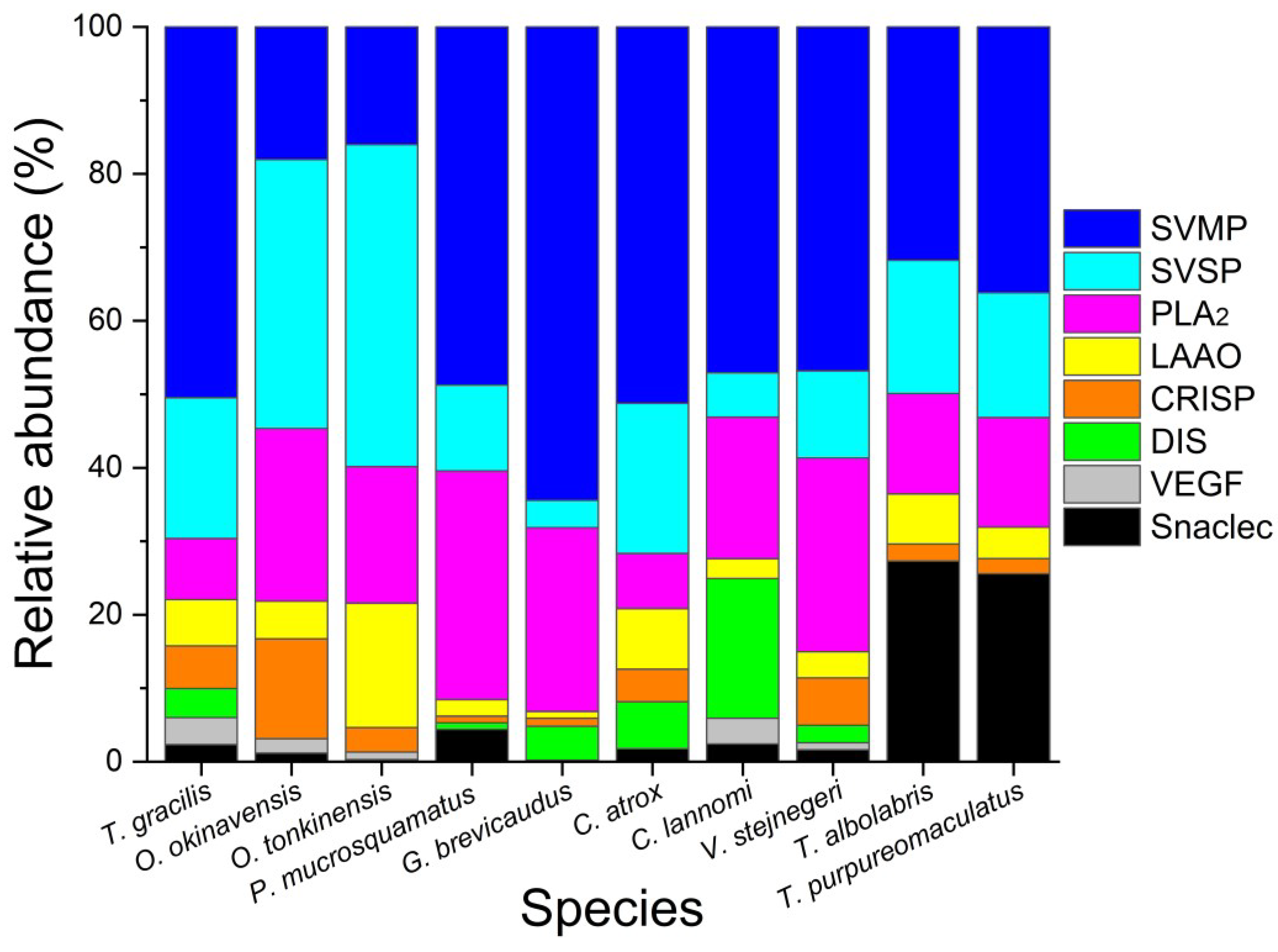
3.2. Comparison of Venom Composition and Toxicity among Closely Related Species
3.3. Sequence Comparison of T. gracilis Venom CRISPs
3.4. Sequence Comparison of T. gracilis Venom VEGFs
4. Conclusions
5. Materials and Methods
5.1. Chemicals
5.2. Animals and Venom
5.3. Determination of Venom Protein Concentration
5.4. Reverse-Phase High-Performance Liquid Chromatography
5.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
5.6. In-Solution Tryptic Digestion and Peptide Identification by Mass Spectrometry
5.7. Molecular Cloning and Sequence Determination of CRISPs and VEGFs
5.8. BLAST Analyses and Sequence Alignments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catalan, J.; Ninot, J.M.; Aniz, M.M. The high mountain conservation in a changing world. In High Mountain Conservation in a Changing World; Catalan, J., Ninot, J.M., Aniz, M.M., Eds.; Springer Nature: Cham, Switzerland, 2017; pp. 3–36. [Google Scholar] [CrossRef]
- Lai, J.S.; Luei, K.Y. Two new Hynobius (Caudata: Hynobiidae) salamanders from Taiwan. Herpetologica 2008, 64, 63–80. [Google Scholar] [CrossRef]
- Schulz, H.M.; Li, C.F.; Thies, B.; Chang, S.C.; Bendix, J. Mapping the montane cloud forest of Taiwan using 12 year MODIS-derived ground fog frequency data. PLoS ONE 2017, 12, e0172663. [Google Scholar] [CrossRef] [PubMed]
- Oshima, S. Notes on the venomous snakes from the islands of Formosa and Riu Kiu. Annu. Rep. Inst. Sci. Gov. Formosa 1920, 2, 1–99. Available online: https://www.biodiversitylibrary.org/item/105191#page/13/mode/1up (accessed on 19 June 2023).
- Zaher, H.; Murphy, R.W.; Arredondo, J.C.; Graboski, R.; Machado-Filho, P.R.; Mahlow, K.; Montingelli, G.G.; Quadros, A.B.; Orlov, N.L.; Wilkinson, M.; et al. Large-scale molecular phylogeny, morphology, divergence-time estimation, and the fossil record of advanced caenophidian snakes (Squamata: Serpentes). PLoS ONE 2019, 14, e0216148. [Google Scholar] [CrossRef]
- Castoe, T.A.; Parkinson, C.L. Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes). Mol. Phylogenet. Evol. 2006, 39, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, A.; Creer, S.; Pook, C.E.; Thorpe, R.S. Inclusion of nuclear intron sequence data helps to identify the Asian sister group of New World pitvipers. Mol. Phylogenet. Evol. 2010, 54, 172–178. [Google Scholar] [CrossRef]
- Wüster, W.; Peppin, L.; Pook, C.E.; Walker, D.E. A nesting of vipers: Phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol. Phylogenet. Evol. 2008, 49, 445–459. [Google Scholar] [CrossRef]
- Lin, C.F.; Tu, M.C. Food habits of the Taiwanese mountain pitviper, Trimeresurus gracilis. Zool. Stud. 2008, 47, 697–703. [Google Scholar]
- Mori, A.; Toda, M.; Ota, H. Winter activity of the Hime-habu (Ovophis okinavensis) in the humid subtropics: Foraging on breeding anurans at low temperatures. In Biology of the Vipers; Schuett, G.W., Höggren, M., Douglas, M.E., Greene, H.W., Eds.; Eagle Mountain Publishing LC: Eagle Mountain, UT, USA, 2002; pp. 329–344. [Google Scholar]
- Tsai, T.S.; Chan, Y.Y.; Huang, S.M.; Chuang, P.C. Case report: Symptoms and prognosis of Trimeresurus gracilis envenomation. Am. J. Trop. Med. Hyg. 2022, 106, 1281–1284. [Google Scholar] [CrossRef]
- Tsai, I.H.; Tsai, T.S.; Wang, Y.M.; Tu, M.C.; Chang, H.C. Cloning and characterization of Trimeresurus gracilis venom phospholipases A2: Comparison with Ovophis okinavensis venom and the systematic implications. Toxicon 2012, 59, 151–157. [Google Scholar] [CrossRef]
- Tsai, T.S.; Wang, Y.M.; Tsai, I.H. Sequence determination and bioinformatic comparison of ten venom serine proteases of Trimeresurus gracilis, a Taiwanese endemic pitviper with controversial taxonomy. Toxicon 2022, 206, 28–37. [Google Scholar] [CrossRef]
- Tsai, T.S.; Tsai, I.H. Full sequencing and comparison of five venom metalloproteases of Trimeresurus gracilis: The PI-enzyme is most similar to okinalysin but the PIII-enzyme is most similar to Crotalus venom enzymes. Toxicon 2023, 225, 107053. [Google Scholar] [CrossRef]
- Aird, S.D.; Watanabe, Y.; Villar-Briones, A.; Roy, M.C.; Terada, K.; Mikheyev, A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genom. 2013, 14, 790. [Google Scholar] [CrossRef]
- Komori, Y.; Murakami, E.; Uchiya, K.; Nonogaki, T.; Nikai, T. Okinalysin, a novel P-I metalloproteinase from Ovophis okinavensis: Biological properties and effect on vascular endothelial cells. Toxins 2014, 6, 2594–2604. [Google Scholar] [CrossRef]
- Alencar, L.R.V.; Quental, T.B.; Grazziotin, F.G.; Alfaro, M.L.; Martins, M.; Venzon, M.; Zaher, H. Diversification in vipers: Phylogenetic relationships, time of divergence and shifts in speciation rates. Mol. Phylogenet. Evol. 2016, 105, 50–62. [Google Scholar] [CrossRef]
- Yang, Z.M.; Guo, Q.; Ma, Z.R.; Chen, Y.; Wang, Z.Z.; Wang, X.M.; Wang, Y.M.; Tsai, I.H. Structures and functions of crotoxin-like heterodimers and acidic phospholipases A2 from Gloydius intermedius venom: Insights into the origin of neurotoxic-type rattlesnakes. J. Proteom. 2015, 112, 210–223. [Google Scholar] [CrossRef]
- Malhotra, A.; Thorpe, R.S. A phylogeny of four mitochondrial gene regions suggests a revised taxonomy for Asian pitvipers (Trimeresurus and Ovophis). Mol. Phylogenet. Evol. 2004, 32, 83–100. [Google Scholar] [CrossRef]
- Modahl, C.M.; Mackessy, S.P. Full-length venom protein cDNA sequences from venom-derived mRNA: Exploring compositional variation and adaptive multigene evolution. PLoS Negl. Trop. Dis. 2016, 10, e0004587. [Google Scholar] [CrossRef]
- Ramazanova, A.S.; Starkov, V.G.; Osipov, A.V.; Ziganshin, R.H.; Filkin, S.Y.; Tsetlin, V.I.; Utkin, Y.N. Cysteine-rich venom proteins from the snakes of Viperinae subfamily–molecular cloning and phylogenetic relationship. Toxicon 2009, 53, 162–168. [Google Scholar] [CrossRef]
- Matsunaga, Y.; Yamazaki, Y.; Hyodo, F.; Sugiyama, Y.; Nozaki, M.; Morita, T. Structural divergence of cysteine-rich secretory proteins in snake venoms. J. Biochem. 2009, 145, 365–375. [Google Scholar] [CrossRef]
- Aoki, N.; Sakiyama, A.; Kuroki, K.; Maenaka, K.; Kohda, D.; Deshimaru, M.; Terada, S. Serotriflin, a CRISP family protein with binding affinity for small serum protein-2 in snake serum. Biochim. Biophys. Acta 2008, 1784, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Tsai, I.H.; Hong, T.M.; Tsai, S.H. Crotalid venom vascular endothelial growth factors has preferential affinity for VEGFR-1. Characterization of Protobothrops mucrosquamatus venom VEGF. Thromb. Haemost. 2005, 93, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Matsunaga, Y.; Tokunaga, Y.; Obayashi, S.; Saito, M.; Morita, T. Snake venom vascular endothelial growth factors (VEGF-Fs) exclusively vary their structures and functions among species. J. Biol. Chem. 2009, 284, 9885–9891. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef]
- Tasoulis, T.; Isbister, G.K. A current perspective on snake venom composition and constituent protein families. Arch. Toxicol. 2023, 97, 133–153. [Google Scholar] [CrossRef]
- Leonardi, A.; Sajevic, T.; Pungercar, J.; Krizaj, I. Comprehensive study of the proteome and transcriptome of the venom of the most venomous european viper: Discovery of a new subclass of ancestral snake venom metalloproteinase precursor-derived proteins. J. Proteome Res. 2019, 18, 2287–2309. [Google Scholar] [CrossRef]
- Ferreira, I.G.; Pucca, M.B.; Oliveira, I.S.; Cerni, F.A.; Jacob, B.; Arantes, E.C. Snake venom vascular endothelial growth factors (svVEGFs): Unravelling their molecular structure, functions, and research potential. Cytokine Growth Factor Rev. 2021, 60, 133–143. [Google Scholar] [CrossRef]
- Gutierrez, J.M.; Rucavado, A.; Chaves, F.; Diaz, C.; Escalante, T. Experimental pathology of local tissue damage induced by Bothrops asper snake venom. Toxicon 2009, 54, 958–975. [Google Scholar] [CrossRef]
- Dawson, C.A.; Ainsworth, S.; Albulescu, L.-O.; Casewell, N.R. Snake venom metalloproteinases. In Handbook of Venoms and Toxins of Reptiles, 2nd ed.; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2021; pp. 363–380. [Google Scholar]
- Sadahiro, S.; Yamauchi, K.; Kondo, S.; Konda, H.; Murata, R. Immunological studies on snake venom I. Comparison of venoms from Genus Trimeresurus inhabiting the Ryukyu islands. Jpn. J. Bacteriol. 1965, 20, 21–26, (In Japanese with English Abstract). [Google Scholar] [CrossRef]
- Tan, C.H.; Palasuberniam, P.; Tan, K.Y. Snake venom proteomics, immunoreactivity and toxicity neutralization studies for the Asiatic Mountain Pit Vipers, Ovophis convictus, Ovophis tonkinensis, and Hime Habu, Ovophis okinavensis. Toxins 2021, 13, 514. [Google Scholar] [CrossRef]
- Gao, J.F.; Wang, J.; He, Y.; Qu, Y.F.; Lin, L.H.; Ma, X.M.; Ji, X. Proteomic and biochemical analyses of short-tailed pit viper (Gloydius brevicaudus) venom: Age-related variation and composition-activity correlation. J. Proteom. 2014, 105, 307–322. [Google Scholar] [CrossRef]
- Villalta, M.; Pla, D.; Yang, S.L.; Sanz, L.; Segura, A.; Vargas, M.; Chen, P.Y.; Herrera, M.; Estrada, R.; Cheng, Y.F.; et al. Snake venomics and antivenomics of Protobothrops mucrosquamatus and Viridovipera stejnegeri from Taiwan: Keys to understand the variable immune response in horses. J. Proteom. 2012, 75, 5628–5645. [Google Scholar] [CrossRef]
- Calvete, J.J.; Fasoli, E.; Sanz, L.; Boschetti, E.; Righetti, P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009, 8, 3055–3067. [Google Scholar] [CrossRef]
- Neri-Castro, E.; Zarzosa, V.; Colis-Torres, A.; Fry, B.G.; Olvera-Rodriguez, A.; Jones, J.; Reyes-Velasco, J.; Zamudio, F.; Borja, M.; Alagon, A.; et al. Proteomic and toxicological characterization of the venoms of the most enigmatic group of rattlesnakes: The long-tailed rattlesnakes. Biochimie 2022, 202, 226–236. [Google Scholar] [CrossRef]
- Anita, S.; Sadjuri, A.R.; Rahmah, L.; Nugroho, H.A.; Mulyadi; Trilaksono, W.; Ridhani, W.; Safira, N.; Bahtiar, H.; Maharani; et al. Venom composition of Trimeresurus albolabris, T. insularis, T. puniceus and T. purpureomaculatus from Indonesia. J. Venom. Anim. Toxins Incl. Trop. Dis. 2022, 28, e20210103. [Google Scholar] [CrossRef]
- LD50men. Available online: https://web.archive.org/web/20120413182323/http://www.venomdoc.com/LD50/LD50men.html (accessed on 20 May 2023).
- De Roodt, A.R.; Desio, M.A.; Lanari, L.C.; Lago, N.R.; Goñi, F.M.; Dozoretz, D.; Calderón, L.; Regner, P.; de Oliveira, V.C.; Damin, C. Paraspecific neutralization of the venom form adults and young Crotalus atrox by paraspecific South American Antivenoms. In Proceedings of the 1st International Electronic Conference on Toxins Session Poster, Online, 16–31 January 2021. [Google Scholar] [CrossRef]
- Choksawangkarn, W.; Sriswasdi, S.; Kalpongnukul, N.; Wongkongkathep, P.; Saethang, T.; Chanhome, L.; Laoungbua, P.; Khow, O.; Sumontha, M.; Chaiyabutr, N.; et al. Combined proteomic strategies for in-depth venomic analysis of the beaked sea snake (Hydrophis schistosus) from Songkhla Lake, Thailand. J. Proteom. 2022, 259, 104559. [Google Scholar] [CrossRef]
- Fry, B.G.; Wuster, W.; Ryan Ramjan, S.F.; Jackson, T.; Martelli, P.; Kini, R.M. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun. Mass Spectrom. 2003, 17, 2047–2062. [Google Scholar] [CrossRef]
- Tadokoro, T.; Modahl, C.M.; Maenaka, K.; Aoki-Shioi, N. Cysteine-rich secretory proteins (CRISPs) from venomous snakes: An overview of the functional diversity in a large and underappreciated superfamily. Toxins 2020, 12, 175. [Google Scholar] [CrossRef]
- Kini, R.M. Proline brackets and identification of potential functional sites in proteins: Toxins to therapeutics. Toxicon 1998, 36, 1659–1670. [Google Scholar] [CrossRef]
- Bernardes, C.P.; Menaldo, D.L.; Zoccal, K.F.; Boldrini-Franca, J.; Peigneur, S.; Arantes, E.C.; Rosa, J.C.; Faccioli, L.H.; Tytgat, J.; Sampaio, S.V. First report on BaltCRP, a cysteine-rich secretory protein (CRISP) from Bothrops alternatus venom: Effects on potassium channels and inflammatory processes. Int. J. Biol. Macromol. 2019, 140, 556–567. [Google Scholar] [CrossRef]
- Shikamoto, Y.; Suto, K.; Yamazaki, Y.; Morita, T.; Mizuno, H. Crystal structure of a CRISP family Ca2+-channel blocker derived from snake venom. J. Mol. Biol. 2005, 350, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Sousa, L.F.; Nicolau, C.A.; Peixoto, P.S.; Bernardoni, J.L.; Oliveira, S.S.; Portes-Junior, J.A.; Mourao, R.H.; Lima-dos-Santos, I.; Sano-Martins, I.S.; Chalkidis, H.M.; et al. Comparison of phylogeny, venom composition and neutralization by antivenom in diverse species of Bothrops complex. PLoS Negl. Trop. Dis. 2013, 7, e2442. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Tan, N.H. A protein decomplexation strategy in snake venom proteomics. Methods Mol. Biol. 2019, 1871, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.Y.; Wong, K.Y.; Tan, N.H.; Tan, C.H. Quantitative proteomics of Naja annulifera (sub-Saharan snouted cobra) venom and neutralization activities of two antivenoms in Africa. Int. J. Biol. Macromol. 2020, 158, 605–616. [Google Scholar] [CrossRef]
- Mullis, K.B.; Faloona, F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987, 155, 335–350. [Google Scholar] [CrossRef]
- Tsai, I.H.; Wang, Y.M.; Huang, K.F. Structures of Azemiops feae venom phospholipases and Cys-rich-secretory protein and implications for taxonomy and toxinology. Toxicon 2016, 114, 31–39. [Google Scholar] [CrossRef]
- Junqueira de Azevedo, I.L.; Farsky, S.H.; Oliveira, M.L.; Ho, P.L. Molecular cloning and expression of a functional snake venom vascular endothelium growth factor (VEGF) from the Bothrops insularis pit viper. A new member of the VEGF family of proteins. J. Biol. Chem. 2001, 276, 39836–39842. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sequence Manipulation Suite. Available online: https://www.bioinformatics.org/sms2/ident_sim.html (accessed on 20 May 2023).
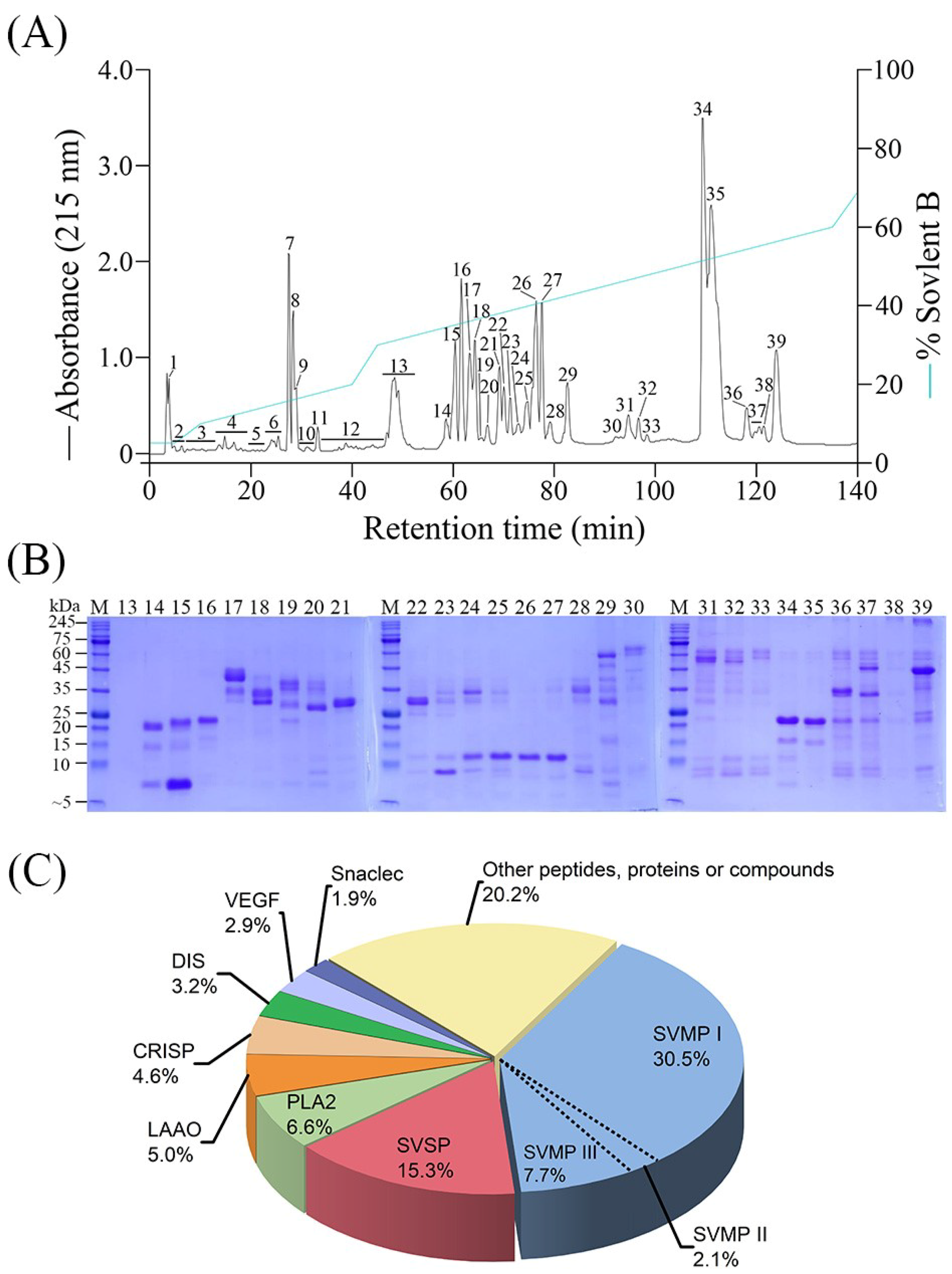
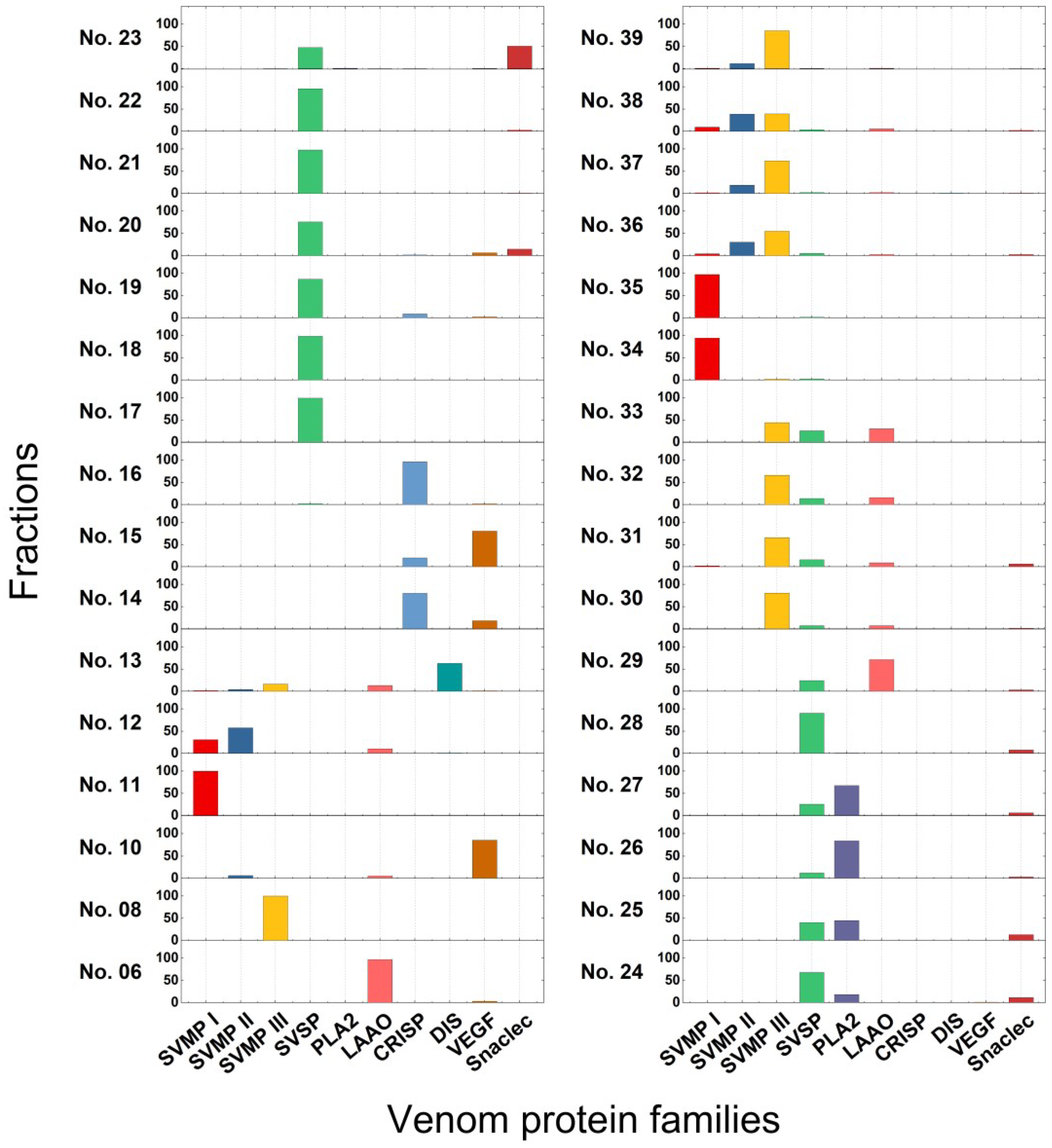
| HPLC Fraction /Toxin Family | Protein (Proteoform) Name | Database Accession (NCBI) | Species | Protein Score | Relative Abundance (%) |
|---|---|---|---|---|---|
| Fraction 6 | |||||
| LAAO | L-amino oxidase | gi|538260091 | Ovophis okinavensis | 69.70 | 1.91 |
| Fraction 8 | |||||
| SVMP III | Metalloprotease PIII [Tgc-PIII] * | gi|335892636 | Trimeresurus gracilis | 34.41 | 0.11 |
| Fraction 10 | |||||
| VEGF | Tgc-VGFb | OQ614864 | Trimeresurus gracilis | 75.04 | 0.28 |
| Fraction 11 | |||||
| SVMP I | Metalloprotease PI [Tgc-MP] | gi|335892630 | Trimeresurus gracilis | 50.20 | 0.63 |
| Fraction 12 | |||||
| SVMP II | Metalloprotease precursor H4, partial | gi|7340946 | Deinagkistrodon acutus | 82.92 | 0.91 |
| SVMP I | Metalloprotease PI [Tgc-MP] | gi|335892630 | Trimeresurus gracilis | 71.95 | 0.49 |
| LAAO | Chain A Amine oxidase | gi|1186227927 | Bothrops atrox | 132.93 | 0.16 |
| Fraction 13 | |||||
| DIS | Metalloprotease PIIb [gracilisin] ** | gi|335892632 | Trimeresurus gracilis | 99.53 | 3.13 |
| LAAO | Chain A Amine oxidase | gi|1186227927 | Bothrops atrox | 93.75 | 0.65 |
| SVMP III | P-III_metalloprotease | gi|547223066 | Ovophis okinavensis | 202.13 | 0.31 |
| SVMP III | Metalloproteinase type III 12b | gi|1041577317 | Agkistrodon conanti | 183.99 | 0.22 |
| SVMP II | Snake venom metalloprotease precursor | gi|2035122236 | Bothrops jararaca | 127.91 | 0.17 |
| SVMP III | Metalloprotease PIII [Tgc-PIII] | gi|335892636 | Trimeresurus gracilis | 147.80 | 0.17 |
| SVMP III | Metalloproteinase (type III) 1a | gi|818935191 | Crotalus adamanteus | 130.40 | 0.11 |
| Fraction 14 | |||||
| CRISP | Serotriflin | gi|1002598708 | Protobothrops mucrosquamatus | 190.38 | 0.46 |
| CRISP | Cysteine-rich seceretory protein Og-CRPb, partial [Tgc-CRb] | gi|190195327 | Trimeresurus gracilis | 347.51 | 0.28 |
| VEGF | Tgc-VGFb | OQ614864 | Trimeresurus gracilis | 207.39 | 0.21 |
| CRISP | CRiSP-Sut-27 | gi|476539526 | Suta fasciata | 52.48 | 0.12 |
| Fraction 15 | |||||
| VEGF | Tgc-VGFc | OQ614865 | Trimeresurus gracilis | 121 | 1.16 |
| VEGF | Tgc-VGFb | OQ614864 | Trimeresurus gracilis | 195.33 | 0.80 |
| CRISP | Cysteine-rich seceretory protein Dr-CRPK | gi|190195321 | Daboia russelii | 155.56 | 0.23 |
| VEGF | Cadam10_VEGF-1 | gi|1178170176 | Crotalus adamanteus | 85.2 | 0.20 |
| CRISP | Cysteine-rich seceretory protein Og-CRPa [Tgc-CRa] | gi|190195325 | Trimeresurus gracilis | 287.81 | 0.18 |
| CRISP | Cysteine-rich secretory protein, partial | gi|2205501413 | Malpolon monspessulanus | 109.71 | 0.13 |
| Fraction 16 | |||||
| CRISP | Cysteine-rich seceretory protein Dr-CRPK | gi|190195321 | Daboia russelii | 176.09 | 0.88 |
| CRISP | Cysteine-rich seceretory protein Bs-CRP | gi|190195305 | Bothriechis schlegelii | 364.81 | 0.69 |
| CRISP | Cysteine-rich secretory protein, partial | gi|2205501413 | Malpolon monspessulanus | 128.48 | 0.61 |
| CRISP | Cysteine-rich secretory protein TRI1 | gi|123898155 | Trimorphodon biscutatus | 53.33 | 0.26 |
| Fraction 17 | |||||
| SVSP | Serine proteinase 12a | gi|1180525223 | Agkistrodon contortrix contortrix | 118.97 | 1.34 |
| SVSP | Thrombin-like enzyme LmrSP-3 | gi|1714612439 | Lachesis muta rhombeata | 59.05 | 0.75 |
| SVSP | Ancrod=thrombin-like alpha-fibrinogenase | gi|247212 | Akistrodon rhodostoma | 89.78 | 0.56 |
| SVSP | Venom thrombin-like enzyme, partial | gi|118430266 | Deinagkistrodon acutus | 109.05 | 0.34 |
| Fraction 18 | |||||
| SVSP | Thrombin-like enzyme collinein-4 | gi|1109550140 | Crotalus durissus collilineatus | 97.45 | 0.39 |
| SVSP | Serine proteinase 8b | gi|1041578893 | Sistrurus tergeminus | 120.13 | 0.39 |
| SVSP | Thrombin-like enzyme bhalternin; Fibrinogen-clotting enzyme | gi|298351882 | Bothrops alternatus | 107.85 | 0.32 |
| SVSP | Snake venom serine protease pallase | gi|158514815 | Gloydius halys | 147.7 | 0.22 |
| SVSP | Ancrod-like protein | gi|1334616 | Calloselasma rhodostoma | 104.21 | 0.20 |
| SVSP | Serine endopeptidase | gi|1333445426 | Crotalus tigris | 157.2 | 0.18 |
| SVSP | Serine proteinase 2 | gi|1041577225 | Agkistrodon conanti | 120.95 | 0.18 |
| SVSP | Agkihpin | gi|484358552 | Gloydius halys | 125.98 | 0.16 |
| SVSP | Plasminogen-activator subtype serine protease (PA1/2) [Tgc-PAH1/2] | gi|2289393718/2289393720 | Trimeresurus gracilis | 304.56 | 0.14 |
| SVSP | Snake venom serine protease precursor | gi|2035122138 | Bothrops jararaca | 172.04 | 0.14 |
| Fraction 21 | |||||
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 277.88 | 0.74 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 156.88 | 0.65 |
| SVSP | Serine proteinase 1 | gi|1041577231 | Agkistrodon conanti | 130.59 | 0.17 |
| Fraction 22 | |||||
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 219.86 | 0.50 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 151.81 | 0.43 |
| SVSP | Serine proteinase 1 | gi|1041577231 | Agkistrodon conanti | 104.59 | 0.23 |
| SVSP | Snake venom serine protease serpentokallikrein-2 isoform X1 | gi|1002585685 | Protobothrops mucrosquamatus | 147.93 | 0.14 |
| Fraction 23 | |||||
| Snaclec | C-type lectin LmsL; Galactose-specific lectin; Mutina | gi|34922643 | Lachesis stenophrys | 220.07 | 0.22 |
| Snaclec | Galactose binding lectin | gi|538260107 | Ovophis okinavensis | 204.87 | 0.16 |
| Snaclec | Galactose binding lectin, partial | gi|538259813 | Protobothrops flavoviridis | 168.47 | 0.16 |
| Snaclec | Chain B Galactose-specific lectin | gi|33357350 | Crotalus atrox | 208.84 | 0.14 |
| SVSP | Serine proteinase 8c | gi|1041578891 | Sistrurus tergeminus | 103.02 | 0.12 |
| Fraction 24 | |||||
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 172.27 | 0.12 |
| SVSP | Plasminogen-activator subtype serine protease (PA3) [Tgc-PA3] | gi|2289393722 | Trimeresurus gracilis | 154.61 | 0.11 |
| Fraction 25 | |||||
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 130.75 | 0.27 |
| PLA2 | Acidic phospholipase A2 [Tgc-E6] | gi|59727071 | Trimeresurus gracilis | 266.65 | 0.23 |
| SVSP | Thrombin-like enzyme | gi|38146946 | Gloydius shedaoensis | 108.67 | 0.14 |
| SVSP | Thrombin-like enzyme halystase | gi|3122187 | Gloydius blomhoffii | 100.35 | 0.14 |
| PLA2 | Phospholipase A2 precursor | gi|743759444 | Protobothrops tokarensis | 66.77 | 0.13 |
| PLA2 | Acidic phospholipase A2 | gi|129420 | Gloydius blomhoffii | 117.08 | 0.13 |
| PLA2 | Phospholipase A2 type IIE | gi|384110782 | Dispholidus typus | 83.97 | 0.13 |
| SVSP | Serine proteinase 19b | gi|1041577233 | Agkistrodon conanti | 112.35 | 0.12 |
| PLA2 | Phospholipase A2 | gi|584481356 | Ovophis makazayazaya | 92.74 | 0.11 |
| Fraction 26 | |||||
| PLA2 | Acidic phospholipase A2 [Tgc-E6] | gi|59727071 | Trimeresurus gracilis | 444.11 | 1.70 |
| PLA2 | Phospholipase A2 isozyme CTs-A3, partial | gi|37785867 | Viridovipera stejnegeri | 136.6 | 1.08 |
| PLA2 | Phospholipase A2, partial | gi|538259861 | Protobothrops flavoviridis | 147.33 | 0.90 |
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 135.89 | 0.25 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 143.16 | 0.19 |
| Fraction 27 | |||||
| PLA2 | Acidic phospholipase A2 [Tgc-E6] | gi|59727071 | Trimeresurus gracilis | 300.99 | 0.79 |
| PLA2 | Phospholipase A2, partial | gi|538259861 | Protobothrops flavoviridis | 121.88 | 0.53 |
| PLA2 | Phospholipase A2 isozyme CTs-A3, partial | gi|37785867 | Viridovipera stejnegeri | 117.83 | 0.40 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 102.54 | 0.20 |
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 121.04 | 0.17 |
| SVSP | Plasminogen-activator subtype serine protease (PA3) [Tgc-PA3] | gi|2289393722 | Trimeresurus gracilis | 144 | 0.15 |
| Fraction 28 | |||||
| SVSP | Plasminogen-activator subtype serine protease (PA3) [Tgc-PA3] | gi|2289393722 | Trimeresurus gracilis | 137.28 | 0.28 |
| SVSP | Serine proteinase 19b | gi|1041577233 | Agkistrodon conanti | 108.99 | 0.27 |
| SVSP | Plasminogen-activator subtype serine protease (PA1/2) [Tgc-PAH1/2] | gi|2289393718/2289393720 | Trimeresurus gracilis | 134.51 | 0.19 |
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 102.15 | 0.17 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 96.4 | 0.13 |
| Fraction 29 | |||||
| LAAO | L-amino-acid oxidase | gi|347602330 | Vipera ammodytes ammodytes | 212.56 | 0.66 |
| LAAO | Chain A Ahp-laao | gi|48425312 | Gloydius halys | 199.24 | 0.51 |
| LAAO | L-amino acid oxidase | gi|538260091 | Ovophis okinavensis | 275.32 | 0.42 |
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 111.55 | 0.16 |
| SVSP | Plasminogen-activator subtype serine protease (PA3) [Tgc-PA3] | gi|2289393722 | Trimeresurus gracilis | 138.9 | 0.13 |
| LAAO | BATXLAAO1 | gi|1127252627 | Bothrops atrox | 176.6 | 0.12 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 119.14 | 0.12 |
| Fraction 30 | |||||
| SVMP III | Metalloproteinase, partial | gi|297593822 | Echis carinatus sochureki | 50.52 | 0.31 |
| SVMP III | Metalloproteinase-disintegrin-like atrolysin-A, partial | gi|1663479917 | Protobothrops mucrosquamatus | 183.98 | 0.25 |
| SVMP III | BATXSVMPIII16 | gi|1127252547 | Bothrops atrox | 74.01 | 0.11 |
| Fraction 31 | |||||
| SVMP III | Metalloprotease PIII [Tgc-PIII] | gi|335892636 | Trimeresurus gracilis | 307.13 | 0.46 |
| SVMP III | Metalloproteinase-disintegrin-like atrolysin-A, partial | gi|1663479917 | Protobothrops mucrosquamatus | 236.99 | 0.40 |
| Fraction 32 | |||||
| SVMP III | Metalloprotease PIII [Tgc-PIII] | gi|335892636 | Trimeresurus gracilis | 202.57 | 0.37 |
| SVMP III | Metalloproteinase-disintegrin-like atrolysin-A, partial | gi|1663479917 | Protobothrops mucrosquamatus | 162.6 | 0.12 |
| LAAO | L-amino acid oxidase | gi|538260091 | Ovophis okinavensis | 181.72 | 0.11 |
| Fraction 34 | |||||
| SVMP I | P-II_metalloprotease *** | gi|547223068 | Ovophis okinavensis | 217.81 | 5.79 |
| SVMP I | Metalloprotease PI [Tgc-MP] | gi|335892630 | Trimeresurus gracilis | 362.99 | 3.88 |
| SVMP I | P-II metalloprotease, partial *** | gi|547223015 | Protobothrops flavoviridis | 189.92 | 0.20 |
| SVMP III | MDC-6d | gi|1829138061 | Crotalus atrox | 167.81 | 0.22 |
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 165.77 | 0.11 |
| Fraction 35 | |||||
| SVMP I | Metalloprotease PI [Tgc-MP] | gi|335892630 | Trimeresurus gracilis | 360.09 | 19.26 |
| SVSP | Kallikrein-like serine protease (KN1) [Tgc-KN1] | gi|2289393706 | Trimeresurus gracilis | 119.16 | 0.15 |
| SVSP | Kallikrein-like serine protease (KN4) [Tgc-KN4] | gi|2289393712 | Trimeresurus gracilis | 105.69 | 0.11 |
| Fraction 36 | |||||
| SVMP III | Metalloproteinase, partial | gi|1773624963 | Dispholidus typus | 62.96 | 0.38 |
| SVMP II | Tgc-PIIc | OK482650 | Trimeresurus gracilis | 308.73 | 0.10 |
| SVMP II | Metalloproteinase type II 6c | gi|1041579264 | Sistrurus miliarius barbouri | 127.08 | 0.10 |
| Fraction 37 | |||||
| SVMP II | Snake venom metalloprotease precursor | gi|2035122167 | Bothrops jararaca | 106.31 | 0.16 |
| SVMP II | Tgc-PIIc | OK482650 | Trimeresurus gracilis | 206.13 | 0.12 |
| SVMP III | Metalloproteinase type III 9b | gi|1041579226 | Sistrurus miliarius barbouri | 75.23 | 0.11 |
| Fraction 38 | |||||
| SVMP II | Tgc-PIIc | OK482650 | Trimeresurus gracilis | 220.3 | 0.11 |
| Fraction 39 | |||||
| SVMP III | Metalloprotease P-III 3, partial | gi|675402421 | Protobothrops flavoviridis | 137.47 | 0.76 |
| SVMP III | Metalloproteinase (type III) 6a | gi|1180525232 | Agkistrodon contortrix contortrix | 222.51 | 0.68 |
| SVMP III | Metalloproteinase type III 12b | gi|1041577317 | Agkistrodon conanti | 206.17 | 0.60 |
| SVMP III | P-III_metalloprotease | gi|547223066 | Ovophis okinavensis | 318.96 | 0.51 |
| SVMP II | Snake venom metalloprotease precursor | gi|2035122236 | Bothrops jararaca | 111.17 | 0.42 |
| SVMP III | Metalloproteinase type III 5a | gi|1041579244 | Sistrurus miliarius barbouri | 230.38 | 0.33 |
| SVMP III | Snake venom metalloprotease precursor | gi|2035122126 | Bothrops jararaca | 115.29 | 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tse, T.-C.; Tsai, I.-H.; Chan, Y.-Y.; Tsai, T.-S. Venom Proteomics of Trimeresurus gracilis, a Taiwan-Endemic Pitviper, and Comparison of Its Venom Proteome and VEGF and CRISP Sequences with Those of the Most Related Species. Toxins 2023, 15, 408. https://doi.org/10.3390/toxins15070408
Tse T-C, Tsai I-H, Chan Y-Y, Tsai T-S. Venom Proteomics of Trimeresurus gracilis, a Taiwan-Endemic Pitviper, and Comparison of Its Venom Proteome and VEGF and CRISP Sequences with Those of the Most Related Species. Toxins. 2023; 15(7):408. https://doi.org/10.3390/toxins15070408
Chicago/Turabian StyleTse, Tsz-Chun, Inn-Ho Tsai, Yuen-Ying Chan, and Tein-Shun Tsai. 2023. "Venom Proteomics of Trimeresurus gracilis, a Taiwan-Endemic Pitviper, and Comparison of Its Venom Proteome and VEGF and CRISP Sequences with Those of the Most Related Species" Toxins 15, no. 7: 408. https://doi.org/10.3390/toxins15070408
APA StyleTse, T.-C., Tsai, I.-H., Chan, Y.-Y., & Tsai, T.-S. (2023). Venom Proteomics of Trimeresurus gracilis, a Taiwan-Endemic Pitviper, and Comparison of Its Venom Proteome and VEGF and CRISP Sequences with Those of the Most Related Species. Toxins, 15(7), 408. https://doi.org/10.3390/toxins15070408





